Abstract
The expression of two Arabidopsis expansin genes (AtEXP7 and AtEXP18) is tightly linked to root hair initiation; thus, the regulation of these genes was studied to elucidate how developmental, hormonal, and environmental factors orchestrate root hair formation. Exogenous ethylene and auxin, as well as separation of the root from the medium, stimulated root hair formation and the expression of these expansin genes. The effects of exogenous auxin and root separation on root hair formation required the ethylene signaling pathway. By contrast, blocking the endogenous ethylene pathway, either by genetic mutations or by a chemical inhibitor, did not affect normal root hair formation and expansin gene expression. These results indicate that the normal developmental pathway for root hair formation (i.e., not induced by external stimuli) is independent of the ethylene pathway. Promoter analyses of the expansin genes show that the same promoter elements that determine cell specificity also determine inducibility by ethylene, auxin, and root separation. Our study suggests that two distinctive signaling pathways, one developmental and the other environmental/hormonal, converge to modulate the initiation of the root hair and the expression of its specific expansin gene set.
INTRODUCTION
Root hairs are polarized outgrowths of root epidermal cells. In Arabidopsis, root hairs normally arise from epidermal cells that contact two underlying cortical cells (the so-called H position), whereas epidermal cells overlying a single cortical cell (in the N position) develop into nonhair cells (Dolan et al., 1993; Galway et al., 1994). This position-dependent hair cell differentiation thus results in a striped pattern of hair cell files along the long axis of the root, which is found in members of Brassicaceae and in a few species of other families (Cormack, 1947; Dolan and Costa, 2001). Root hair development in Arabidopsis can be divided into three phases: cell specification, initiation, and elongation. Cell specification refers to the fate determination of epidermal cells into hair cells and nonhair cells, depending on position. Initiation refers to the formation of a protrusion or bulge at the site of hair outgrowth. Elongation refers to the process of sustained tip growth that normally follows initiation. Numerous experimental observations indicate that these three phases involve different cellular and genetic processes (for reviews, see Schiefelbein, 2000; Foreman and Dolan, 2001).
Several genes that control root epidermal cell specification have been identified. Loss-of-function mutations in TTG (TRANSPARENT TESTA/GLABROUS) or GL2 (GLABRA2) result in root hairs in both H and N positions (Galway et al., 1994; Masucci et al., 1996), indicating that TTG (a protein with WD40 repeats) and GL2 (a homeodomain transcription factor) function as negative regulators of the differentiation of nonhair cells to hair cells. Mutations in another MYB transcription factor, WER (WEREWOLF), also generate root hairs in almost every root epidermal cell, because WER positively regulates GL2 expression (Lee and Schiefelbein, 1999). On the other hand, cpc (caprice) mutants have only a few root hairs, indicating that CPC, a MYB-like protein, functions as a positive regulator for root hair cell differentiation (Wada et al., 1997). A recent study demonstrated the interactions among these regulatory genes (Lee and Schiefelbein, 2002). In the N position, WER positively regulates the expression of CPC and GL2. CPC (or its downstream signal) appears to move to cells in the H position and inhibits the expression of WER, CPC, and GL2, which leads the cell to initiate hair formation.
Root hair initiation, which is genetically downstream of GL2 (Masucci and Schiefelbein, 1996), is regulated by another set of genes and is sensitive to hormonal and environmental factors (Schiefelbein, 2000). The auxin-resistant mutant (axr2) develops few root hair bulges (Wilson et al., 1990), and the defect of root hair initiation in root hair defective (rhd6) can be reversed by treatment with auxin or the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) (Masucci and Schiefelbein, 1994, 1996). ACC treatment of wild-type plants induces root hairs in the N position, as do the constitutively ethylene-responsive ctr1 and ethylene-overproducing eto mutants (Dolan et al., 1994; Masucci and Schiefelbein, 1996; Cao et al., 1999). ACC has been suggested as a factor that determines the developmental fate of cells in the H position (Tanimoto et al., 1995). Also implicating ethylene involvement in root hair initiation, the ethylene biosynthesis inhibitor aminoethoxyvinylglycine (AVG) and silver ion (an inhibitor of ethylene perception) have been found to inhibit root hair formation (Masucci and Schiefelbein, 1994, 1996; Tanimoto et al., 1995). However, the role of ethylene in root hair formation is questioned because the ethylene-insensitive mutants etr1 and ein2 maintained normal root hair numbers (Masucci and Schiefelbein, 1996). Additionally, environmental factors such as nutrients (Peterson and Stevens, 2000), light, and separation of the root from the agar medium (Okada and Shimura, 1994) also affect root hair development. It has been suggested that hormones and environmental factors affect root hair initiation through a pathway distinctive from the normal development-associated pathway (Okada and Shimura, 1994; Schiefelbein, 2000), but experimental confirmation for this is needed.
Elongation of the root hair is achieved by tip growth (Schiefelbein, 2000). Hair elongation likely is governed by genetic components distinct from those that govern hair initiation, but root hair elongation is influenced by auxin, ethylene, and environmental factors as well (Okada and Shimura, 1994; Pitts et al., 1998; Schiefelbein, 2000).
Spatial regulation of cell wall expansion is critical for cell morphogenesis in plants (Fowler and Quatrano, 1997). Thus, outgrowth of the root hair from the epidermal cell is expected to accompany localized cell wall loosening at the correct position. Bibikova et al. (1998) demonstrated localized wall acidification at the site of root hair initiation. This acidification could activate expansins. Expansins are cell wall–loosening proteins capable of mediating cell wall extension in acidic conditions without hydrolytic breakage of major structural components of the cell wall (McQueen-Mason et al., 1992; for recent reviews, see Cosgrove, 2000; Lee et al., 2001). Expansin genes are found throughout the entire plant kingdom (Cosgrove, 1999; Li et al., 2002), and their pattern of expression indicates that they are related closely to cell growth and tissue differentiation (for review, see Cho, 2001). Alteration of endogenous expansin gene expression modulates leaf growth and pedicel abscission in Arabidopsis (Cho and Cosgrove, 2000) and leaf morphology and phylotaxy in tobacco (Pien et al., 2001). Two families of expansins are recognized at present (Cosgrove, 2000), α- and β-expansins, and Arabidopsis has 26 α- and 5 β-expansin genes (see http://www.bio.psu.edu/expansins). In the course of analyzing the expression of these genes in Arabidopsis, two α-expansin genes, AtEXP7 and AtEXP18, were found to be expressed specifically in root hair cells (D.M. Durachko and D.J. Cosgrove, unpublished data).
In this study, we examined in detail the expression patterns of these two root hair–specific expansin genes in various root hair mutants as well as under hormonal (auxin and ethylene) and environmental (separation of the root from the medium) treatments. In particular, the role of endogenous ethylene in root hair development was studied closely. Promoter analyses of the two expansin genes, in conjunction with the effect of root hair–inducing factors, also were conducted to elucidate the regulation of expression of these root hair–specific genes. Our results show that the expression of these expansin genes is linked tightly to root hair initiation and subsequent elongation. Moreover, we find that, although ethylene mediates the effects of auxin and root separation on root hair initiation, it is not essential for the normal (or default) development of root hairs in wild-type plants. These results alter current views of ethylene involvement in root hair development.
RESULTS
Root Hair Cell–Specific Expression of AtEXP7 and AtEXP18
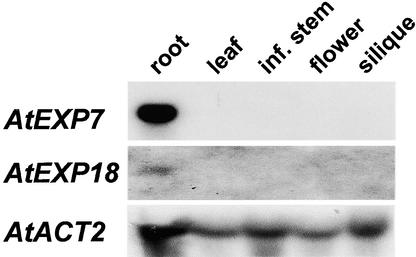
RNA gel blot and promoter-reporter gene expression analyses were performed to investigate the organ- and tissue-specific expression patterns of AtEXP7 and AtEXP18. The transcripts of both expansin genes were found in the root but were undetectable in other major plant organs (Figure 1). Wild-type plants harboring the AtEXP7 promoter::β-gluc-uronidase (GUS) or AtEXP7 promoter::green fluorescent protein (GFP) construct showed staining (or fluorescence) solely in root hair cell files (Figures 2A to 2D). No reporter gene expression was found in other cell types of the root or other organs except a weak expression in the inner layer of the seed coat. Different ecotypes, Columbia and Wassilewskija, showed the same reporter gene expression pattern. The expression of AtEXP7 occurred approximately one cell before the root hair bulges appeared (Figure 2B), indicating the gene's close temporal expression with the hair initiation process. Plants harboring the AtEXP18 promoter::reporter construct also showed the same expression pattern as plants with the AtEXP7 promoter::reporter construct (data not shown). However, the level of AtEXP18 expression was lower than that of AtEXP7. Promoter analyses, as described below, showed that the average promoter activity of AtEXP18 was ∼60% of AtEXP7 promoter activity. In this study, the expression pattern of AtEXP7 is described in greater detail, but the results also hold for AtEXP18.
Figure 1.
Expression of AtEXP7 and AtEXP18 in Different Tissues.
Total RNA was isolated from seedling roots, young leaves, growing inflorescence (inf.) stems, whole floral organs, and young green siliques of Columbia wild-type Arabidopsis plants. Twenty micrograms of total RNA was analyzed per lane. The transcript levels of Arabidopsis actin2 (AtACT2) served as a loading control.
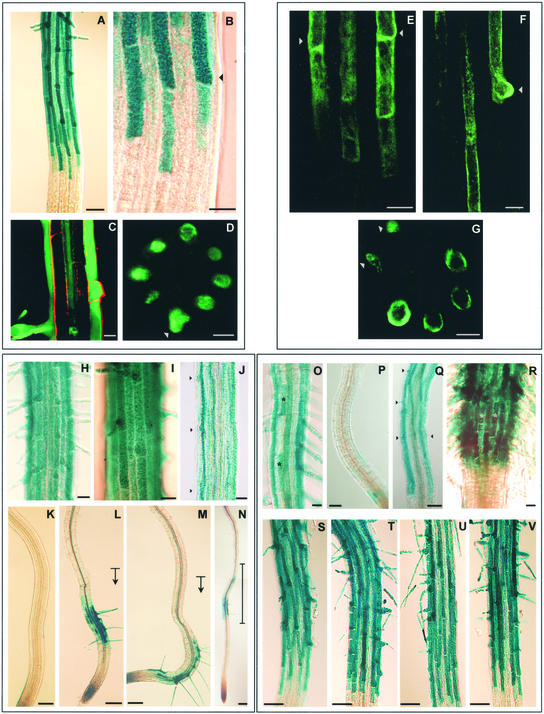
Figure 2.
Root Hair Cell–Specific Expression Pattern of AtEXP7 in the Arabidopsis Root.
(A), (B), and (H) to (V) show AtEXP7 promoter::GUS expression; (C) and (D) show AtEXP7 promoter::GFP expression; and (E) to (G) show AtEXP7 promoter::genomic AtEXP7-GFP expression.
(A) to (D) In the wild-type root, reporter gene expression occurs in the root hair cell files. The weaker blue staining between the strong stains are from the hair cell files of the opposite side. (C) shows an optical longitudinal section demonstrating GFP expression at the root hair cell files. The red area from propidium iodide indicates the cell boundary. (D) shows an optical cross-section of the root demonstrating gene expression at the eight root hair cells. The arrowheads in (B) and (D) indicate emerging root hair bulges.
(E) to (G) Expression of the AtEXP7-GFP fusion protein shows the same pattern as expression of GUS or GFP alone. (G) shows an optical cross-section. Arrowheads indicate emerging root hair bulges.
(H) and (I) In the ttg-1 (H) and gl2-1 (I) backgrounds, reporter gene expression is observed in cells from both the H and N positions.
(J) axr2-1 background. Arrowheads indicate some root hair bulges.
(K) to (N) rhd6 background with no treatment (K) or with 5 μM ACC (L), 30 nM IAA (M), or separation of the root from the medium (N). The bases of the arrows in (L) and (M) indicate the approximate starting points of hormone treatments. The vertical bar in (N) indicates where the root was separated from agar.
(O) to (Q) Wild-type roots treated with 5 μM ACC (O), 5 μM AVG (P), or 50 μM silver ion (Q). Stars in (O) indicate ectopic expression of GUS in the N positions.
(R) ctr1-1 background. Stars indicate ectopic expression of GUS in the N positions.
(S) to (U) Dominant ethylene receptor mutants etr1-1 (S), ein4 (T), and ers2-1 (U).
(V) ein2-1 background.
Bars = 100 μm in (K) to (N), 50 μm in (A), (P), (Q), and (S) to (V), and 20 μm in (B) to (J), (O), and (R).
The AtEXP7 protein expression pattern also was examined by expressing the AtEXP7-GFP fusion protein driven by the AtEXP7 promoter. The cell-type specificity and the timing of protein expression were almost identical with the expression pattern of the reporter gene alone (Figures 2E to 2G). The fluorescence from the fusion protein was highest in regions of root hair initiation and elongation. Although the AtEXP7-GFP fusion protein tended to localize more at the emerging root hair tip and to distribute peripherally in the root hair cell (Figures 2E to 2G), it was detected predominantly inside the plasma membrane upon plasmolysis (data not shown). This finding indicates that the fusion protein was not secreted to the cell wall.
We have searched for mutants defective in AtEXP7 or AtEXP18. An Arabidopsis line that includes a T-DNA insertion in the second intron of AtEXP7 was identified, but the homozygous line still expressed transcripts of the correct size, albeit at a lower level than in the wild type. This line did not show obvious alterations in the root hair, most likely as a result of the leakiness of the mutation and functional redundancy by AtEXP18 and perhaps other expansin genes.
Effect of Root Hair–Regulating Factors on the Expression of Root Hair Expansin Genes
Root hair formation in Arabidopsis is regulated by developmental regulators, hormones, and environmental factors. Because AtEXP7 is a root hair–specific gene and is thought to function in root hair formation, we investigated whether AtEXP7 expression is modulated by various root hair–regulating factors. For this purpose, the AtEXP7 promoter::GUS reporter construct was introduced into root hair mutants, and the reporter gene expression pattern was monitored.
In ttg and gl2 mutants, which have hairs in both the H and N positions, AtEXP7 promoter::GUS was expressed in both positions (Figures 2H and 2I), suggesting that TTG and GL2 negatively regulate the expression of AtEXP7, just as they negatively regulate root hair formation in the N position of the wild-type plant (Galway et al., 1994; Masucci et al., 1996).
The axr2 mutant is defective in hair elongation and partially in hair initiation; thus, it produces few root hair bulges (Masucci and Schiefelbein, 1994, 1996) (Figure 2J). The spatial pattern of AtEXP7 promoter::GUS expression was not changed in this mutant (Figure 2J) compared with that in wild-type plants. However, the expression level of AtEXP7 was much lower in the mutant than in the wild type (Figure 3). Because auxin positively regulates root hair formation and AtEXP7 expression (see below), AXR2 likely downregulates the expression of AtEXP7 and partially inhibits root hair formation.
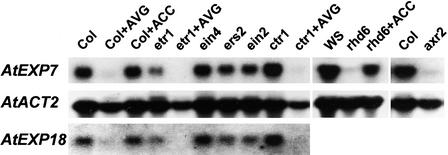
Figure 3.
RNA Gel Blot Analyses of AtEXP7 and AtEXP18 Transcripts in Different Mutant Backgrounds and under Treatment with Ethylene Precursor and Inhibitor.
Total RNA was prepared from roots of 4-day-old wild-type and mutant seedlings. For ACC (5 μM) and AVG (5 μM) treatments, the seedlings were transferred to chemical-containing plates on day 3. Ten micrograms of total RNA, except for Wassilewskija and rhd6 (30 μg), was analyzed. The transcript level of Arabidopsis actin2 (AtACT2) served as a loading control. Col, Columbia wild type; WS, Wassilewskija wild type.
The mutant rhd6 also is defective in root hair initiation, but unlike axr2, it develops almost no root hair bulges (Masucci and Schiefelbein, 1994, 1996) (Figure 2K). AtEXP7 expression in rhd6 was blocked almost completely, as shown by GUS expression and by transcript analysis (Figures 2K and 3). In rhd6, treatment with the ethylene precursor ACC or auxin, or separation of the root from the agar medium induced normal root hair formation (Masucci and Schiefelbein, 1994, 1996) (Table 1). In agreement with their effects on root hair formation, all of these treatments induced AtEXP7 expression in rhd6 roots (Figures 2L to 2N and 3). AtEXP18 expression in rhd6, as described below, also was inducible by these treatments. These results indicate that RHD6 is a positive regulator of AtEXP7 and AtEXP18 expression.
Table 1.
Root Hair Number in Wild-Type and Mutant Plants with Ethylene Precursor or Inhibitor Treatment
| Percent of Total Root Hair Cellsa
|
Percent of Root Hair Cells in the N positionb
|
|||||
|---|---|---|---|---|---|---|
| Plant | No Treatment | ACC (5 μM) | AVG (5 μM) | 1-MCP (1 μL/L) | No Treatment | ACC (5 μM) |
| Columbia | 51.1 ± 3.3 | 65.9 ± 5.8 | 1.3 ± 2.3 | 44.0 ± 4.2 | 1.1 ± 3.3 | 15.9 ± 5.8 |
| etr1-1 | 45.8 ± 6.7 | 48.1 ± 3.7 | 0 ± 0 | 43.0 ± 2.7 | 2.7 ± 3.3 | 2.5 ± 4.6 |
| etr2 | 51.3 ± 2.3 | 51.5 ± 3.4 | 0.6 ± 1.8 | 48.3 ± 2.5 | 1.3 ± 2.3 | 1.5 ± 3.4 |
| ers1 | 55.4 ± 6.2 | 52.5 ± 4.2 | 0.6 ± 1.8 | 49.5 ± 2.7 | 5.4 ± 6.2 | 2.5 ± 4.2 |
| ers2-1 | 50.0 ± 3.0 | 55.0 ± 8.2 | 0 ± 0 | 44.1 ± 3.8 | 0.8 ± 1.9 | 6.4 ± 8.5 |
| ein4 | 50.4 ± 1.4 | 57.2 ± 8.7 | 0 ± 0 | 42.3 ± 3.5 | 0.4 ± 1.4 | 7.2 ± 8.7 |
| ein2-1 | 48.8 ± 3.8 | 45.8 ± 2.0 | 0 ± 0 | N.D.c | 2.5 ± 3.4 | 0 ± 0 |
| ctr1-1 | 65.0 ± 6.4 | N.D. | 0 ± 0 | N.D. | 15.0 ± 6.4 | N.D. |
| eto2 | 63.8 ± 4.8 | N.D. | 0 ± 0 | N.D. | 13.8 ± 4.8 | N.D. |
| etr1-7 | 50.7 ± 5.1 | N.D. | N.D. | N.D. | 0.7 ± 5.1 | N.D. |
| rhd6 | 0 ± 0 | 37.6 ± 13.9 | N.D. | N.D. | 0 ± 0 | 3.7 ± 6.6 |
Values shown are means ± sd (n = 140 to 260).
Percentage of root hair–bearing epidermal cells among total epidermal cells counted, including cells in both the H and N positions.
Percentage of root hair–bearing epidermal cells at the N position among total epidermal cells counted.
N.D., not determined.
Exogenous Ethylene Is a Positive Effector for the Expression of Root Hair Expansin Genes in Concert with Root Hair Formation
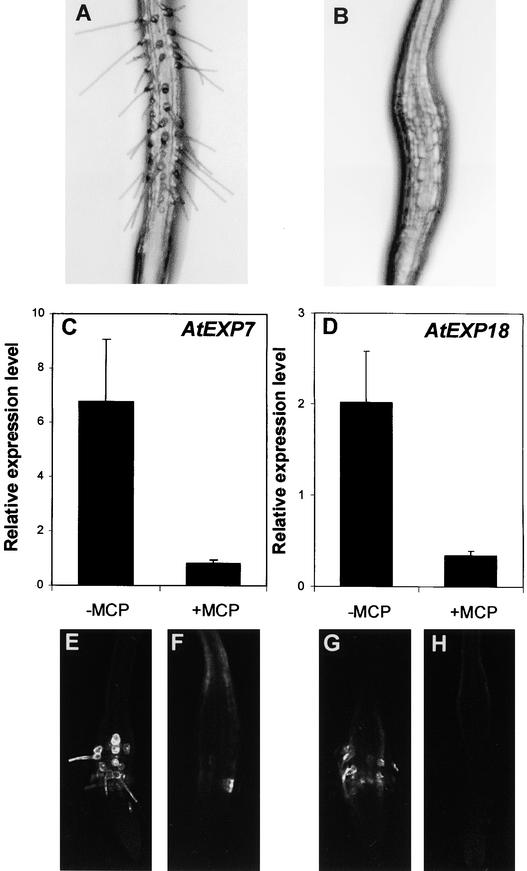
Because ethylene is a positive effector of root hair formation (Masucci and Schiefelbein, 1994, 1996; Tanimoto et al., 1995), we investigated whether ethylene coordinately regulates the expression of root hair expansin genes with root hair formation. The ethylene precursor ACC (5 μM) induced root hair formation and AtEXP7 expression in the N position of the wild-type root (Figure 2O). Mutation in CTR1, which showed constitutive ethylene effects and thus induced the formation of root hairs in the N position (Table 1), likewise activated AtEXP7 expression in root hairs in the N position (Figure 2R) and increased the transcript level by 36% relative to that of the wild type (Figure 3). Compared with the wild type, the root hair–defective rhd6 mutant had only ∼10% of the AtEXP7 transcript (Figure 3), which could derive from the occasional root hairs in the rhd6 root. Treatment of the mutant with 5 μM ACC restored 78% of the transcript level and 74% of the root hair number (Table 1). Ethylene gas (1 μL/L) treatment also induced a similar level of root hairs in the rhd6 root, as did 5 μM ACC (data not shown), and the effect of exogenous ethylene or ACC could be blocked completely by 1-methylcyclopropene (1-MCP), the competitive inhibitor of ethylene binding to the receptors. We chose 1-MCP as an antagonist of ethylene action because of its high specificity of action and lack of deleterious side effects (Sisler et al., 1996; Hall et al., 2000). At 1 μL/L, 1-MCP almost completely abolished ACC-induced root hair formation and the expression of AtEXP7 and AtEXP18 in rhd6 (Figure 4).
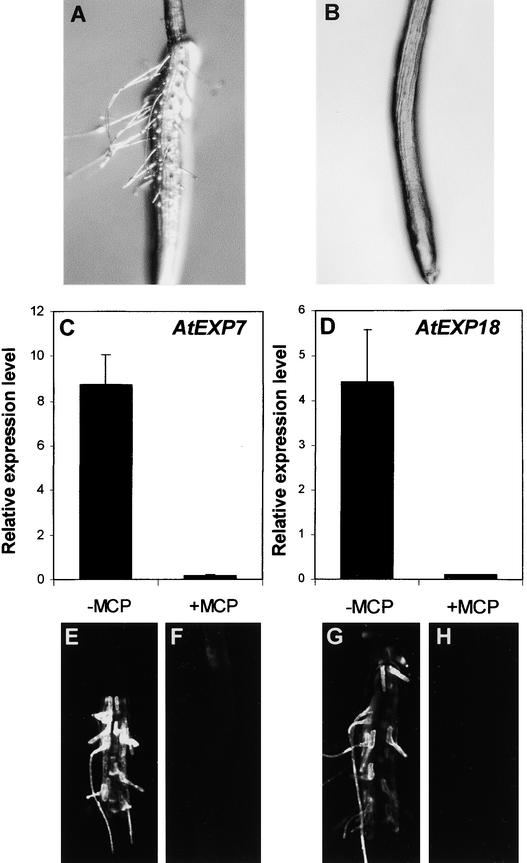
Figure 4.
Effect of 1-MCP on ACC-Induced Root Hair Formation and Expansin Gene Expression in the rhd6 Root.
(A) and (B) Bright-field microscopy images of roots grown in 5 μM ACC without (A) or with (B) 1 μL/L 1-MCP.
(C) and (D) Relative expression levels of AtEXP7 (C) and AtEXP18 (D) in the root when induced by 5 μM ACC without (−MCP) or with (+MCP) 1-MCP. Relative expression levels were evaluated from GFP expression (fluorescence) driven by the AtEXP7 promoter or the AtEXP18 promoter. Bars indicate standard errors (n = 11 to 18).
(E) to (H) Confocal microscopy images of the roots harboring AtEXP7 promoter::GFP ([E] and [F]) and AtEXP18 promoter::GFP ([G] and [H]). Seedlings were incubated in 5 μM ACC without ([E] and [G]) or with ([F]) and [H]) 1-MCP.
1-MCP Inhibits Auxin- or Root Separation–Induced Root Hair Formation and Expression of Root Hair Expansin Genes
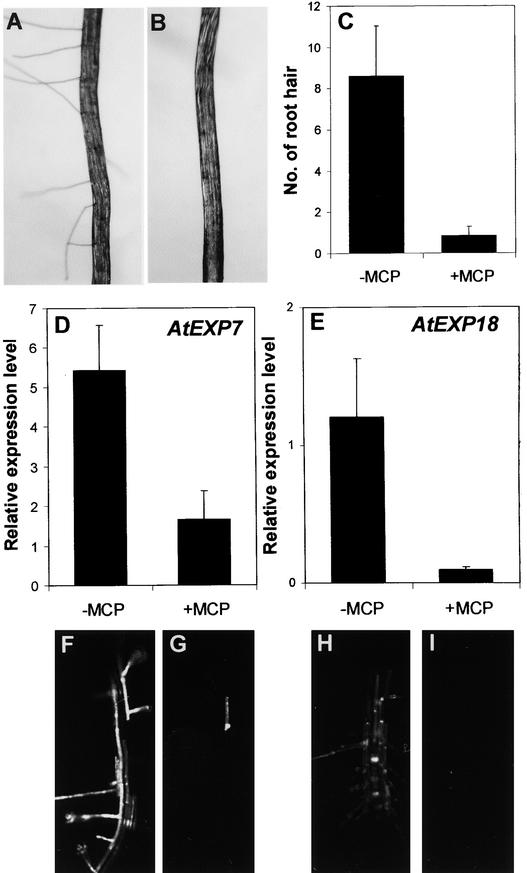
To investigate the possible involvement of ethylene receptors in root hair formation and expansin gene expression induced by auxin or root separation from the agar medium, the antagonism of these effectors by 1-MCP was investigated in the rhd6 background. Auxin- or root separation–induced root hair formation was greatly inhibited by 1-MCP (1 μL/L). No root hair bulges or elongated root hairs were observed in mutant seedlings treated with indole 3-acetic acid (IAA; 30 nM) together with 1-MCP (Figure 5B). Similarly, 1-MCP inhibited 90% of the root hair formation induced by root separation (Figures 6B and 6C). Consistent with these results, 1-MCP inhibited 70 to 90% of IAA- or root separation–induced expression of AtEXP7 and AtEXP18 (Figures 5C to 5H and 6D to 6I). These results show that the coordinate induction of root hairs and expansin gene expression by auxin and root separation requires ethylene sensing, most likely because ethylene is part of the signaling pathway for these effects.
Figure 5.
Effect of 1-MCP on IAA-Induced Root Hair Formation and Expansin Gene Expression in the rhd6 Root.
(A) and (B) Bright-field microscopy images of the roots grown in 30 nM IAA without (A) or with (B) 1 μL/L 1-MCP.
(C) and (D) Relative expression levels of AtEXP7 (C) and AtEXP18 (D) in the root when induced by IAA without (−MCP) or with (+MCP) 1-MCP. Relative expression levels were evaluated from GFP expression (fluorescence) driven by the AtEXP7 promoter or the AtEXP18 promoter. Bars indicate standard errors (n = 7 to 12).
(E) to (H) Confocal microscopy images of the roots harboring AtEXP7 promoter::GFP ([E] and [F]) and AtEXP18 promoter::GFP ([G] and [H]). Seedlings were incubated in IAA without ([E] and [G]) or with ([F] and [H]) 1-MCP.
Figure 6.
Effect of 1-MCP on Root Separation-Induced Root Hair Formation and Expansin Gene Expression in the rhd6 Root.
(A) and (B) Bright-field microscopy images of the roots separated from the agar medium without (A) or with (B) 1 μL/L 1-MCP.
(C) Effect of 1-MCP on root hair number in separation-treated roots. Total root hairs were counted from the separated region of the root. Bars indicate standard errors (n = 13 to 19).
(D) and (E) Relative expression levels of AtEXP7 (D) and AtEXP18 (E) in the root when induced by separation of the root without (−MCP) or with (+MCP) 1-MCP. Relative expression levels were evaluated from GFP expression (fluorescence) driven by the AtEXP7 promoter or the AtEXP18 promoter. Bars indicate standard errors (n = 11 to 15).
(F) to (I) Confocal microscopy images of roots harboring AtEXP7 promoter::GFP ([F] and [G]) and AtEXP18 promoter::GFP ([H] and [I]). Seedlings whose roots were separated from the medium were incubated without ([F] and [H]) or with ([G] and [I]) 1-MCP.
Endogenous Ethylene Is Not Involved in Normal (Default) Root Hair Formation and Expression of Root Hair Expansin Genes in the Wild Type
To verify the role of endogenous ethylene during root hair formation and expression of root hair expansin genes, we examined the effects of dominant mutations of ethylene receptors and inhibitors of ethylene action. Here, we use the term “endogenous ethylene” to designate the internal ethylene level in the plant without any mutations or treatments that would induce the overproduction of ethylene.
Our results showed that mutations in the ethylene signaling pathway failed to inhibit root hair formation and expansin gene expression. None of the five dominant-negative ethylene receptor mutants showed a significant reduction in root hair density (Table 1). The ein2 mutant, which is known to exhibit the strongest ethylene phenotype, also had a normal number of root hairs, consistent with a previous report (Masucci and Schiefelbein, 1996). AtEXP7 expression also was patterned normally in roots of the ethylene mutants (Figures 2S to 2V), and expression levels were not reduced greatly in the mutant backgrounds (Figure 3).
Aminoethoxyvinylglycine (AVG), an inhibitor of ethylene biosynthesis, has been used to test the role of ethylene in root hair formation (Masucci and Schiefelbein, 1994, 1996; Tanimoto et al., 1995). Our results showed that 5 μM AVG almost completely blocked root hair formation and AtEXP7 expression in the wild type (Figures 2P and 3, Table 1). However, surprisingly, AVG (5 μM) almost completely inhibited root hair formation in the constitutively ethylene-responsive mutant ctr1-1 (Table 1), even though this mutant should not respond to AVG inhibition of ethylene synthesis. AVG markedly repressed the expression of AtEXP7 in ctr1-1 and other genotypes, but it also reduced actin gene expression (AtACT2; Figure 3). Although it was reported that ACC could partially restore root hair formation in the AVG-treated root (Masucci and Schiefelbein, 1994, 1996), our results indicate that AVG has significant deleterious effects on root hair development. Toxicity of AVG also is reported in root formation (Jackson, 1991) and somatic embryogenesis (Meijer, 1989). This may occur because AVG, functioning as an inhibitor of pyridoxal phosphate–dependent enzymes (Abel, 1985), probably interferes with other biochemical processes that are vulnerable to the inhibitor, not only ethylene biosynthesis.
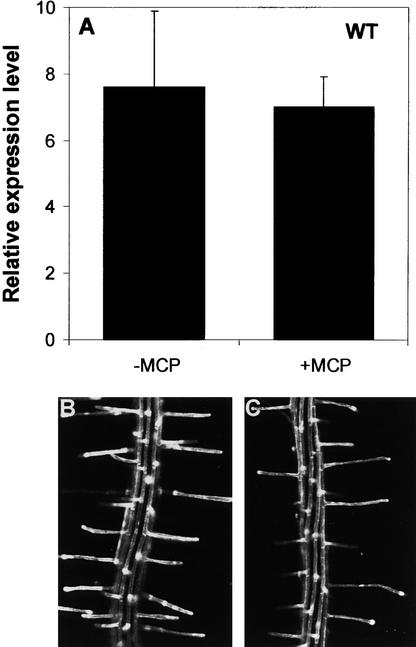
To further test the role of endogenous ethylene during root hair formation in wild-type plants, we used 1-MCP, which binds to multiple ethylene receptors (Hall et al., 2000). Thus, we expected that 1-MCP would strongly inhibit root hair formation in wild-type plants if endogenous ethylene were involved. However, root hair formation in the wild-type root was inhibited very little by 1 μL/L 1-MCP (Table 1) or even by 10 μL/L (data not shown). By contrast, 0.22 μL/L 1-MCP showed saturated inhibitory effects on both ethylene binding to the receptors and the triple response (Hall et al., 2000). 1-MCP also did not significantly inhibit the expression of AtEXP7 and AtEXP18 in the wild type (Figure 7). 1-MCP is not able to reverse the constitutive ethylene-responsive phenotype of ctr1 (Hall et al., 2000), in contrast to the deleterious effect of AVG on the ctr1 root.
Figure 7.
Effect of 1-MCP on Expansin Gene Expression and Root Hair Formation in the Wild Type.
(A) Relative expression levels of AtEXP7 without (−MCP) or with (+MCP) 1 μL/L 1-MCP. Relative expression levels were evaluated from GFP expression (fluorescence) driven by the AtEXP7 promoter. Bars indicate standard errors (n = 7 to 10). WT, wild type.
(B) and (C) Confocal microscopy images of roots harboring AtEXP7 promoter::GFP without (B) or with (C) 1 μL/L 1-MCP.
Silver ion (an inhibitor of ethylene perception) at 50 μM did not abolish root hair formation and AtEXP7 expression, although it completely inhibited hair elongation (Figure 2Q). A previous study reported that silver ion (1 μM) greatly reduced root hair number (Tanimoto et al., 1995), but it is not clear whether small bulges were counted. The effects of ACC, AVG, and mutations in ethylene signaling on the expression of AtEXP18 also resembled those on AtEXP7 expression, as shown by RNA gel blot analysis (Figure 3).
Endogenous Ethylene Affects Root Hair Elongation
In contrast to root hair initiation, ethylene showed an unambiguous effect on root hair elongation, consistent with a previous report (Pitts et al., 1998). Root hair length was decreased significantly in four dominant ethylene receptor mutants (Table 2). Treatment with 1-MCP also greatly decreased root hair elongation in the wild type. Considering the effect of each dominant mutation on root hair length, we can assess the cell type–specific roles of the five ethylene receptors. ETR1 seems to play the most significant role in root hair elongation, followed by ERS1 ≥ ERS2 > ETR2. EIN4 appears to have no function in root hair elongation.
Table 2.
Root Hair Length in Wild-Type and Ethylene Mutant Plants with Ethylene Precursor or Inhibitor Treatment
| Plant | No Treatment | ACC (5 μM) | 1-MCP (1 μL/L) |
|---|---|---|---|
| Columbia | 0.91 ± 0.22 | 1.07 ± 0.14 | 0.51 ± 0.19 |
| etr1-1 | 0.26 ± 0.14 | 0.21 ± 0.20 | 0.27 ± 0.16 |
| etr2 | 0.72 ± 0.20 | 0.66 ± 0.08 | 0.65 ± 0.09 |
| ers1 | 0.40 ± 0.14 | 0.41 ± 0.12 | 0.39 ± 0.13 |
| ers2-1 | 0.47 ± 0.20 | 0.47 ± 0.19 | 0.35 ± 0.11 |
| ein4 | 0.92 ± 0.12 | 0.95 ± 0.12 | 0.56 ± 0.13 |
| ein2-1 | 0.04 ± 0.02 | 0.05 ± 0.03 | N.D.a |
| etr1-7 | 0.82 ± 0.07 | N.D. | N.D. |
| ctr1-1 | 1.22 ± 0.18 | N.D. | N.D. |
Values shown are means ± sd in mm (n = 35).
N.D., not determined.
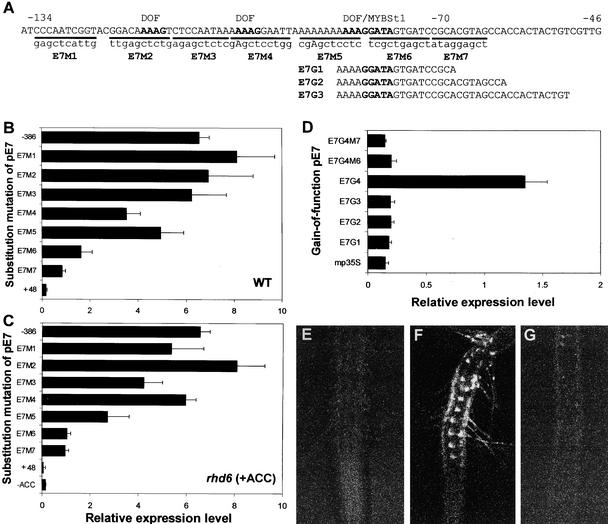
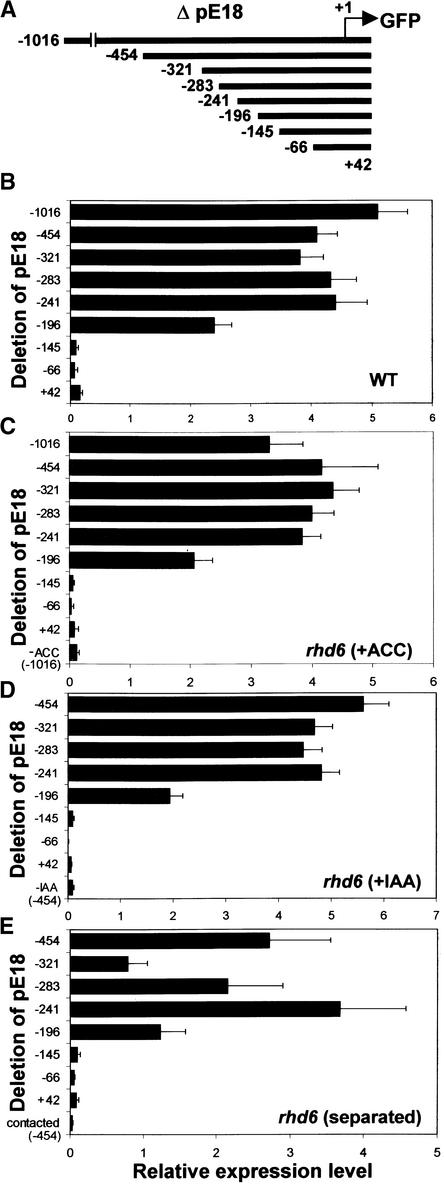
Promoter Analyses of AtEXP7 and AtEXP18
AtEXP7 and AtEXP18 are expressed specifically in the root hair cell and are induced by ethylene, auxin, and separation of the root from the medium. To define the regulatory elements for the hair cell specificity and effector inducibility of the promoter, we performed promoter analyses of the genes by sequential deletion of the 5′ regions, nucleotide substitution, and gain of function of the cis elements. The deleted or substituted promoters were fused directly to the GFP coding sequence, and the gain-of-function cis elements were combined with the 35S minimal promoter region of Cauliflower mosaic virus (−64 35S promoter; Eyal et al., 1995) that was followed by the GFP sequence. For unambiguous evaluation of the promoter activities, the promoter::GFP constructs were introduced stably into plants (wild type and rhd6) by Agrobacterium transformation. To assess the inducibility of promoter activities by ethylene, auxin, and root separation, we treated the transformed rhd6 plants with 5 μM ACC, 30 nM IAA, or separation of the root. Promoter activity was evaluated by confocal laser scanning microscopy to measure GFP fluorescence in roots of the first generation of transformants (9 to 62 independent T1 lines per construct, with an average of 28). The histogram function of Adobe Photoshop was used to quantify the relative GFP fluorescence.
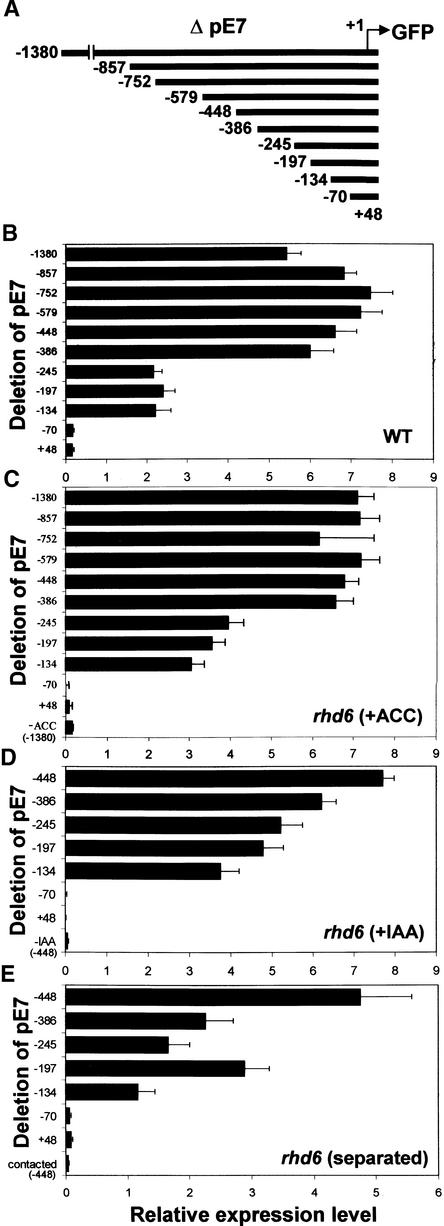
For 5′ deletion analysis of the AtEXP7 promoter, sequential deletions from −1380 to +48 bp, relative to the predicted transcription initiation site, were generated (Figure 8A). Deletions to −386 bp did not significantly affect the promoter activity in either the wild type or rhd6 treated with ACC (Figures 8B and 8C). Further deletion to −245 bp decreased promoter activity by 50 to 70% in both backgrounds, and this level continued through additional deletions to −134 bp. In auxin-treated rhd6 transformants, the promoter activity decreased gradually in deletions from −386 to −134 bp, where ∼50% of the activity remained (Figure 8D). Root separation treatment of rhd6 also gave a similar result, except that the deletion to −386 bp decreased the promoter activity significantly (Figure 8E).
Figure 8.
Deletion Analysis of the AtEXP7 Promoter.
(A) Deletions of the AtEXP7 promoter (ΔpE7) that are fused to the coding region of GFP. Numbers indicate nucleotide positions relative to the transcription initiation site (+1).
(B) Relative activities (GFP expression) of the truncated AtEXP7 promoters in the wild-type (WT) root. Bars indicate standard errors.
(C) to (E) Relative activities of the truncated AtEXP7 promoters in the rhd6 root. For gene induction, the transformed mutant seedlings were treated with 5 μM ACC (C) or 30 nM IAA (D) or roots were separated from agar to expose them to air (E) for 1 day before observation. Bars indicate standard errors.
In (B) to (E), n = 27 to 62.
Although the promoter activity was reduced considerably by deletion to −245 bp, both the cell specificity and the inducibility by effectors were maintained until deletion to −134 bp, and no novel expression patterns were observed in other tissues (data not shown). The cell specificity and the inducibility by effectors disappeared completely with deletion to −70 bp. Although elements for auxin (TGTCTC; −808 bp) and ethylene (AATTCAAA; −615 bp) response are located on the AtEXP7 promoter, deletions of those elements did not affect the responsiveness of the promoter to these hormones (Figures 8C and 8D). Deletion analysis of the AtEXP7 promoter suggested that the elements for cell specificity and inducibility by these effectors are located between −134 and −70 bp. In this region (Figure 9A) are three repeats of a core binding sequence (AAAG) for the DOF zinc finger protein (Yanagisawa and Schmidt, 1999) and one core motif (GGATA) for MYBSt1, a MYB-like protein (Baranowskij et al., 1994). The distal region between −386 and −245 bp likely contains some enhancing elements, because deletion of this region reduced promoter activity significantly. A MYBSt1 core motif also is found in this distal promoter region (−281 to −276 bp).
Figure 9.
Substitution and Gain-of-Function Analyses of the AtEXP7 Promoter.
(A) The proximal promoter region of AtEXP7 between −134 and −46 bp. For substitution mutations (E7M1∼E7M7), the underlined regions were replaced by the nucleotides shown in lowercase letters. These substitution mutations were generated from the region between −386 and +48 bp. E7G1∼E7G3 are the gain-of-function promoter fragments. The substituted promoters were fused to the coding region of GFP, and the gain-of-function promoter fragments were connected to the minimal 35S promoter of Cauliflower mosaic virus (mp35S) before the GFP gene. The putative DOF (AAAG) and MYBSt1 (GGATA) core motifs are indicated.
(B) and (C) Relative activities (GFP expression) of the substituted AtEXP7 promoters in the wild-type (WT) root (B) and in the rhd6 root with 5 μM ACC treatment (C). Bars indicate standard errors (n = 15 to 32).
(D) Relative activities of the gain-of-function AtEXP7 promoters in the wild-type root. Bars indicate standard errors (n = 9 to 14). E7G4 contains the −134/−46 region (wild-type promoter), and E7G4M6 and E7G4M7 are the same as the E7G4 construct but with E7M6 and E7M7 substitution mutations, respectively.
(E) to (G) Confocal microscopy images of roots harboring the gain-of-function AtEXP7 promoters mp35S (E), E7G4 (F), and E7G4M6 (G) (a similar expression pattern was observed with E7G4M7).
To define the cis-regulatory elements in the proximal region (−134 to −70 bp) of the AtEXP7 promoter, seven 9- to 10-bp-long substitution mutations were introduced into this region. To acquire the greatest mutational effects, an A/T base pair was changed to G/C or C/G. Substitution mutations by ∼9 to 10 bp are small enough to localize the controlling elements with reasonable precision (Carey and Smale, 2000). These substitutions replaced the DOF and MYBSt1 core elements and their flanking regions. The substitution mutations E7M1∼E7M7 were produced from the −386-bp deletion so that the wild-type promoter had full activity (Figure 9A). Although promoter activity fluctuated between 50 and 130% compared with wild-type (−386 bp) activity, the substitutions E7M1∼E7M5 did not greatly diminish promoter activity in either wild-type or ACC-treated rhd6 roots (Figures 9B and 9C). However, both E7M6 (which includes the MYBSt1 core element) and E7M7 (flanking E7M6) decreased the activity to ∼13 to 26%. Similar results were obtained by treatment with auxin or root separation (data not shown). These results suggest that the 19-bp motif containing the MYBSt1 element (hereafter called the −80/−62 element) is most important for both hair cell specificity and inducibility by ethylene, auxin, and root separation.
A gain-of-function analysis was performed to confirm that the identified elements are able, in isolation, to direct hair cell specificity. E7G1∼E7G3 are short sequences that contain the proximal MYBSt1 core with different 3′ extensions, and E7G4 includes the entire proximal region between −134 and −46 bp. E7G4M6 and E7G4M7 are the same as E7G4 except that they harbor E7M6 and E7M7 substitution mutations, respectively (Figure 9A). The results shown in Figures 9D to 9G are from the wild-type background, but similar results were obtained with ACC-treated rhd6 (data not shown). The 35S minimal promoter (mp35S) alone did not show GFP expression (Figure 9E). The gain-of-function promoter constructs E7G1∼E7G3 showed promoter activity as weak as that of mp35S (Figure 9D), but 20 to 30% of T1 lines from these constructs showed very low and irregular GFP fluorescence in root hair cells (see supplemental data online), which was undetectable in mp35S roots. No significant differences in promoter strength among E7G1∼E7G3 were found. By contrast, E7G4 could direct strong hair cell–specific expression of the reporter gene (Figures 9D and 9F). However, the substitution mutation of the E7G4 promoter fragment at the E7M6 or E7M7 site eliminated the promoter activity almost completely (Figures 9D and 9G). This gain-of-function promoter analysis demonstrates that the −80/−62 element confers the hair cell specificity of the AtEXP7 promoter. However, some additional elements in the proximal region, particularly between −134 and −81 bp, seem to be required for strong promoter activity. These additional elements could be functionally redundant, because the individual substitution mutations (E7M1∼E7M5) elsewhere than in the −80/−62 region did not reduce promoter activity substantially (Figures 9B and 9C).
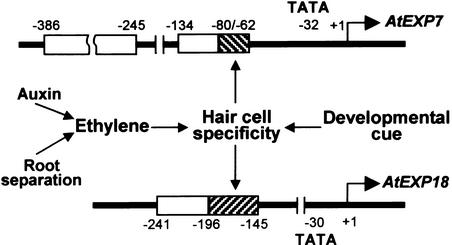
For analysis of the AtEXP18 promoter, deletions between −1016 and +42 bp were generated (Figure 10A). The AtEXP18 promoter activity maintained its full strength until the deletion to −241 bp and showed an ∼50% decrease by further deletion to −196 bp in both wild-type and ACC- or IAA-treated rhd6 seedlings (Figures 10B to 10D). The deletions beyond −145 bp completely eliminated the promoter activity in both the wild type and rhd6 with effector treatments. A similar change of promoter activity was observed in root separation–treated rhd6, except that the deletion to −321 bp reduced the activity significantly (Figure 10E). Deletion analysis of the AtEXP18 promoter indicated that the −196/−145 region contains elements for hair cell specificity and the −241/−196 region may include some enhancing elements relevant to promoter strength. The −196/−145 region of AtEXP18, resembling the −80/−62 element of AtEXP7, is likely the target of signals from ethylene, auxin, and root separation, because these effectors all require this region for gene induction (Figure 11). However, the −196/−145 region of AtEXP18 does not contain the MYBSt1 binding motif or similar sequences found in the −80/−62 element of AtEXP7. This difference indicates that the cell specificity of the two promoters probably is determined by different transcription factors, which nevertheless are regulated similarly by developmental factors, auxin, ethylene, and root separation.
Figure 10.
Deletion Analysis of the AtEXP18 Promoter.
(A) Deletions of the AtEXP18 promoter (ΔpE18) that are fused to the coding region of GFP. Numbers indicate nucleotide positions relative to the transcription initiation site (+1).
(B) Relative activities (GFP expression) of the truncated AtEXP18 promoters in the wild-type (WT) root. Bars indicate standard errors.
(C) to (E) Relative activities of the truncated AtEXP18 promoters in the rhd6 root. For gene induction, the transformed mutant seedlings were treated with 5 μM ACC (C) or 30 nM IAA (D) or roots were separated from the agar medium (E) for 1 day before observation. Bars indicate standard errors.
In (B) to (E), n = 25 to 40.
Figure 11.
Summary of Promoter Analyses of AtEXP7 and AtEXP18.
The hatched boxes represent elements for hair cell specificity, and the open boxes represent elements that are likely to be relevant to promoter strength. The environmental (root separation) and hormonal signals converge on the elements for hair cell specificity. Numbers indicate nucleotide positions relative to the transcription initiation site (+1). TATA indicates the TATA box.
DISCUSSION
Ethylene and Root Hair Development
Recent studies have contributed significantly to our understanding of cell fate determination in the Arabidopsis root epidermis. However, the morphogenetic process of root hair development, which is regulated by hormones and environmental factors, has remained less characterized. In this study, we examined the role of endogenous ethylene and the hierarchical relationship between ethylene, auxin, and an environmental factor (root separation from the agar medium) in root hair initiation. To understand the action of these factors at the gene regulation level, we adopted two expansin genes, AtEXP7 and AtEXP18, whose expression is linked tightly to root hair initiation, as molecular markers.
The involvement of ethylene in root hair formation has been demonstrated in genetic and pharmacological studies. Treatment with the ethylene precursor ACC and mutations of ctr1 and eto induced additional root hairs from the cells in the N position (Dolan et al., 1994; Masucci and Schiefelbein, 1994, 1996; Tanimoto et al., 1995; Cao et al., 1999) (Table 1), and these factors also could restore root hairs in the root hair–defective rhd6 mutant (Masucci and Schiefelbein, 1996). Although these results clearly show that ethylene is a positive effector of root hair formation, they are indicative of the effect of a constitutive ethylene response and excessive (or exogenous) ethylene but not of the effect of the normal endogenous ethylene level. The mutation of CTR1 causes constitutive ethylene responses regardless of the absence or presence of ethylene, and the eto mutants produce excessive ethylene from 2- to 100-fold (Kieber et al., 1993).
The ethylene biosynthesis inhibitor AVG has been used to show the role of endogenous ethylene, which greatly inhibits root hair formation (Masucci and Schiefelbein, 1994, 1996; Tanimoto et al., 1995) (Table 1). However, AVG likely has toxicity to root hair development, because it completely inhibited root hair formation and the expression of AtEXP7 and AtEXP18, even in the ctr1 mutant (Figure 3, Table 1).
The role of endogenous ethylene in the wild type can be assessed by the use of mutations that block the ethylene responses. A previous study reported that the dominant ethylene receptor mutant etr1 maintains normal root hair density (Masucci and Schiefelbein, 1996), thereby raising doubt about the role of endogenous ethylene during the normal (default) process of root hair formation. In Arabidopsis, there are five ethylene receptors whose physiological function, in terms of the triple response, is similar. Dominant mutations in these receptors negatively regulate ethylene responses by constitutively activating CTR1, the negative regulator of downstream ethylene responses. Thus, a dominant mutation in any one of the receptors is able to suppress ethylene responses (Hua and Meyerowitz, 1998). In spite of this genetic principle, inhibition by the dominant mutation shows a dosage-dependent response according to the number of mutant loci and also shows different degrees of phenotypic effect among the five receptors (Hall et al., 1999). Therefore, the contribution of ethylene receptors to root hair formation might depend on receptor species and their temporal/spatial expression pattern.
To determine whether ethylene receptors other than ETR1 are involved in root hair formation, we examined the effect of dominant mutations in all five ethylene receptors. Furthermore, because multiple receptors might be involved in root hair formation, the specific ethylene antagonist 1-MCP was used to simultaneously inhibit ethylene binding by different ethylene receptors. Our results showed that neither the dominant mutations of ethylene receptors nor 1-MCP treatment substantially reduced root hair numbers and the expression of AtEXP7 and AtEXP18 (Figures 3 and 7, Table 1), indicating that endogenous ethylene is not required for normal (default) root hair formation in the wild type.
Ethylene, however, is likely to mediate auxin- or root separation–induced root hair formation. Blocking the ethylene perception by 1-MCP almost completely inhibited auxin- or root separation–induced root hair formation and expression of AtEXP7 and AtEXP18 (Figures 5 and 6). Auxin and certain biotic/abiotic factors, such as pathogens, wounding, chilling, hypoxia, and water stress, are well-known stimulators of ethylene biosynthesis (McKeon et al., 1995). Localized water stress could develop in the root when it is separated from the agar medium or exposed to air, a treatment that is known to stimulate root hair elongation (Okada and Shimura, 1994). Therefore, auxin and root separation may induce root hair initiation through an increase in ethylene production, although we do not exclude the possibility that these treatments affect components of ethylene signaling.
A previous study suggested that ethylene and auxin take separate pathways to affect root hair development. Auxin restored root hairs in the AVG-treated root and in the aux1 etr1 double mutant (Masucci and Schiefelbein, 1996). However, the latter case indicates a complicated aspect of root hair development, because ACC significantly suppressed root hair formation in the double mutant rather than simply having no effect on the restoration of root hairs. A similar perplexing result from the same study is that ACC also inhibited root hair formation considerably in the rhd6 ein2 double mutant. It appears that excessive ethylene (or its pre-cursor) inhibits the ethylene responses of ethylene-insensitive mutants in certain conditions.
The dominant mutant axr2 maintains 64% of root hairs compared with the wild type, and ACC or auxin only partially restores the root hair number in the mutant (74 to 81% compared with the wild type) (Masucci and Schiefelbein, 1996). The axr2 mutant carries the gain-of-function mutation in an Aux/IAA transcriptional repressor (IAA7) so that the mutant molecule is resistant to the auxin-mediated degradation process (Nagpal et al., 2000; Tiwari et al., 2001). The axr2 plant can be less sensitive to ACC if AXR2/IAA7 represses expression of the components of ethylene signaling or if the gain-of-function mutant protein finds new targets, such as genes required for the root hair initiation machinery, as a result of its acquired durability and increased concentration in the nucleus. The epistatic effect of axr2 over ttg or gl2 (Masucci and Schiefelbein, 1996) could be acquired if the latter case occurs.
In contrast to root hair initiation, root hair elongation is dependent on endogenous ethylene. Blocking ethylene perception by gain-of-function mutations of the ethylene receptors or by 1-MCP markedly inhibited root hair elongation (Table 2). The difference in ethylene action on the initiation and elongation of root hairs leads us to propose that the two responses have different sensitivities to ethylene. Root hair initiation may require a higher ethylene level than does the root hair elongation process. Treatment with ACC, auxin, or other stimuli is required to exceed the ethylene concentration needed to stimulate root hair initiation, whereas the lower endogenous ethylene level is sufficient to regulate root hair elongation. Alternatively, it is conceivable that ethylene biosynthesis increases during root hair elongation. Genetic studies indicate that different sets of gene products are instrumental for the root hair initiation and root hair elongation steps (Parker et al., 2000; Schiefelbein, 2000). This finding implies that the two ethylene-dependent responses in a single root hair cell result from the activation of different genetic pathways by different ethylene levels.
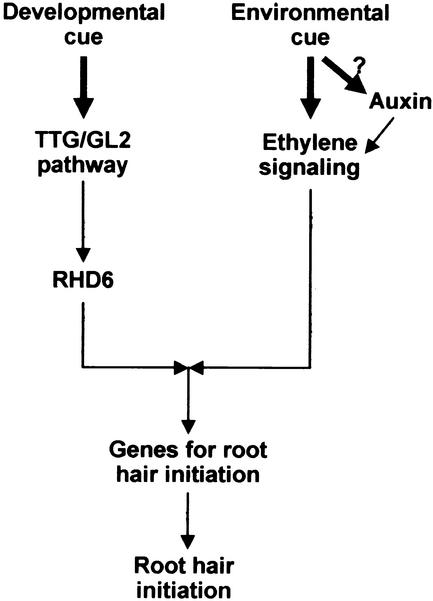
Two Different Pathways Manipulate Root Hair Initiation
RHD6 is likely to be a major regulator in the developmental pathway (through TTG/GL2) for root hair formation. Defects in the negative regulator TTG or GL2 induce root hairs from the cells in the N position as well as in the H position. However, root hair numbers in ttg and gl2 mutants are reduced greatly by the defect in RHD6, indicating that RHD6 is an important downstream regulator of the TTG/GL2 pathway (Masucci and Schiefelbein, 1996). The fact that auxin, ethylene, and root separation can restore root hairs in rhd6 led us to a scheme, illustrated in Figure 12, whereby the separate environmental/hormonal signaling pathway converges with the normal developmental pathway downstream of RHD6. We show the environmental signal (root separation from the medium) as separate from the auxin pathway because root separation restores normal root hair growth in the aux1 mutant (Okada and Shimura, 1994).
Figure 12.
Model Illustrating How Two Separate Signaling Pathways from Developmental and Environmental Cues Merge to Regulate Root Hair Initiation in Arabidopsis.
Arrows designate the information flow.
However, the environmental/hormonal pathway appears to have a differential influence on the two epidermal positions (H and N). This is seen clearly in the rhd6 background (Table 1), in which 5 μM ACC stimulated root hair formation in the H position but had negligible effect in the N position. Even higher levels of ACC (50 μM), as well as the ctr1 and eto mutations, induced only some of the cells in the N position to form root hairs (Dolan et al., 1994; Masucci and Schiefelbein, 1996; Cao et al., 1999) (Table 1). This differential response could result from a lower ethylene (or ACC) sensitivity of cells in the N position compared with cells in the H position (Dolan, 1997; Cao et al., 1999).
Because the occasional root hairs in rhd6 emerge in abnormal cell positions, RHD6 was implicated in the control of hair cell polarity (Masucci and Schiefelbein, 1994). Cell specification seems to be normal in the rhd6 mutant, because the distinctive cytoplasmic characteristics between H- and N-positioned cells are the same as in the wild type; apparently, only the hair-inducing machinery is impaired (Masucci and Schiefelbein, 1996). We found that the rhd6 mutation inhibited the expression of both AtEXP7 and AtEXP18 almost completely (Figures 2K, 3, 8, and 10), suggesting that the molecular function of RHD6 is to regulate gene expression in the root hair cell either as a transcriptional regulator or as its upstream component. RHD6 may regulate the expression of the hair cell genes necessary for hair initiation, such as those involved in cytoskeletal dynamics, localized secretion, wall loosening, and wall synthesis. However, RHD6 probably does not target AtEXP7 and AtEXP18 directly, because expansin gene expression in rhd6 can be restored by hormonal and environmental treatments. Our promoter analyses of the two expansin genes showed that ethylene, auxin, and root separation signals require the same promoter elements that control cell specificity (Figure 11). Thus, we propose that the signals from the developmental and environmental/hormonal pathways are merged at or before the transcription regulators that direct the hair cell specificity of the expansin genes. Identification and characterization of these transcription regulators will be important for understanding the mechanism of pattern formation in the root epidermis.
METHODS
Plant Materials
Arabidopsis thaliana was the model plant in this study. Unless indicated otherwise, the wild type was the Columbia ecotype. The mutant seeds of ttg-1 (CS89), gl2-1 (CS65), eto2 (CS8059), etr1-1 (CS237), ers2-1 (CS8854), ein4 (CS8053), ctr1-1 (CS8057), and ein2-1 (CS3071) were obtained from the ABRC (Columbus, OH). rhd6 seeds were obtained from J.W. Schiefelbein (University of Michigan, Ann Arbor, MI). Seeds of the gain-of-function mutants etr2 and ers1 (Hua et al., 1995) were obtained from J. Hua (Cornell University, Ithaca, NY), and seeds of the loss-of-function mutant etr1-7 were from E. Schaller (University of New Hampshire, Durham, NH). The seeds were sowed on agar plates including 4.3 g/L Murashige and Skoog (1962) nutrient mix (Sigma), 1% Suc, 0.5 g/L Mes, pH 5.7, with KOH, and 0.8% phytagar. After vernalization for 3 days, the seeds were germinated at 23°C under continuous light. For pharmacological experiments, 3-day-old seedlings were transferred to new plates containing growth regulators or antagonists and incubated for 1 additional day, after which root hairs and reporter gene expression patterns were examined. Transformed plants were selected on hygromycin-containing plates (10 μg/mL).
RNA Gel Blot Analyses
Total RNA preparation and RNA gel blot analyses were conducted as described previously (Cho and Kende, 1997). Gene-specific probes for AtEXP7 and AtEXP18 were generated from 3′ untranslated regions of the genes. To confirm equal amounts of RNA loading, the membranes were rehybridized with the Arabidopsis actin2 probe. Transcript levels were quantified from autoradiographs using Adobe Photoshop (Adobe Systems, San Jose, CA) as described previously (Cho and Cosgrove, 2000).
Reporter Gene Constructs
For the reporter gene constructs, the AtEXP7 promoter region (between −1380 and +48 bp relative to the predicted transcription initiation site) from Arabidopsis BAC F5O11 was inserted into HindIII-XbaI sites of the binary vector pGPTV-HYG (Becker et al., 1992), which resulted in the AtEXP7 promoter::uidA (β-glucuronidase [GUS]) construct. For the AtEXP7 promoter::green fluorescent protein (GFP) construct, the uidA gene was replaced with the gene for GFP. The coding region of GFP was obtained from the pEGFP vector (Clontech, Palo Alto, CA) by PCR using primers 5′-AGTTGGAGC-TCTCGAGTCGC-3′ (with the SacI site) and 5′-ATCCCCGGGTACCGGTC-3′ (with the SmaI site). This fragment of the GFP coding region replaced the uidA gene between the SacI and SmaI sites of the AtEXP7 promoter::GUS construct. For the AtEXP7 promoter:: AtEXP7-GFP construct, in which the AtEXP7 promoter directs the expression of the AtEXP7-GFP fusion protein, the coding region of AtEXP7 was amplified from the genomic AtEXP7 clone by PCR using primers 5′-CCTAAGAATCTAGAAAAAGAGGCTAGAATG-3′ (with the XbaI site) and 5′-AAAAGCCCGGGTAACACGGAAATTAGC-3′ (with the SmaI site). This fragment was inserted into XbaI-SmaI sites of the AtEXP7 promoter::GFP construct. All of the constructs were confirmed by DNA sequencing. The constructs were introduced into Arabidopsis plants by Agrobacterium tumefaciens strain C58C1 (pMP90) using the vacuum infiltration method (Bechtold and Pelletier, 1998).
Detection of Reporters
GUS staining was performed as described previously (Cho and Cosgrove, 2000). For the detection of GFP, fluorescence from the seedling root was observed with a confocal laser scanning microscope (LSM-410; Carl Zeiss, Jena, Germany). For the cross-sectional view, 1- to 2-mm root sections were made after embedding the root in 1% agarose. To outline the cell boundary in some samples, the root was stained with propidium iodide (10 μg/mL). Green fluorescence was detected by excitation at 488 nm and emission at 543 nm. Red fluorescence from propidium iodide was detected by excitation at 568 nm and emission at 617 nm. Fluorescence images of the separate channels were digitized with LSM software version 3.5 (New Freedom, PA) and merged using Adobe Photoshop. The false red and green colors were adopted for propidium iodide and GFP fluorescence, respectively.
Observation of Root Hair Number and Length
The number of root hairs was determined using a differential interference contrast microscope according to Masucci and Schiefelbein (1996) with some modifications. For each seedling root, 5 consecutive epidermal cells from the same cell file were observed, and a total of 20 cells from two hair cell files and the adjacent two nonhair cell files were counted. Seven to 13 roots (for a total of 140 to 260 cells) per treatment or genotype were scored. Any protrusion was scored as the presence of the root hair, regardless of the length. In the root separated from the agar medium, total root hairs from the separated region were counted. For root separation, the agar medium immediately below the root tip was cut out, and the root was left to grow to the air. Root hair length was measured using a stereomicroscope when the root hair reached the maximum length. Seven root hairs per plant and five plants per genotype or treatment (for a total of 35 root hairs) were scored.
Treatment of 1-Methylcyclopropene
SmartFresh (0.14% 1-methylcyclopropene [1-MCP]) was obtained from H. Warner at Rohm and Haas (Spring House, PA). 1-MCP gas was produced by mixing the powder with water in a tightly sealed container according to the manufacturer's protocol. The gas was administered to the seedlings so that the final concentration was 1 or 10 μL/L in the container.
Promoter Analyses
The mutated AtEXP7 promoters with 5′ deletions were prepared by PCR using the same reverse primer (5′-GGACCCATTCTAGAC-TCTTT-3′, containing the XbaI site) from the 3′ end (+48 bp) and the forward primers (containing the HindIII site) from the various 5′ ends, as indicated in Figure 8A, with the Arabidopsis BAC F5O11 clone as a template. Deletion of the AtEXP18 promoter was performed similarly by PCR using a reverse primer (5′-TTTACTCTAGATTCTTGAGGGCGCCT-3′, containing the XbaI site) from the 3′ end (+42 bp) and the forward primers (containing the HindIII site) from the 5′ ends, as shown in Figure 10A, with the Arabidopsis BAC F16P17 clone as a template.
Substitution mutagenesis of the proximal region (−134 to −70 bp) of the AtEXP7 promoter, designated E7M1∼E7M7 in Figure 9A, was performed using the “megaprimer PCR” method (Barik, 1995). The megaprimers were amplified using the forward primer 5′-TAGTTA-AGCTTTGGAAACGTAA-3′ (located at −386 bp and containing the HindIII site) and the mutagenized reverse primers from the regions indicated in Figure 9A. The second PCR was performed with these megaprimers and the same reverse primer that was used for the deletion analysis.
The gain-of-function promoters of AtEXP7 were made by associating diverse lengths of proximal promoter parts with the minimal 35S promoter of Cauliflower mosaic virus (mp35S). The mp35S region (−64 35S promoter; Eyal et al., 1995) was produced by PCR using the forward primer 5′-AAGGGTCTAGACACAATCCCACTA-3′ (containing the XbaI site) and the reverse primer 5′-GACCACCCGGGG-ATCCACTA-3′ (containing the SmaI site) from the pBI121 vector (Clontech). This PCR product was inserted between the XbaI and SmaI sites of pGPTV-HYG so that mp35S was followed by the GFP reporter gene. The gain-of-function AtEXP7 promoters E7G1 to E7G3, as shown in Figure 9A, were prepared by complementing the sense and antisense oligonucleotides, which contained HindIII and XbaI sites at their 5′ and 3′ flanking regions, respectively. The E7G4 gain-of-function promoter was produced by PCR amplification of the region between −134 and −46 bp. E7G4M6 and E7G4M7 were made from the same region from which E7G4 was made except that PCR was performed with the E7M6 and E7M7 constructs as templates, respectively.
The truncated or substituted promoter fragments were inserted between the HindIII and XbaI sites of the pGPTV-HYG vector so that the promoters were followed by the GFP reporter gene, and the gain-of-function promoters were inserted between the HindIII and XbaI sites of the pGPTV-HYG vector containing mp35S::GFP. The constructs were introduced into Arabidopsis plants (either the wild type or rhd6) using Agrobacterium as described above. The first generation of transformants (T1, 9 to 62 independent lines per construct) was used to quantify the relative expression levels of GFP in the root. After selection for 5 days on hygromycin-containing plates, the transformants were transferred to new plates without effectors for the wild-type background or with 1-aminocyclopropane-1-carboxylic acid (5 μM), indole-3-acetic acid (30 nM), or separation of the root from the agar medium for the rhd6 mutant, and GFP expression was observed 1 day after the treatments.
To evaluate the promoter activity (GFP expression), fluorescence images of roots were taken digitally using a confocal laser scanning microscope. Relative brightness from the digital images was quantified using the histogram function in Adobe Photoshop. For the histogram analysis, a rectangular marquee (4 × 3 of the root diameter) was located around the root, where GFP fluorescence is maximal, and the mean value was read from the histogram window. The final relative brightness was calculated by subtracting the background values.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession numbers for the genes described in this article are AC025416 (AtEXP7), AC011000 (AtEXP18), and U41998 (AtACT2).
Supplementary Material
Acknowledgments
We thank Daniel M. Durachko for technical assistance and Cheryl Granger for kind advice with confocal microscopy imaging. We also are grateful to Drs. Jian Hua (Cornell University), John Schiefelbein (University of Michigan), and Eric Schaller (University of New Hampshire) for kindly sending the mutant seeds and to Dr. H. Warner (Rohm and Haas) for the generous gift of 1-MCP. We also thank the reviewers for helpful comments and suggestions on the manuscript. This research was supported by a grant from the National Science Foundation to D.J.C.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006437.
Footnotes
Online version contains Web-only data.
References
- Abel, F.B. (1985). Ethylene and plant development: An introduction. In Ethylene and Plant Development, J.A. Roberts and G.A. Tucker, eds (London: Butterworths), pp. 1–8.
- Baranowskij, N., Frohberg, C., Prat, S., and Wilmitzer, L. (1994). A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 13, 5283–5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik, S. (1995). Site-directed mutagenesis in vitro by megaprimer PCR. Methods Mol. Biol. 57, 203–215. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Arabidopsis Protocols, J.M. Martinez-Zapater and J. Salinas, eds (Totowa, NJ: Humana Press), pp. 259–266. [DOI] [PubMed]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20, 1195–1197. [DOI] [PubMed] [Google Scholar]
- Bibikova, T.N., Jacob, T., Dahse, I., and Gilroy, S. (1998). Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125, 2925–2934. [DOI] [PubMed] [Google Scholar]
- Cao, X.F., Linstead, P., Berger, F., Kieber, J., and Dolan, L. (1999). Differential ethylene sensitivity of epidermal cells is involved in the establishment of cell pattern in the Arabidopsis root. Physiol. Plant. 106, 311–317. [DOI] [PubMed] [Google Scholar]
- Carey, M., and Smale, S.T. (2000). Transcriptional Regulation in Eukaryotes. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Cho, H.-T. (2001). Modulation of plant organ growth by expansins, the cell wall loosening proteins. AgBiotechNet 3, ABN 069 (http://www.agbiotechnet.com/reviews/available.asp).
- Cho, H.-T., and Cosgrove, D.J. (2000). Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 9783–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H.-T., and Kende, H. (1997). Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 9, 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack, R.G.H. (1947). A comparative study of developing epidermal cells in white mustard and tomato roots. Am. J. Bot. 34, 310–314. [Google Scholar]
- Cosgrove, D.J. (1999). Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 391–417. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2000). Loosening of plant cell walls by expansins. Nature 407, 321–326. [DOI] [PubMed] [Google Scholar]
- Dolan, L. (1997). The role of ethylene in the development of plant form. J. Exp. Bot. 48, 201–210. [Google Scholar]
- Dolan, L., and Costa, S. (2001). Evolution and genetics of root hair stripes in the root epidermis. J. Exp. Bot. 52, 413–417. [DOI] [PubMed] [Google Scholar]
- Dolan, L., Duckett, C.M., Grierson, C., Linstead, P., Schneider, K., Lawson, E., Dean, C., Poethig, S., and Roberts, K. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120, 2465–2474. [Google Scholar]
- Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., and Scheres, B. (1993). Cellular organization of the Arabidopsis root. Development 119, 71–84. [DOI] [PubMed] [Google Scholar]
- Eyal, Y., Curie, C., and McCormick, S. (1995). Pollen specificity elements reside in 30 bp of the proximal promoters of two pollen-expressed genes. Plant Cell 7, 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman, J., and Dolan, L. (2001). Root hairs as a model system for studying plant cell growth. Ann. Bot. 88, 1–7. [Google Scholar]
- Fowler, J.E., and Quatrano, R.S. (1997). Plant cell morphogenesis: Plasma membrane interactions with the cytoskeleton and cell wall. Annu. Rev. Cell Dev. Biol. 13, 697–743. [DOI] [PubMed] [Google Scholar]
- Galway, M.E., Masucci, J.D., Lloyd, A.M., Walbot, V., Davis, R.W., and Schiefelbein, J.W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, 740–754. [DOI] [PubMed] [Google Scholar]
- Hall, A.E., Chen, Q.G., Findell, J.L., Schaller, G.E., and Bleecker, A.B. (1999). The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiol. 121, 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A.E., Findell, J.L., Schaller, G.E., Sisler, E.C., and Bleecker, A.B. (2000). Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol. 123, 1449–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, J., Chang, C., Sun, Q., and Meyerowitz, E.M. (1995). Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269, 1712–1714. [DOI] [PubMed] [Google Scholar]
- Hua, J., and Meyerowitz, E.M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94, 261–271. [DOI] [PubMed] [Google Scholar]
- Jackson, M. (1991). Ethylene in root growth and development. In The Plant Hormone Ethylene, A.K. Mattoo and J.C. Suttle, eds (Boca Raton, FL: CRC Press), pp. 159–181.
- Kieber, J.J., Roman, G., Feldmann, K.A., and Ecker, J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72, 427–441. [DOI] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. [DOI] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Choi, D., and Kende, H. (2001). Expansins: Ever-expanding numbers and functions. Curr. Opin. Plant Biol. 4, 527–532. [DOI] [PubMed] [Google Scholar]
- Li, Y., Darley, C.P., Ongaro, V., Fleming, A., Schipper, O., Baldauf, S.L., and McQueen-Mason, S.J. (2002). Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol. 128, 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J., and Schiefelbein, J.W. (1994). The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 106, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J.D., Rerie, W.G., Foreman, D.R., Zhang, M., Galway, M.E., Marks, M.D., and Schiefelbein, J.W. (1996). The homeobox gene GLABRA 2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260. [DOI] [PubMed] [Google Scholar]
- Masucci, J.D., and Schiefelbein, J.W. (1996). Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon, T.A., Fernández-Maculet, J.C., and Yang, S.-F. (1995). Biosynthesis and metabolism of ethylene. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Press), pp. 118–139.
- McQueen-Mason, S., Durachko, D.M., and Cosgrove, D.J. (1992). Two endogenous proteins that induce cell wall expansion in plants. Plant Cell 4, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, E.G.M. (1989). Developmental aspects of ethylene biosynthesis during somatic embryogenesis in tissue cultures of Medicago sativa. J. Exp. Bot. 40, 479–484. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the AUX/IAA protein family. Plant Physiol. 123, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, K., and Shimura, Y. (1994). Modulation of root growth by physical stimuli. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 665–684.
- Parker, J.S., Cavell, A.C., Dolan, L., Roberts, K., and Grierson, C.S. (2000). Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 12, 1961–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, R.L., and Stevens, K.J. (2000). Evidence for the uptake of non-essential ions and essential nutrient ions by root hairs and their effect on root hair development. In Root Hairs, R.W. Ridge and A.M.C. Emons, eds (Tokyo: Springer), pp. 179–195.
- Pien, S., Wyrzykowska, J., McQueen-Mason, S., Smart, C., and Fleming, A. (2001). Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc. Natl. Acad. Sci. USA 98, 11812–11817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts, R.J., Cernac, A., and Estelle, M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16, 553–560. [DOI] [PubMed] [Google Scholar]
- Schiefelbein, J.W. (2000). Constructing a plant cell: The genetic control of root hair development. Plant Physiol. 124, 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisler, E.C., Dupille, E., and Serek, M. (1996). Effect of 1-methylcyclopropene and methylenecyclopropane on ethylene binding and ethylene action on cut carnations. Plant Growth Regul. 18, 79–86. [Google Scholar]
- Tanimoto, M., Roberts, K., and Dolan, L. (1995). Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J. 8, 943–948. [DOI] [PubMed] [Google Scholar]
- Tiwari, S.B., Wang, X.-J., Hagen, G., and Guilfoyle, T.J. (2001). AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13, 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. [DOI] [PubMed] [Google Scholar]
- Wilson, A.K., Pickett, F.B., Turner, J.C., and Estelle, M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222, 377–383. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, S., and Schmidt, R.J. (1999). Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 17, 209–214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.