Abstract
E2F transcription factors regulate genes expressed at the G1/S boundary of the cell division cycle in higher eukaryotes. Although animal E2F proteins and their target promoters have been studied extensively, little is known about how these factors regulate plant promoters. An earlier study identified two E2F consensus binding sites in the promoter of a Nicotiana benthamiana gene encoding proliferating cell nuclear antigen (PCNA) and showed that the proximal element (E2F2) is required for the full repression of PCNA expression in mature leaves. In this study, we examined the distal element (E2F1) and how it interacts with the E2F2 site to regulate the PCNA promoter. Gel shift assays using plant nuclear extracts or purified Arabidopsis E2F and DP proteins showed that different complexes bind to the two E2F sites. Mutation of the E2F1 site or both sites differentially altered PCNA promoter function in transgenic plants. As reported previously for the E2F2 mutation, the E2F1 and E2F1+2 mutations partially relieved the repression of the PCNA promoter in mature leaves. In young tissues, the E2F1 mutation resulted in a threefold reduction in PCNA promoter activity, whereas the E2F1+2 mutation had no detectable effect. The activity of E2F1+2 mutants was indistinguishable from that of E2F2 mutants. These results demonstrate that both E2F elements contribute to the repression of the PCNA promoter in mature leaves, whereas the E2F1 site counters the repression activity of the E2F2 element in young leaves.
INTRODUCTION
In higher eukaryotes, the E2F family of transcription factors plays an important role in both the positive and negative regulation of the cell cycle (Black and Azizkhan-Clifford, 1999; Harbour and Dean, 2000; Muller and Helin, 2000; Trimarchi and Lees, 2002). E2F family members, which heterodimerize with DP proteins to form functional transcription factor complexes, interact with the retinoblastoma gene product (pRb) during G1. This interaction leads to the transcriptional repression of cell cycle–regulated genes that encode proteins required for DNA replication and progression through S-phase (Lavia and Jansen-Durr, 1999). pRb represses transcription by sequestering free E2F and blocking its ability to activate transcription (Zacksenhaus et al., 1996) or by interacting with E2F bound to DNA and recruiting histone deacetylases and SWI/SNF-like enzymes for chromatin remodeling (Zhang and Dean, 2001). In late G1, phosphorylation of pRb by cyclin-dependent kinases disrupts its association with E2F and allows the expression of genes required for S-phase.
Many components of the pRb-E2F pathway have been found in plants. pRb homologs (pRBR) have been identified in maize, tobacco, Arabidopsis, and Chenopodium (Grafi et al., 1996; Xie et al., 1996; Ach et al., 1997; Fountain et al., 1999; Nakagami et al., 1999). Like its animal homologs, pRBR is phosphorylated by cyclin-dependent kinases associated with G1 and S. pRBR levels are high in differentiated tissues, consistent with its involvement in the repression of genes required for cell proliferation (Ach et al., 1997; Nakagami et al., 1999; Boniotti and Gutierrez, 2001). E2F and DP homologs have been isolated from wheat, tobacco, carrot, rice, and Arabidopsis (Ramirez-Parra et al., 1999; Sekine et al., 1999; Albani et al., 2000; Magyar et al., 2000; Ramirez-Parra and Gutierrez, 2000; de Jager et al., 2001; Kosugi and Ohashi, 2002a; Mariconti et al., 2002; Vandepoele et al., 2002). As in mammals, the six E2F proteins of Arabidopsis can be separated into two groups that function primarily as activators or repressors of gene expression (Kosugi and Ohashi, 2002b; Mariconti et al., 2002).
E2F consensus sites have been found in the promoters of a number of plant genes, but there are limited functional data regarding their roles. Purified plant E2F/DP proteins bind in vitro to an E2F site in the Arabidopsis CDC6 gene (Castellano et al., 2001; de Jager et al., 2001). E2F consensus binding sites in the tobacco RNR genes and the Arabidopsis MCM3 gene are required for activation in cultured cells and intact plants (Chaboute et al., 2000, 2002; Stevens et al., 2002). E2F sites also have been implicated in developmental expression of the PCNA (proliferating cell nuclear antigen) genes from rice and tobacco (Egelkrout et al., 2001; Kosugi and Ohashi, 2002a). Overexpression of Arabidopsis E2Fa and DPa proteins induces mature leaf cells to reenter S-phase (Rossignol et al., 2002). Arabidopsis plants that ectopically express AtE2Fa also show sustained cell proliferation in cotyledons and hypocotyls and extra DNA replication in endoreduplicating cells (De Veylder et al., 2002).
There are several important differences between the pRb/E2F networks of plants and animals. pRb is a member of a multigene family in mammals but is represented by a single RBR gene in Arabidopsis (Durfee et al., 2000). Naturally occurring mutations in the RBR gene have not been identified, and there is no known link between pRBR and tumor formation in plants. The DNA binding domains of plant E2F homologs show the highest conservation with animal E2F family members. However, in some Arabidopsis E2F homologs, this binding domain is duplicated and these proteins can bind to DNA in the absence of a DP partner (Kosugi and Ohashi, 2002b; Mariconti et al., 2002). Arabidopsis E2F family members associated with transcriptional activation are dependent on their DP partners for nuclear localization, whereas their human counterparts can translocate into the nucleus independent of their DP partners (Kosugi and Ohashi, 2002c).
Many mammalian DNA tumor viruses disrupt the pRb/E2F pathway (Nevins, 1992; Weinberg, 1995) and activate genes required for DNA replication, leading to untimely entry into S-phase and cell transformation. Geminiviruses are a diverse family of single-stranded DNA viruses that infect plants and replicate in nuclei using host replication machinery (Gutierrez, 2000; Hanley-Bowdoin et al., 2000). The geminivirus Tomato golden mosaic virus (TGMV) causes accumulation of the DNA polymerase δ processivity factor, PCNA, in mature plant tissues by altering transcription of the PCNA gene (Nagar et al., 1995; Egelkrout et al., 2001). The PCNA promoter of Nicotiana benthamiana contains two E2F consensus binding sites. Mutation of the proximal E2F2 site in the N. benthamiana PCNA gene abrogates TGMV induction of the PCNA promoter in infected leaves (Egelkrout et al., 2001), and induction of PCNA transcription depends on interactions between pRBR and the TGMV replication protein, AL1 (Ach et al., 1997; Kong et al., 2000). Together, these results suggest that geminiviruses also disrupt the pRBR/E2F pathway in plants.
Many E2F-responsive genes, including those that encode human Htf9-a/RanBP1, human p107, and Drosophila PCNA (Yamaguchi et al., 1995; Zhu et al., 1995; Di Fiore et al., 1999), contain multiple E2F sites that cooperate to regulate their expression. The tobacco RNR2 gene has two E2F sites (Chaboute et al., 2000), both of which are required for activation during S-phase and one that negatively regulates RNR2 expression outside of S-phase. One of the E2F sites in the Arabidopsis MCM3 gene is required for meristematic expression in seedlings, whereas the other site negatively regulates the promoter during G2 in cultured cells (Stevens et al., 2002). The Nicotiana and rice PCNA promoters also contain two E2F sites, but their relative contributions to gene regulation are not known (Egelkrout et al., 2001; Kosugi and Ohashi, 2002a). In a previous study, we showed that the proximal E2F site (E2F2) in the N. benthamiana PCNA promoter is required for repression in mature leaves (Egelkrout et al., 2001). Here, we examined the role of the distal E2F site (E2F1) and its interaction with the E2F2 site in the regulation of PCNA expression. Our data are consistent with a model in which the E2F sites interact in both young and mature tissues to regulate PCNA expression during plant development.
RESULTS
Two E2F Sites Display Different Binding Properties in Vitro
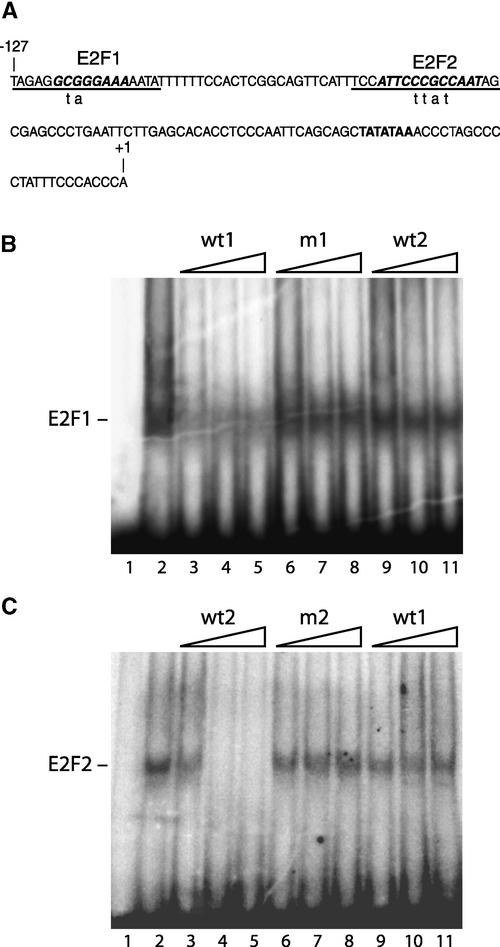
The PCNA promoter from N. benthamiana contains two elements that have strong homology with the consensus binding site for the E2F family of transcription factors (Figure 1A) (Egelkrout et al., 2001). The upstream site (E2F1) consists of a single copy of the consensus sequence (Herwig and Strauss, 1997; Black and Azizkhan-Clifford, 1999). The downstream element (E2F2) is an inverted repeat of the consensus (Wade et al., 1995) and also shows homology with the CDE/CHR bipartite repressor element (Zwicker et al., 1995; Zwicker and Muller, 1997). A previous study showed that the E2F2 site is bound specifically by a factor(s) in nuclear extracts from N. benthamiana suspension cultures (Egelkrout et al., 2001). In Figure 1B, we used electrophoretic mobility shift assays to determine if the E2F1 motif also binds to a nuclear protein(s) in vitro. A single shifted product was detected in assays containing a radiolabeled, double-stranded DNA corresponding to the E2F1 site and a N. benthamiana nuclear extract from asynchronous cycling cells (Figure 1B, lane 2). Formation of the E2F1 protein complex was competed by excess unlabeled E2F1 DNA (Figure 1B, lanes 3 to 5) but not by an oligonucleotide carrying a 2-bp mutation in the consensus site (lanes 6 to 8). Interestingly, the E2F1 binding activity also was not competed by excess unlabeled E2F2 DNA (Figure 1B, lanes 9 to 11). Binding to radiolabeled E2F2 DNA also was not competed by excess unlabeled E2F1 DNA (Figure 1C, lanes 9 to 11), even though competition was observed in the presence of unlabeled E2F2 DNA (lanes 3 to 5). Together, these results suggest that the two E2F elements in the N. benthamiana PCNA promoter bind to different proteins in the nuclear extract even though they form complexes of similar electrophoretic mobilities (data not shown).
Figure 1.
PCNA E2F Sites Show Different Binding Properties with N. benthamiana Nuclear Extracts.
(A) The sequence of the PCNA promoter from nucleotide −127 to the transcription start site (+1). The two E2F consensus sites are shown in boldface italic type. The TATA box is shown in boldface type. The promoter sequences included in the probes used for gel shift analysis ([B] and Figure 2) are underlined. Mutations in E2F sites are shown with lowercase letters beneath the probes.
(B) and (C) Oligonucleotides corresponding to the E2F1 or E2F2 sites were annealed, labeled with 32P, and assayed for binding to nuclear proteins from N. benthamiana suspension cultures. The sequences of the probe and competitor DNAs were as follows: E2F1 (wt1), 5′-gatcTAGAGGCGGGAAAAATAgatc-3′; mE2F1 (m1), 5′-gatcTAGAGGtaGGAAAAATAgatc-3′; E2F2 (wt2), 5′-gatcTCCATTCC-CGCCAATAGgatc-3′; and mE2F2 (m2), 5′-gatcTCCATTCttatCAA-TAGgatc-3′. Lowercase letters represent mutations in the E2F consensus sequences or unrelated flanking sequence. The reactions in (B) contained the E2F1 (wt1) probe, whereas those in (C) contained the E2F2 (wt2) probe. Lanes 1 show no shift in the absence of nuclear extract. Lanes 2 show a shifted complex in the presence of nuclear extract. In lanes 3 to 11, increasing amounts (10-, 100-, and 250-fold excess) of unlabeled competitor DNAs were added as indicated at top. The bound complexes are identified at left.
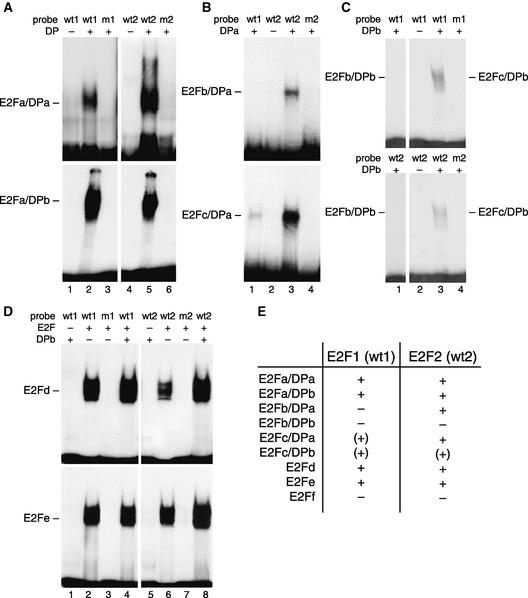
The Arabidopsis genome encodes six members of the E2F family (E2Fa, E2Fb, E2Fc, E2Fd, E2Fe, and E2Ff) and the two DP homologs (DPa and DPb) (Magyar et al., 2000). Recently, purified recombinant proteins corresponding to Arabidopsis E2F and DP family members were produced and shown to bind to the canonical E2F consensus sequence in vitro (Mariconti et al., 2002). Because the homologous N. benthamiana proteins have not been isolated or expressed in vitro, we examined the binding of the recombinant Arabidopsis proteins to the N. benthamiana PCNA promoter E2F elements. E2F1 and E2F2 DNAs displayed similar binding patterns with E2Fa (Figure 2A), E2Fd, and E2Fe (Figure 2D), and neither site bound to E2Ff (data not shown). Both DNAs bound to E2Fa in combination with either DPa or DPb (Figure 2A, lanes 2 and 5) but not to E2Fa alone (lanes 1 and 4). By contrast, the E2F1 and E2F2 probes bound to E2Fd and E2Fe independent of the presence of a DP partner (Figure 2D, lanes 2, 4, 6, and 8). However, the presence of DPb may have enhanced the binding of E2Fd and E2Fe to the E2F2 site (Figure 2D, cf. lanes 6 and 8). DNAs corresponding to mutant E2F1 and E2F2 sites did not bind to E2Fa/DP (Figure 2A, lanes 3 and 6), E2Fd, and E2Fe (Figure 2D, lanes 3 and 7), thereby establishing the specificity of the interactions.
Figure 2.
Electrophoretic Mobility Shift Analysis with Recombinant E2F and DP Proteins.
(A) to (D) Representative electrophoretic mobility shift assays (EMSAs) are shown with the probes indicated at top. The probe designations (see Figure 1A) are E2F1 (wt1), mE2F1 (m1), E2F2 (wt2), and mE2F2 (m2). E2F proteins were included in all of the reactions except where indicated in (D). The presence (+) or absence (−) of DP is indicated at the top of each panel. The bound complexes are marked.
(A) EMSA with E2Fa and DPa (top, lanes 2, 3, 5, and 6) or DPb (bottom, lanes 2, 3, 5, and 6) and the indicated probes. Lanes 1 and 4 show E2Fa and probe without DP.
(B) EMSA with E2Fb and DPa (top, lanes 1, 3, and 4) or E2Fc and DPa (bottom, lanes 1, 3, and 4) and the indicated probes. Lane 2 shows E2F and probe without DP.
(C) EMSA with E2Fb (lane 1) and E2Fc (lanes 3 and 4) with DPb and the indicated probes. Lane 2 shows E2Fc and probe without DP.
(D) EMSA with E2Fd (top) or E2Fe (bottom) and DPb with the indicated probes (lanes 4 and 8). Lanes 2, 3, 6, and 7 show E2F and probe without DP; lanes 1 and 5 show DP and probe without E2F. Binding of E2Fd and E2Fe in the presence of DPa was not tested.
(E) Summary of the binding properties of combinations of recombinant Arabidopsis E2F and DP proteins for the E2F1 and E2F2 sites of the N. benthamiana PCNA promoter. The data for E2Ff are not shown. +, strong interactions; (+), weak interactions; −, no detectable interaction.
The PCNA E2F sites showed distinct binding patterns with Arabidopsis E2Fb and E2Fc (Figures 2B and 2C). The E2F1 DNA was unable to recruit E2Fb in the presence of DPa (Figure 2B, top gel, lane 1), whereas E2F2 formed a stable complex with E2Fb/DPa (lane 3). Neither DNA bound to E2Fb alone (Figure 2B, lane 2) or in combination with DPb (Figure 2C, lane 1). E2F1 formed a faint complex with E2Fc in the presence of DPa (Figure 2B, bottom gel, lane 1), whereas E2F2 gave a strong band (lane 3). Both E2F1 and E2F2 bound weakly to E2Fc in combination with DPb (Figure 2C, lane 3) but not to E2Fc alone (lane 2). As summarized in Figure 2E, the binding characteristics of the E2F1 and E2F2 sites showed differences, consistent with the data presented in Figure 1, indicating that they interact with different proteins from nuclear extracts.
E2F Elements Differentially Regulate the PCNA Promoter
Because multiple E2F sites within a promoter can interact to regulate gene expression (Lavia and Jansen-Durr, 1999), we assessed the relative roles of the PCNA promoter E2F elements in transgenic plants. A series of promoter constructs with mutations (Figure 1B) in E2F1, E2F2, or both sites were generated in the background of the N. benthamiana −633 PCNA promoter fragment shown previously to be regulated developmentally in plants (Egelkrout et al., 2001). The promoter constructs were fused to the luciferase reporter gene and introduced into N. benthamiana via Agrobacterium-mediated transformation.
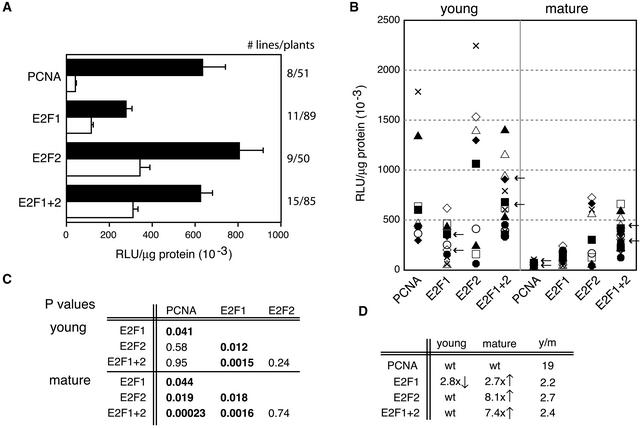
We compared the impact of the individual and double mutations on PCNA promoter activity in T1 plants selected on kanamycin. The average reporter activities in young (leaves 1 and 2) and mature (leaf 9) leaves for the PCNA promoter constructs are shown in Figure 3A. Figure 3B shows a scatterplot of average reporter activities for individual transgenic lines for each construct. Our previous results with the wild-type promoter and the E2F2 site mutant are included for comparison (Egelkrout et al., 2001). The results of two-tailed Student's t tests comparing mean reporter activities for each of the four constructs is shown in Figure 3C. In mature tissue, there was a statistically significant increase in reporter activity for each of the three E2F mutant PCNA promoter constructs relative to the wild-type promoter, indicative of a partial loss of repression. The activities of E2F2 and E2F1+2 promoters both increased sevenfold to eightfold relative to that of the wild-type PCNA promoter, whereas the activity of the E2F1 mutant was threefold greater than that of the wild type. In young leaves, the activities of the E2F2 and E2F1+2 mutant promoters were similar to those of the wild type, whereas the E2F1 promoter showed a threefold decrease in activity compared with the wild-type PCNA promoter. These changes resulted in a decrease in the ratio of reporter activity in young versus mature leaves. For the E2F1, E2F2, and E2F1+2 mutant constructs, the ratios were 2.2, 2.7, and 2.4, respectively, whereas the ratio was 19 for the wild-type promoter (Figure 3D).
Figure 3.
Mutation of the E2F Sites Alters the Developmental Regulation of the PCNA Promoter.
(A) The average luciferase specific activity in young (leaves 1 and 2; black bars) and mature (leaf 9; white bars) leaves of N. benthamiana plants carrying the wild-type, E2F1 mutant, E2F2 mutant, or E2F1+2 mutant PCNA:luciferase construct. The transgene is shown at left, and the number of lines and plants examined for each construct is given at right. RLU, relative light units. Error bars correspond to 2 sd.
(B) Scatterplot showing luciferase specific activity in young (left) and mature (right) leaves for individual transgenic lines carrying the constructs (bottom) described in (A). Each symbol represents the average activity of a minimum of three plants for each transgenic line. The arrows indicate the transgenic lines analyzed in Figure 4.
(C) Statistical analysis of the data shown in (B) using a two-tailed Student's t-test. Boldface type indicates significant comparisons (P < 0.05).
(D) Summary of significant fold changes in reporter activities for young and mature leaves of E2F1 mutant, E2F2, and E2F1+2 mutant lines relative to the wild-type PCNA promoter. Wild-type levels of reporter activities are designated as wt. The average ratios of luciferase specific activity in young versus mature leaves also are given.
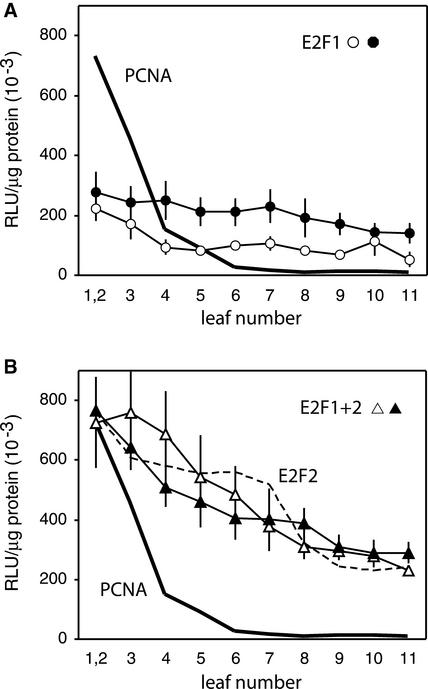
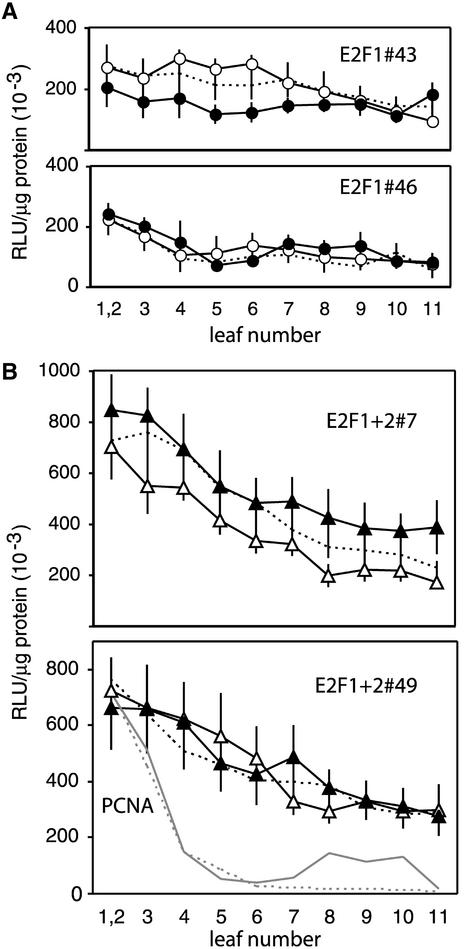
We also analyzed the expression of the PCNA promoter–luciferase constructs in every leaf of transgenic plants. Our previous study showed a sharp decrease in PCNA expression during development from high activity in the youngest leaves (leaves 1 to 3) to low activity by leaf 5 (Egelkrout et al., 2001). Figure 4 shows a similar developmental analysis for two plant lines carrying either the E2F1 (A) or the E2F1+2 (B) PCNA promoter construct and displaying reporter gene activities near the overall mean for each construct. Our previous results for the wild-type and E2F2 mutant promoters are included for comparison. As observed with the E2F2 mutant, the activities of both the E2F1 and E2F1+2 constructs declined more gradually over the course of development than those of the wild type. However, expression levels for the E2F1 mutant construct were lower than those for the other two mutant constructs throughout development. Surprisingly, the expression pattern for the E2F1+2 double-mutant construct was similar to that for the E2F2 construct and not a composite of the patterns seen for the individual E2F1 and E2F2 mutants.
Figure 4.
Developmental Profiles of the PCNA Promoter Carrying E2F1 or E2F1+2 Mutations.
Luciferase specific activity (left) was measured in each leaf of transgenic plants carrying the E2F1 mutant and E2F1+2 mutant PCNA:luciferase constructs. The lines used for these studies are marked with arrows in Figure 3B. A minimum of four plants was analyzed for each line. RLU, relative light units. The vertical lines represent 2 se.
(A) Average luciferase specific activities for E2F1 mutant plant lines 46 (open circles) and 43 (closed circles) are shown compared with the average activity supported by the wild-type promoter (solid line).
(B) Average luciferase specific activity is shown for E2F1+2 lines 7 (open triangles) and 49 (closed triangles). The activity supported by the wild-type (solid line) and the E2F2 mutant (dashed line) promoters are shown for comparison.
These results suggested that both E2F sites contribute to the repression of PCNA activity in mature tissue, with the E2F2 site playing the major role. Analysis of the single-site mutants indicated that the E2F1 site is required for full activity of the PCNA promoter in young tissue, whereas the E2F2 site has no apparent involvement in transcription in young tissue. However, our results do not exclude a role for the E2F2 site at a specific stage of the cell cycle that cannot be detected in an asynchronous cell population, as occurs in young leaves. Analysis of E2F1+2 mutant lines revealed that the E2F1 site is required in young tissues only when the E2F2 site is intact. These data are consistent with the idea that the E2F2 site also functions as a negative regulatory element in young tissues but that this activity is countered by the E2F1 element in the wild-type promoter. The E2F1 mutant lines uncovered the negative regulatory potential of the E2F2 element, whereas the E2F1+2 mutants revealed that E2F1 acts as an antirepressor or a strong activator to overcome repression through the E2F2 element.
We also examined the impact of the E2F mutations on PCNA promoter activity in transient assays using N. tabacum BY2 and N. benthamiana suspension cells. These experiments failed to detect any statistically significant impact of the mutations on PCNA promoter activity in asynchronous cultured cells (data not shown).
Geminivirus Infection Has No Effect on the E2F Mutant Constructs
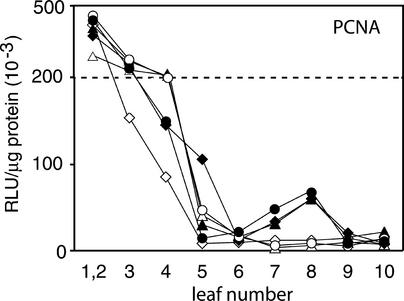
Because E2F mutations interfere with the correct developmental regulation of the PCNA promoter, we hypothesized that the same mutations might affect geminivirus-mediated activation of the promoter in mature tissues (Nagar et al., 1995; Egelkrout et al., 2001). We monitored luciferase activity in consecutive leaves starting at the apex during infection to determine if PCNA promoter constructs carrying the E2F1 or the E2F1+2 mutation could be activated by TGMV. The activity of the E2F1 mutant construct was very similar for healthy untreated, mock-inoculated, and TGMV-infected leaves at different stages of development (Figure 5A). Promoter activity also was comparable in untreated, mock-inoculated, and infected plants carrying the E2F1+2 mutant construct (Figure 5B). We previously found that TGMV infection has no detectable effect on the activity of the E2F2 mutant promoter (Egelkrout et al., 2001). Thus, the presence of intact E2F sites is necessary for the induction of PCNA expression over basal levels in infected mature tissue. However, the basal levels of reporter activity in healthy mature tissues of the various E2F mutant plants were threefold to eightfold higher than those of uninfected wild-type plants. Given that TGMV induction of the wild-type PCNA promoter ranged from twofold to sevenfold (Figure 5B, bottom), we cannot exclude the possibility that there is a response to geminivirus infection that is masked by the high basal levels of reporter gene expression in mature leaves of the mutant lines.
Figure 5.
E2F Mutant Promoters Are Not Activated by Geminivirus Infection.
Plants carrying the E2F1 mutant or E2F1+2 mutant PCNA:luciferase constructs were infected with TGMV by agroinoculation, and luciferase activity was measured in soluble protein extracts from each leaf of mature plants. Average luciferase specific activity in each leaf of healthy (dotted line), mock-inoculated (open circles or triangles), or TGMV-infected (closed circles or triangles) plants is shown for E2F1 lines 43 and 46 (A) and E2F1+2 lines 7 and 49 (B). A minimum of four plants were analyzed for each line and treatment. The vertical lines represent 2 se. The gray lines in the bottom graph in (B) correspond to healthy (dotted) and TGMV-infected (solid) profiles of plants carrying the wild-type PCNA:luciferase construct. RLU, relative light units.
PCNA Promoter Induction Is Characteristic of Begomovirus Infection
We asked if other geminiviruses in addition to TGMV activate the PCNA promoter during infection. For these experiments, we used Cabbage leaf curl virus (CbLCV), another geminivirus of the begomovirus genus. CbLCV and TGMV have similarly arranged bipartite genomes and share a common host in N. benthamiana (Hill et al., 1998). However, CbLCV is representative of a small group of viruses whose replication protein has diverged significantly from that of the majority of begomoviruses, including TGMV (our unpublished observation). Transgenic plants with the wild-type PCNA promoter fused to luciferase were infected with CbLCV, and PCNA promoter activity was monitored 11 days after infection (Figure 6). Infected plants showed a sixfold increase in PCNA promoter activity in leaf 8 relative to the equivalent mock-inoculated leaf. The CbLCV induction profile was similar to that observed for TGMV infection of the same transgenic line (Figure 5B, bottom) (Egelkrout et al., 2001). We also detected PCNA mRNA in mature leaves of CbLCV-infected plants (J.T. Ascencio-Ibañez and L. Hanley-Bowdoin, unpublished data), thereby providing independent verification of PCNA promoter activation by CbLCV.
Figure 6.
CbLCV Infection Activates the Wild-Type PCNA Promoter in Mature Leaves.
Transgenic plants carrying the wild type −633 PCNA:luciferase construct were infected with CbLCV by agroinoculation, and luciferase activity was measured in soluble protein extracts from each leaf of three infected (closed circles, diamonds, and triangles) and three mock-inoculated (open circles, diamonds, and triangles) plants. RLU, relative light units.
DISCUSSION
E2F transcription factors interact with each other and with other components of the transcription apparatus to regulate gene expression during the cell cycle and development. In animal systems, it is not uncommon for promoters to contain multiple E2F binding sites that act synergistically or antagonistically to regulate transcription (Yamaguchi et al., 1995; Zhu et al., 1995; Di Fiore et al., 1999). The N. benthamiana PCNA promoter contains two E2F consensus sequences (Egelkrout et al., 2001). We showed that both sequences, albeit to different degrees, contribute to the repression of the PCNA promoter in mature leaves. In young plant tissues, one of the elements can function as a negative regulator, but its activity is masked by the other element, which acts either as an antirepressor or as a strong activator to ensure high levels of PCNA expression in cycling cells. Our results and the results of others demonstrated that the regulation of PCNA transcription in plants is mediated by an intricate set of interactions involving different E2F complexes as well as other transcription factors (Kosugi et al., 1995; Kosugi and Ohashi, 1997; Egelkrout et al., 2001). Our studies also established the efficacy of using intact plants to study the mechanisms underlying the developmental regulation of cell cycle–associated genes in higher eukaryotes.
Our results differ significantly from the expression studies of the tobacco PCNA gene by Kosugi and Ohashi (2002a), who reported that mutations in either E2F site or in both sites together reduced the activity of the tobacco PCNA promoter in both young and mature tissues of transgenic tobacco. This finding was surprising given that N. benthamiana and N. tabacum PCNA genes are 92% identical over a 321-bp region encompassing both the proximal promoter and the leader, and because their E2F sites are identical in sequence and position. However, there are several key differences between the transgenic plants analyzed by Kosugi and Ohashi (2002a) and in the experiments reported here. Different Nicotiana species and reporter genes were used. In addition, different lengths of the PCNA promoters were fused to the reporters. We examined the E2F mutations in the context of a 743-bp fragment (a 677-bp promoter plus a 66-bp transcribed sequence) of the N. benthamiana gene, whereas Kosugi and Ohashi tested their mutations in a 321-bp fragment (a 208-bp promoter plus a 113-bp transcribed sequence) of the tobacco gene. The tobacco −208 promoter construct displayed low activity in transgenic plants (Kosugi and Ohashi, 2002a), and a N. benthamiana −275 promoter construct was 60% less active than the longer −946 and −633 constructs in cultured cells (Egelkrout et al., 2001). Together, these results showed that upstream sequences lacking in the −208 and −275 constructs are necessary for full PCNA promoter activity. There is evidence in animals that the function of E2F sites is dependent on the promoter context and interactions with other transcription factors (Fry et al., 1997; van Ginkel et al., 1997) whose recognition sites may be absent in the shorter PCNA promoter constructs.
Alternatively, the choice of mutations may have influenced the results. Both studies mutated the same two nucleotides in the E2F1 site and obtained similar results when differences in promoter strength are taken into consideration. By contrast, the E2F2 site was modified at only two positions in the tobacco promoter, whereas four positions were changed in the N. benthamiana promoter. In vitro binding studies verified that the 4-bp mutation prevents E2F binding, but there is no information regarding the impact of the 2-bp mutation on binding. Both types of mutations had minimal impact in young leaves when experimental scatter is considered, but only the 4-bp mutation resulted in the relief of repression in mature leaves. Because the E2F2 site consists of two overlapping consensus sequences (Wade et al., 1995), it is possible that the 2-bp mutation was not sufficient to block E2F binding. In this case, the prediction would be that the tobacco E2F2 mutant would resemble the wild type, whereas the double mutant would resemble the E2F1 mutant, as reported previously (Kosugi and Ohashi, 2002a).
Many eukaryotic promoters contain multiple E2F sites, but whether the sites have synergistic or opposing activities varies among promoters (Yamaguchi et al., 1995; Zhu et al., 1995; Di Fiore et al., 1999). Two lines of evidence suggest that the two E2F elements in the N. benthamiana PCNA promoter perform different functions. First, the two elements have distinct sequences, with E2F1 consisting of a single copy of the consensus motif and E2F2 containing inverted, overlapping motifs. In animal systems, the binding affinities of these types of elements differ, with the overlapping motif binding to E2F/DP heterodimers more strongly (Wade et al., 1995). Second, in vitro binding assays using nuclear extracts or recombinant E2F and DP proteins showed that the two sites can interact with different protein complexes. In particular, the E2F1 and E2F2 sites did not compete with each other in assays containing nuclear extracts. In addition, only the E2F2 element bound to recombinant E2Fb/DPa, and the E2F2 site bound E2Fc/DPa more strongly than the E2F1 site. There is precedent in animal promoters for different E2F sites to interact with different E2F/DP complexes (Zhu et al., 1995; Di Fiore et al., 1999), but this type of selectivity has not been described for plant E2F elements.
Mutations in the E2F sites affected PCNA promoter activity differently in transgenic plants. In young tissues, the E2F1 mutation resulted in a threefold decrease in promoter activity, whereas the E2F2 mutation had no detectable effect. Mutations in either element derepressed the PCNA promoter in mature leaves, but the E2F2 mutation was significantly stronger than the E2F1 mutation (eightfold versus threefold increase). The different results in young versus mature tissues may reflect developmental regulation of the factors that bind to the E2F1 and E2F2 sites. This idea, which is supported by RNA data indicating that the various E2F and DP family members have distinct developmental and tissue-specific expression patterns (Magyar et al., 2000; Kosugi and Ohashi, 2002b), provides an explanation for why the E2F1 element activates transcription in young tissues and represses it in mature tissues. In addition, the sequence and context of an E2F element can influence it binding properties (Fry et al., 1997; van Ginkel et al., 1997), thereby providing a basis for why the E2F2 site recruits a strong repressor and the E2F1 site binds to a weak repressor in mature tissues.
Analysis of the E2F1+2 double mutant revealed that PCNA transcriptional repression is complex and that the two E2F sites do not act independently. Failure of the E2F1+2 mutation to fully derepress the PCNA promoter in mature leaves indicated that other cis elements or irreversible changes in chromatin structure also contribute to repression. The role for chromatin structure is supported by the observation that the activities of the E2F mutant promoters were not altered significantly in transient assays. These assays used plasmid-based reporter cassettes, and it is unlikely that the chromatin remodeling factors recruited by E2F/pRb complexes affect episomal and chromosomal DNA similarly. Another possibility is that transcriptional activators in mature tissues are not as effective as those in young tissues at activating the PCNA promoter, even in the absence of repression.
Comparison of the E2F1+2 double mutant with individual mutants showed that the E2F1 site is required in young leaves only when a functional E2F2 element is present. This result is consistent with a model in which the E2F1 site overcomes the repressive activity of the E2F2 site to ensure high PCNA promoter activity in young tissues. One possibility is that E2F1 recruits a strong transcriptional activator that counters the activity of a repressor bound to E2F2. Alternatively, E2F1 may recruit an antirepressor that interferes directly with the activity of the repressor but is not itself a transcriptional activator. The best-characterized example of an antirepressor element associated with the cell division cycle is the overlapping CDE/CHR motif in the genes that encode cdc25C, cdc2, cyclin A, and B-myb (Liu et al., 1998). Interestingly, the E2F2 element in the N. benthamiana PCNA promoter shows homology with the CHR. There is evidence that competitive binding of E2F and an E2F-unrelated transcriptional repressor to the CDE/CHR site regulates promoter activity (Liu et al., 1997, 1998). However, the separation of the E2F1 and E2F2 sites in the N. benthamiana PCNA promoter suggests that the two sites function via a different mechanism.
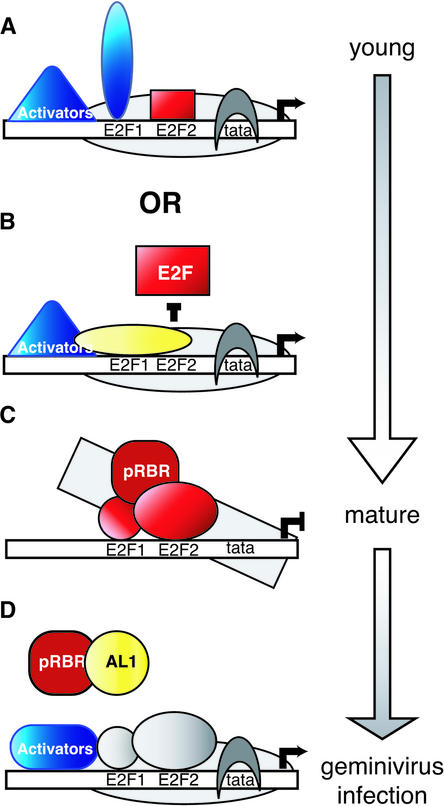
Plant PCNA promoters are likely to be controlled by a combination of transcriptional activation, repression, and antirepression (Figure 7). Our data showed that PCNA expression is repressed via the conserved E2F1 and E2F2 sites in the proximal promoter region in mature tissues. By analogy to animal E2F-regulated promoters, these sites act by recruiting E2F/pRBR complexes that alter chromatin structure and block access to the promoter (Figure 7C). Like mammalian DNA tumor antigen proteins, the geminivirus AL1 protein relieves repression through its interaction with pRBR, leading to the disruption of repressive E2F/pRBR complexes (Figure 7D). In young plant tissues, high levels of PCNA expression are achieved by the combined action of transcriptional activators and the antirepression activity of factors recruited to the E2F1 site (Figures 7A and 7B). Antirepression activity may reflect factors bound to the E2F1 site that block the binding of the repressor to the E2F2 site, interfering with the ability of a bound repressor to contact other components of the transcription apparatus or masking repression through the recruitment of a strong activator. The different activities of the E2F1 and E2F2 elements at the same developmental stages and their changing activities during leaf development may reflect the binding of different E2Fs and their partners, which also are likely to be subject to developmental regulation. Future experiments to characterize the protein complexes recruited to the N. benthamiana PCNA promoter as well as their interactions and occupancy in young and mature leaves will address these possibilities and provide insight into the functions of pRBR and different E2F family members during plant development.
Figure 7.
Model for the Regulation of a Plant PCNA Promoter.
A model for PCNA promoter regulation is shown, with activators indicated in blue, repressors indicated in red, and antirepressors indicated in yellow. E2F1 and E2F2 may be occupied by different family members at different stages, as reflected by the different shapes. The sizes of the shapes reflect the relative strengths of the factors occupying the sites.
(A) In young tissue, E2F1 may recruit a strong activator that masks the activity of a weak repressor bound to the E2F site. Other transcription factors also contribute to the full activation of the promoter.
(B) Alternatively, in young tissue, binding of an antirepressor to the E2F1 site prevents the binding of a repression complex to the E2F2 site or interferes with protein interactions required for repression. Other transcription factors are responsible for the activation of the promoter.
(C) As plant tissues mature, changes in the E2F1 binding complex allow recruitment of a pRBR/E2F complex to the E2F2 site, leading to the repression of the PCNA promoter. Factors binding to the E2F1 site also contribute to repression, albeit to a lesser extent.
(D) In geminivirus-infected cells, AL1 interacts with pRBR to induce the release of the repressive complex bound to the E2F sites and the reassembly of an active promoter. It is not known if factors bound to the E2F1 and E2F2 sites function as activators during infection.
METHODS
E2F Mutant Constructs and Transient Transfection Assays
The proliferating cell nuclear antigen (PCNA) E2F1 and E2F1+2 mutant constructs were generated by two-step overlap extension PCR, as described previously (Egelkrout et al., 2001). The gene-specific primers for the E2F1 construct were E2F1 #1 (5′-CCAAAATAGAGGtaGGAAAAATATTTTTTCCAC-3′) and E2F1 #2 (5′-ATATTTTTCCtaCCTCTATTTTGGGC-3′), with mutations shown in lowercase letters. The PCR product was gel purified, repaired with Klenow, and cloned into the SmaI site of pUC119 to give pNSB925. To prepare the E2F1+2 mutant, pNSB925 was used as a template for PCR mutagenesis of the E2F2 site using the gene-specific primers E2F2 #1 and E2F2 #2 (Egelkrout et al., 2001). The PCR product was cloned into pUC119 to produce pNSB927. Mutant promoter fragments from pNSB925 and pNSB927 with BamHI and trimmed SacI ends were fused to the luciferase coding sequence in pMON8796 (Eagle et al., 1994) with BglII and trimmed PstI ends to produce pNSB939 and pNSB940, respectively. Preparation of protoplasts from Nicotiana benthamiana and N. tabacum BY2 cells, DNA transfections, and enzyme assays were as described previously (Eagle et al., 1994).
Generation of Transgenic Plants and Reporter Assays
The PCNA promoter–luciferase fusions from pNSB939 and pNSB940 were cloned as NotI expression cassettes into NotI-digested pMON721 (Lanahan et al., 1994) to generate pNSB936 and pNSB938, respectively. The constructs were introduced stably into N. benthamiana plants via Agrobacterium tumefaciens–mediated transformation using standard protocols (Horsch et al., 1985). The transgene promoters were verified in planta by PCR amplification of chromosomal DNA using the primers pMON721 (5′-TCGAAGCCGTGTGCGAGA-GACACC-3′) and LUCSEQ (5′-GGCGTATCTCTTCATAGCCTTATG-C-3′) followed by DNA sequencing. Plants were grown in a controlled-environment chamber at 25°C with a 16-h/8-h light/dark cycle and 65% RH. Preparation of crude extracts, infection of plants by agroinoculation, luciferase reporter assays, and statistical analysis were as described previously (Egelkrout et al., 2001). In all cases, plants were of the T1 generation and selected on kanamycin for the presence of the transgene.
Electrophoretic Mobility Shift Assays
N. benthamiana nuclear extracts were prepared from log-phase cultured cells as described previously (Albani et al., 2000; Egelkrout et al., 2001) except that pelleted nuclei were frozen at −80°C overnight before lysis in some cases. Electrophoretic mobility shift assays with nuclear extracts (1.9 μg per reaction) were as described previously (Egelkrout et al., 2001) except that 2.5 × 104 or 2.5 × 105 cpm/probe was used for the E2F2 and E2F1 experiments, respectively. Production of recombinant Arabidopsis E2F and DP proteins and electrophoretic mobility shift assays with purified proteins were as described (Albani et al., 2000; Mariconti et al., 2002) except that the probes indicated in Figure 1A were used. Each assay contained 100 ng of each recombinant protein and 5 × 104 cpm/probe.
Cabbage leaf curl virus Clones and Infectivity Assays
The plasmids pCpCLCVA.003.2 and pCpCLCVB.002.2, carrying single copies of the Cabbage leaf curl virus (CbLCV) A and B components, respectively, have been described (Turnage et al., 2002). Single copies of the A and B components were isolated from these clones as KpnI-EcoRI and XbaI-EcoRI fragments. The resulting fragments were ligated into the Agrobacterium transformation vector pMON721 (Lanahan et al., 1994) and digested with the same enzymes to produce pNSB1087 and pNSB1088. pNSB1087 was digested with EcoRI and BamHI and ligated to the EcoRI-BamHI fragment from pCpCLCVA.003.2 to generate a partial tandem copy of the CbLCV A component in pNSB1090. pNSB1088 digested with EcoRI was ligated to the EcoRI fragment from pCpCLCVB.002.2 to generate a partial tandem copy of the CbLCV B component in pNSB1091. Transformation into Agrobacterium, plant infection by syringe inoculation, preparation of protein extracts, and reporter assays all were as described (Egelkrout et al., 2001) except that tissue was harvested 11 days after inoculation. The −633 PCNA:luciferase line L-39 (Egelkrout et al., 2001) was used in the CbLCV infectivity assays.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Tara Nash for her valuable technical support. This research was supported by grants from the National Research Initiative Competitive Grants Program of the U.S. Department of Agriculture (98-01392 to L.H.-B. and D.R.), the National Science Foundation (MCB-9809953 and MCB-0110536 to L.H.-B.), and the Ministero dell'Istruzione, dell'Università e della Ricerca (to L.M. and R.C).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006403.
References
- Ach, R.A., Durfee, T., Miller, A.B., Taranto, P., Hanley-Bowdoin, L., Zambryski, P.C., and Gruissem, W. (1997). RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol. Cell. Biol. 17, 5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani, D., Mariconti, L., Ricagno, S., Pitto, L., Moroni, C., Helin, K., and Cella, R. (2000). DcE2F, a functional plant E2F-like transcriptional activator from Daucus carota. J. Biol. Chem. 275, 19258–19267. [DOI] [PubMed] [Google Scholar]
- Black, A.R., and Azizkhan-Clifford, J. (1999). Regulation of E2F: A family of transcription factors involved in proliferation control. Gene 237, 281–302. [DOI] [PubMed] [Google Scholar]
- Boniotti, M.B., and Gutierrez, C. (2001). A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. 28, 341–350. [DOI] [PubMed] [Google Scholar]
- Castellano, M.M., del Pozo, J.C., Ramirez-Parra, E., Brown, S., and Gutierrez, C. (2001). Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13, 2671–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboute, M.E., Clement, B., and Philipps, G. (2002). S phase and meristem-specific expression of the tobacco RNR1b gene is mediated by an E2F element located in the 5′ leader sequence. J. Biol. Chem. 277, 17845–17851. [DOI] [PubMed] [Google Scholar]
- Chaboute, M.E., Clement, B., Sekine, M., Philipps, G., and Chaubet-Gigot, N. (2000). Cell cycle regulation of the tobacco ribonucleotide reductase small subunit gene is mediated by E2F-like elements. Plant Cell 12, 1987–2000. [PMC free article] [PubMed] [Google Scholar]
- de Jager, S.M., Menges, M., Bauer, U.M., and Murray, J.A. (2001). Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Mol. Biol. 47, 555–568. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., de Almeida Engler, J., Ormenese, S., Maes, S., Naudts, M., Van Der Schueren, E., Jacqmard, A., Engler, G., and Inze, D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21, 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore, B., Guarguaglini, G., Palena, A., Kerkhoven, R.M., Bernards, R., and Lavia, P. (1999). Two E2F sites control growth-regulated and cell cycle-regulated transcription of the Htf9-a/RanBP1 gene through functionally distinct mechanisms. J. Biol. Chem. 274, 10339–10348. [DOI] [PubMed] [Google Scholar]
- Durfee, T., Feiler, H.S., and Gruissem, W. (2000). Retinoblastoma-related proteins in plants: Homologues or orthologues of their metazoan counterparts? Plant Mol. Biol. 43, 635–642. [DOI] [PubMed] [Google Scholar]
- Eagle, P.A., Orozco, B.M., and Hanley-Bowdoin, L. (1994). A DNA sequence required for geminivirus replication also mediates transcriptional regulation. Plant Cell 6, 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout, E.M., Robertson, D., and Hanley-Bowdoin, L. (2001). Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13, 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountain, M.D., Murray, J.A.H., and Beck, E. (1999). Isolation of a full-length cDNA encoding a retinoblastoma (accession No. AJ011681) protein from suspension cultured photoautotrophic Chenopodium rubrum L. cells. Plant Physiol. 119, 363.10232959 [Google Scholar]
- Fry, C.J., Slansky, J.E., and Farnham, P.J. (1997). Position-dependent transcriptional regulation of the murine dihydrofolate reductase promo-ter by the E2F transactivation domain. Mol. Cell. Biol. 17, 1966–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafi, G., Burnett, R.J., Helentjaris, T., Larkins, B.A., DeCaprio, J.A., Sellers, W.R., and Kaelin, W.G., Jr. (1996). A maize cDNA encoding a member of the retinoblastoma protein family: Involvement in endoreduplication. Proc. Natl. Acad. Sci. USA 93, 8962–8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez, C. (2000). DNA replication and cell cycle in plants: Learning from geminiviruses. EMBO J. 19, 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin, L., Settlage, S.B., Orozco, B.M., Nagar, S., and Robertson, D. (2000). Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol. Biol. 35, 105–140. [PubMed] [Google Scholar]
- Harbour, J.W., and Dean, D.C. (2000). The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 14, 2393–2409. [DOI] [PubMed] [Google Scholar]
- Herwig, S., and Strauss, M. (1997). The retinoblastoma protein: A master regulator of cell cycle, differentiation and apoptosis. Eur. J. Biochem. 246, 581–601. [DOI] [PubMed] [Google Scholar]
- Hill, J.E., Strandberg, J.O., Hiebert, E., and Lazarowitz, S.G. (1998). Asymmetric infectivity of pseudorecombinants of cabbage leaf curl virus and squash leaf curl virus: Implications for bipartite geminivirus evolution and movement. Virology 250, 283–292. [DOI] [PubMed] [Google Scholar]
- Horsch, R.B., Fry, J.E., Hoffman, N.L., Wallroth, M., Eicholtz, D., Rogers, S.G., and Fraley, R.T. (1985). A simple and general method for transferring cloned genes into plants. Science 227, 1229–1231. [DOI] [PubMed] [Google Scholar]
- Kong, L.J., Orozco, B.M., Roe, J.L., Nagar, S., Ou, S., Feiler, H.S., Durfee, T., Miller, A.B., Gruissem, W., Robertson, D., and Hanley-Bowdoin, L. (2000). A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19, 3485–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9, 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2002. a). E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J. 29, 45–59. [DOI] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2002. b). E2Ls, E2F-like repressors of Arabidopsis that bind to E2F-sites in a monomeric form. J. Biol. Chem. 277, 16553–16558. [DOI] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2002. c). Interaction of the Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol. 128, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S., Suzuka, I., and Ohashi, Y. (1995). Two of three promoter elements identified in a rice gene for proliferating cell nuclear antigen are essential for meristematic tissue-specific expression. Plant J. 7, 877–886. [DOI] [PubMed] [Google Scholar]
- Lanahan, M.B., Yen, H.-C., Giovannoni, J.J., and Klee, H.J. (1994). The never ripe mutation blocks ethylene perception in tomato. Plant Cell 6, 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavia, P., and Jansen-Durr, P. (1999). E2F target genes and cell-cycle checkpoint control. Bioessays 21, 221–230. [DOI] [PubMed] [Google Scholar]
- Liu, N., Lucibello, F.C., Engeland, K., and Muller, R. (1998). A new model of cell cycle-regulated transcription: Repression of the cyclin A promoter by CDF-1 and anti-repression by E2F. Oncogene 16, 2957–2963. [DOI] [PubMed] [Google Scholar]
- Liu, N., Lucibello, F.C., Korner, K., Wolfraim, L.A., Zwicker, J., and Muller, R. (1997). CDF-1, a novel E2F-unrelated factor, interacts with cell cycle-regulated repressor elements in multiple promoters. Nucleic Acids Res. 25, 4915–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar, Z., Atanassova, A., De Veylder, L., Rombauts, S., and Inze, D. (2000). Characterization of two distinct DP-related genes from Arabidopsis thaliana. FEBS Lett. 486, 79–87. [DOI] [PubMed] [Google Scholar]
- Mariconti, L., Pellegrini, B., Cantoni, R., Stevens, R., Bergounioux, C., Cella, R., and Albani, D. (2002). The E2F family of transcription factors from Arabidopsis thaliana: Novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 277, 9911–9919. [DOI] [PubMed] [Google Scholar]
- Muller, H., and Helin, K. (2000). The E2F transcription factors: Key regulators of cell proliferation. Biochim. Biophys. Acta 1470, M1–M12. [DOI] [PubMed] [Google Scholar]
- Nagar, S., Pedersen, T.J., Carrick, K.M., Hanley-Bowdoin, L., and Robertson, D. (1995). A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7, 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami, H., Sekine, M., Murakami, H., and Shinmyo, A. (1999). Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. Plant J. 18, 243–252. [DOI] [PubMed] [Google Scholar]
- Nevins, J.R. (1992). E2F: A link between the Rb tumor suppressor protein and viral oncoproteins. Science 258, 424–429. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra, E., and Gutierrez, C. (2000). Characterization of wheat DP, a heterodimerization partner of the plant E2F transcription factor which stimulates E2F-DNA binding. FEBS Lett. 486, 73–78. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra, E., Xie, Q., Boniotti, M.B., and Gutierrez, C. (1999). The cloning of plant E2F, a retinoblastoma-binding protein, reveals unique and conserved features with animal G(1)/S regulators. Nucleic Acids Res. 27, 3527–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol, P., Stevens, R., Perennes, C., Jasinski, S., Cella, R., Tremousaygue, D., and Bergounioux, C. (2002). AtE2F-a and AtDP-a, members of the E2F family of transcription factors, induce Arabidopsis leaf cells to re-enter S phase. Mol. Genet. Genomics 266, 995–1003. [DOI] [PubMed] [Google Scholar]
- Sekine, M., Ito, M., Uemukai, K., Maeda, Y., Nakagami, H., and Shinmyo, A. (1999). Isolation and characterization of the E2F-like gene in plants. FEBS Lett. 460, 117–122. [DOI] [PubMed] [Google Scholar]
- Stevens, R., Mariconti, L., Rossignol, R., Perennes, C., Cella, R., and Bergounioux, C. (2002). Two E2F sites in the Arabidopsis MCM3 promoter have different roles in cell cycle activation and meristematic expression. J. Biol. Chem. 277, 32978–32984. [DOI] [PubMed] [Google Scholar]
- Trimarchi, J.M., and Lees, J.A. (2002). Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3, 11–20. [DOI] [PubMed] [Google Scholar]
- Turnage, M.A., Muangsan, N., Peele, C.G., and Robertson, D. (2002). Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J., 29, 1–9. [DOI] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouze, P., Rombauts, S., and Inze, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ginkel, P.R., Hsiao, K.M., Schjerven, H., and Farnham, P.J. (1997). E2F-mediated growth regulation requires transcription factor cooperation. J. Biol. Chem. 272, 18367–18374. [DOI] [PubMed] [Google Scholar]
- Wade, M., Blake, M.C., Jambou, R.C., Helin, K., Harlow, E., and Azizkhan, J.C. (1995). An inverted repeat motif stabilizes binding of E2F and enhances transcription of the dihydrofolate reductase gene. J. Biol. Chem. 270, 9783–9791. [DOI] [PubMed] [Google Scholar]
- Weinberg, R.A. (1995). The retinoblastoma protein and cell cycle control. Cell 81, 323–330. [DOI] [PubMed] [Google Scholar]
- Xie, Q., Sanz-Burgos, A.P., Hannon, G.J., and Gutierrez, C. (1996). Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 15, 4900–4908. [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, M., Hayashi, Y., and Matsukage, A. (1995). Essential role of E2F recognition sites in regulation of the proliferating cell nuclear antigen gene promoter during Drosophila development. J. Biol. Chem. 270, 25159–25165. [DOI] [PubMed] [Google Scholar]
- Zacksenhaus, E., Jiang, Z., Phillips, R.A., and Gallie, B.L. (1996). Dual mechanisms of repression of E2F1 activity by the retinoblastoma gene product. EMBO J. 15, 5917–5927. [PMC free article] [PubMed] [Google Scholar]
- Zhang, H.S., and Dean, D.C. (2001). Rb-mediated chromatin structure regulation and transcriptional repression. Oncogene 20, 3134–3138. [DOI] [PubMed] [Google Scholar]
- Zhu, L., Zhu, L., Xie, E., and Chang, L.S. (1995). Differential roles of two tandem E2F sites in repression of the human p107 promoter by retinoblastoma and p107 proteins. Mol. Cell. Biol. 15, 3552–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker, J., Lucibello, F.C., Wolfraim, L.A., Gross, C., Truss, M., Engeland, K., and Muller, R. (1995). Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 14, 4514–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker, J., and Muller, R. (1997). Cell-cycle regulation of gene expression by transcriptional repression. Trends Genet. 13, 3–6. [DOI] [PubMed] [Google Scholar]