Abstract
Positional cloning of the hcf109 (high chlorophyll fluorescence) mutation in Arabidopsis has identified a nucleus-encoded, plastid-localized release factor 2–like protein, AtprfB, indicating that the processes of translational termination in chloroplasts resemble those of eubacteria. Control of atprfB expression by light and tissues is connected to chloroplast development. A point mutation at the last nucleotide of the second intron causes a new splice site farther downstream, resulting in a deletion of seven amino acid residues in the N-terminal region of the Hcf109 protein. The mutation causes decreased stability of UGA-containing mRNAs. Our data suggest that transcripts with UGA stop codons are terminated exclusively by AtprfB in chloroplasts and that AtprfB is involved in the regulation of both mRNA stability and protein synthesis. Furthermore, sequence data reveal a +1 frameshift at an internal in-frame TGA stop codon in the progenitor prfB gene of cyanobacteria. The expression pattern and functions of atprfB could reflect evolutionary driving forces toward the conservation of TGA stop codons exclusively in plastid genomes of land plants.
INTRODUCTION
Although of cyanobacterial origin, the regulation of gene expression in chloroplasts differs from that in eubacteria and the cytoplasmic system in eukaryotes (Mayfield et al., 1995). Therefore, chloroplast gene expression represents a unique chimeric system assembled from multiple origins during endocytobiosis (Bruick and Mayfield, 1999). To date, little attention has been paid to the role of chloroplast stop codons in the processes of termination, especially the regulation of translation (Zerges, 2000). Recognition of the three stop codons and peptidyl-tRNA hydrolysis require a single omnipotent release factor, eRF and aRF, for the eukaryotic cytosolic system and archaebacteria, respectively. By contrast, two peptide chain release factors are present in eubacteria: prfA (RF1) decodes UAA and UAG, whereas prfB (RF2) decodes UAA and UGA (Scolnick et al., 1968; Nakamura and Ito, 1998).
The guanine and adenine of UGA stop codons are recognized by the first and third amino acids in the highly conserved tripeptide motif Ser-Pro-Phe of PrfB in Escherichia coli (Ito et al., 2000; Nakamura et al., 2000). A GGQ motif present in all known release factors is essential for the catalysis of peptidyl-tRNA hydrolysis (Frolova et al., 1999) and represents a structural counterpart to the CCA-3′ acceptor stem of the tRNA–amino acyl group (Frolova et al., 2000). An in-frame UGA codon inside the prfB gene allows many eubacteria to autoregulate exquisitely the synthesis of their own release factors by a +1 frameshift in the recoding site (Craigen and Caskey, 1986). Only recently, the crystal structures of the human eukaryotic release factor eRF1 and the bacterial RF2 confirmed the proposed tRNA–protein mimicry hypothesis, in which release factor anticodon determinants play a crucial role in stop codon recognition (Moffat and Tate 1994; Ito et al., 1996; Tate et al., 1996; Song et al., 2000; Vestergaard et al., 2001).
Mutations in prfB are known to lead to nonsense suppression, most often causing inefficient reading or misreading of UGA stop signals by incorporating alternative amino acids, increased frameshifting, and temperature-sensitive or toxic phenotypes (Mikuni et al., 1991; Adamski et al., 1993). On the other hand, tRNA suppressor mutations allow the incorporation of nondecoded amino acids at UGA signals in competition with the termination of translation (Eggertsson and Söll, 1988).
Previously, we characterized the pleiotropic hcf109 mutant in Arabidopsis, with severe lesions in thylakoid membrane complexes, predominantly in photosystem II (Meurer et al., 1996a, 1996b). The mutant hcf109 is affected primarily in the stability of distinct plastid mRNAs. Positional cloning of the defective gene identified the first prfB-like peptide chain release factor gene, atprfB, which is eukaryotic in its composition with resemblance to eubacterial homologs. The nuclear gene encodes a chloroplast protein containing a cleavable transit peptide. Functional analyses revealed that the atprfB gene product is required for the proper translation and stability of UGA-containing transcripts in chloroplasts. To the best of our knowledge, this effect has not been observed in any of the eubacterial prfB mutants analyzed to date and provides additional evidence for differences in gene expression between the two lineages, eubacteria and chloroplasts. During the evolution of most algae, the TGA stop codon tends to get lost, and in some cases, it is lost already. However, unlike in land plants, the significantly higher number of plastid TGA stop codons suggests the divergence of ribosomal release even among algae and vascular plants.
RESULTS
High-Resolution Mapping Identified a Peptide Chain Release Factor of Eubacterial Origin
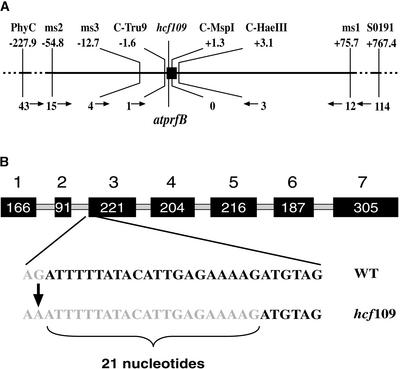
The hcf109 mutant has been mapped on chromosome 5 with the pARMS set and the microsatellite marker PhyC (Meurer et al., 1996a). One previously existing marker, SO191 (TAIR database; http://www.arabidopsis.org/search/), and six newly developed molecular cleaved amplified polymorphic sequence (C-Tru9, C-MspI, and C-HaeIII) and simple sequence length polymorphism (ms1, ms2, and ms3) markers (see Methods) located close to the mutation enabled very high-resolution mapping of hcf109 (Figure 1A). The final mapping population used for this study consisted of 1684 F2 individuals derived from backcrosses to the accession Landsberg. The lethal mutation was confirmed by following the F3 offspring. Markers ms1 and ms2 localized hcf109 on BAC MAB16 with 12 and 15 recombinations, respectively. The mutant locus was enclosed between the two 4.7-kb-distant markers, C-Tru9 and C-HaeIII, with one and three recombinations, respectively. No recombinational event has been identified between the mutation and the cleaved amplified polymorphic sequence marker C-MspI, which was enclosed by C-Tru9 and C-HaeIII (Figure 1A). The C-MspI marker is located directly within a gene that has striking homology with prfB in eubacteria; therefore, it was named atprfB (see below).
Figure 1.
High-Resolution Mapping and Gene Structure of atprfB.
(A) The distance between molecular markers used in our analysis in kb (top) is shown with the number of recombinant events (bottom). The hcf109 locus corresponds to atprfB, the only gene between the two markers C-Tru9 and C-HaeIII.
(B) The seven exons are shown as black boxes, and introns are shown as gray lines. The hcf109 mutation is based on a G-to-A transition of the last nucleotide of the second intron. The mutation results mainly in the splicing of 21 nucleotides farther downstream. WT, wild type.
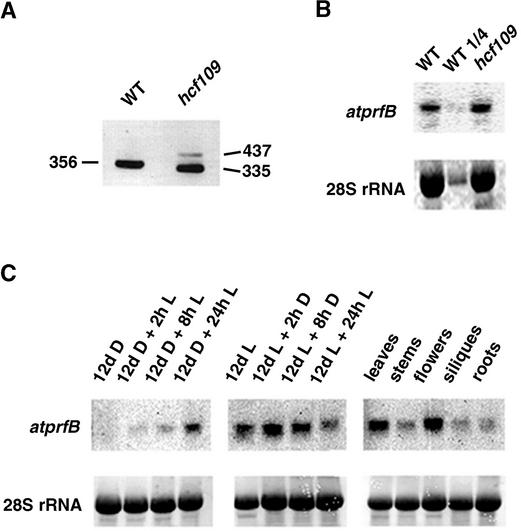
Screening of a cDNA library resulted in the identification of nine positive clones. The atprfB gene contains six introns, and the ATG context consists of the nucleotides AAC-ATGCCA, which are conserved among plant translational start sites (Lutcke et al., 1987). The full-length cDNA of atprfB encodes a protein of 456 amino acids. Sequencing of the complete mutant locus revealed a single G-to-A transition at the last nucleotide of the second intron (Figure 1B). Extensive reverse transcriptase–mediated (RT) PCR analysis of atprfB followed by the sequencing of 29 individual clones confirmed two mRNA species of different abundance in the mutant (Figure 2A). The 437-bp band of very low abundance resulted from the failure of splicing in intron 2. Consequently, the mutation generated a premature in-frame UGA stop codon present in this intron, resulting in a truncated protein of only 92 amino acids. The abundant fragment of 335 bp is characterized by a new 3′ splice site of the second intron, which is 21 nucleotides downstream of the wild-type splice site (Figure 1B). As a consequence, seven amino acids are deleted in the N-terminal part of the mutated protein in hcf109. Using wild-type tissue for RT-PCR generated only one product, of 356 bp, containing the complete and correct spliced form.
Figure 2.
RT-PCR and RNA Gel Blot Analysis of atprfB.
(A) Specific primers of exon 1 and exon 3 (ex1-f and ex3-r, respectively) were used for RT-PCR of mutant and wild-type leaf mRNA. DNA fragments of the indicated sizes (bp) were separated on an agarose gel and visualized with ethidium bromide staining. The top band in the mutant lane represents the unspliced form of atprfB, and the bottom band represents the incorrectly spliced form of atprfB. WT, wild type.
(B) Expression levels of atprfB from 3-week-old mutant and wild-type leaves of plants grown under identical conditions and analyzed by RNA gel blot hybridization. Levels of 28S rRNA are shown to demonstrate uniform loading.
(C) The effect of light (L) and dark (D) incubation on the expression of atprfB in seedlings and the levels of atprfB in different tissues were analyzed in the wild type using RNA gel blots, as described in Methods.
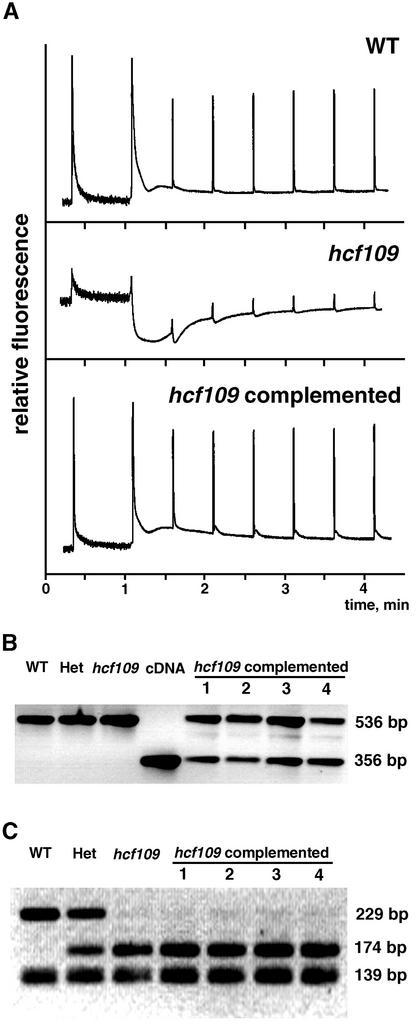
Complementation to the wild-type phenotype was confirmed by expressing the full-length cDNA of atprfB, by photoautotrophic growth, and by fertility in 11 independently transformed mutant plants (Figure 3). The presence of the wild-type cDNA and homozygosity in complemented lines was confirmed by PCR analysis, sequence analysis, and a restriction fragment length polymorphism that had been induced by the mutation (Figures 3B and 3C). The mutant is characterized by a high initial level of chlorophyll fluorescence reflecting a disconnection between the antenna and the reaction center of photosystem II. The fluorescence decreased far below the initial fluorescence during induction, followed by an increase in fluorescence emission (Figure 3A). Complemented mutant plants exhibited fluorescence behavior identical to that of wild-type plants. The ratio of variable to maximal fluorescence was 0.8 (compared with 0.19 in mutants), initial fluorescence was as low as in the wild type based on leaf area, and the fluorescence quenching characteristics were identical to those of the wild type (Figure 3A), suggesting that the deficiencies in hcf109 are caused by the point mutation in atprfB.
Figure 3.
Complementation of hcf109 with the atprfB cDNA.
(A) Chlorophyll fluorescence induction of wild-type (WT) and mutant plants and complemented lines. Fluorescence characteristics of complemented mutant lines were identical to those of the wild type.
(B) Amplification of genomic DNA (using primers ex1-f and ex3-r [see Methods]) for wild-type plants, heterozygous plants (Het), mutant plants (hcf109), sulfadiazine-resistant transformants 1 to 4, and the corresponding cDNA region of atprfB. Transformant lines 1 to 4 show the presence of the cDNA in the genome.
(C) Tru9I-digested PCR products (with primers in2-f and ex4-r [see Methods]) of atprfB from pooled DNA in wild-type, heterozygous, and mutant plants and individually selected transformant plants 1 to 4. Viable lines 1 to 4 were homozygous for the hcf109 mutation. Several restriction products of <50 bp are not shown.
Light-Regulated and Tissue-Specific Expression of atprfB
No differences in atprfB mRNA level were detected between wild-type and hcf109 leaves (Figure 2B). A transcript of 1.45 kb accumulated in light-grown seedlings but was barely detectable in dark-grown seedlings (Figure 2C). Illumination of dark-grown seedlings induced a slow but continuous increase in the transcript level over 24 h. Subjecting seedlings to dark conditions after light growth significantly reduced the atprfB transcript levels after a dark period of 24 h. To our surprise, high atprfB transcript levels were detected in flowers, and low transcript levels were detected in stem, silique, and root tissues (Figure 2C).
atprfB Encodes a Soluble Chloroplast Protein
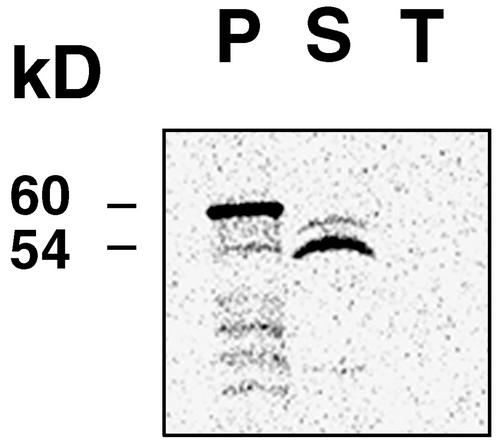
Analysis of the N-terminal 57 amino acids of AtprfB revealed sequence similarities with chloroplast transit peptides (von Heijne, 1990). The presence of a putative cleavable transit peptide was predicted with high confidence using the algorithms ChloroP and TargetP (Emanuelsson et al., 2000), Predotar (http://www.inra.fr/Internet/Produits), and PSORT (http://psort.nibb.ac.jp/). Import experiments confirmed the localization of AtprfB in the soluble fraction of the chloroplast (Figure 4).
Figure 4.
Import of AtprfB into the Chloroplast.
The AtprfB protein was synthesized and labeled with 35S in vitro. The precursor protein (P) of 60 kD was imported into isolated chloroplasts, and a mature protein of 54 kD was detected in the stroma fraction (S). AtprfB was not present in the thylakoid membranes (T).
+1 Frameshift of prfB in Synechocystis and Comparison of AtprfB and Corresponding Eubacterial Forms
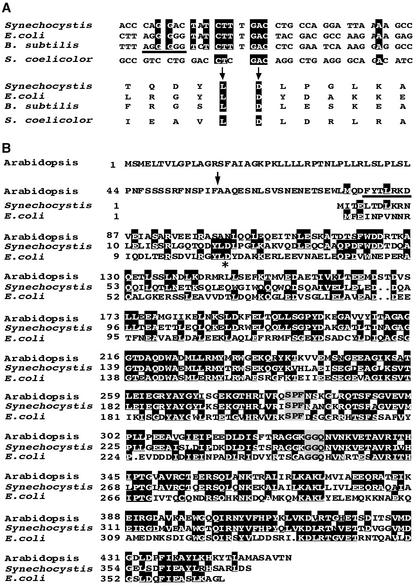
Sequence data of prfB genes from eubacteria revealed that the annotated sequence of Synechocystis sp PCC6803 appears to be incorrect because a frameshift site has been ignored. We sequenced the prfB gene of Synechocystis and did not find any mistakes in the published sequence mentioned above. If a +1 frameshift at an internal in-frame UGA termination signal is taken into consideration, a number of conserved motifs characteristic of eubacterial PrfB proteins also can be detected in the Synechocystis protein. (1) For a large number of bacteria, a +1 frameshift has been described at a highly conserved distance relative to the start codon (Figure 5A) (Craigen et al., 1985). (2) The N-terminal part of the +1 frameshifted prfB from Synechocystis shows significant homology with prfB genes from other eubacteria, atprfB, and prfA from Synechocystis (Figure 5B and data not shown). (3) The annotated PrfB protein is predicted to start with an unusual GTG codon, and the protein has a length of 289 amino acids, which is at least 80 amino acid residues shorter than all other known eubacterial release factors. The proposed frameshifting site in Synechocystis is identical to that of genes in which frameshifting has been shown (Figures 5A and 5B). (4) The presence of a characteristic purine-rich Shine-Dalgarno–like stretch directly upstream of the frameshift site in Synechocystis provides additional evidence for a read-through by a +1 frameshift (Figure 5A). The nonframeshifted prfB from Streptomyces coelicolor (Ogawara et al., 1995) is missing the typical features mentioned above (Figure 5A). Thus, we propose that prfB in Synechocystis contains an internal in-frame UGA stop codon and encodes a protein with 373 amino acids that is able to discriminate between UGA frameshifting sites and release signals.
Figure 5.
Frameshifting of prfB in Synechocystis and Sequence Alignments with AtprfB.
(A) Conserved prfB frameshift sites in E. coli and B. subtilis were compared with that proposed in Synechocystis at the DNA (top) and protein (bottom) levels. The sequences of S. coelicolor, which does not perform frameshifting, are included. Conserved sequences are highlighted. The purine-rich stretch upstream of the frameshift site is underlined.
(B) Comparison of eubacterial release factors with AtprfB in Arabidopsis. The asterisk indicates the conserved eubacterial frameshifting site, and the arrow indicates the predicted cleavage site of the plastid target sequence of AtprfB. The deletion of seven amino acid residues in hcf109 is underlined. Conserved motifs responsible for stop codon recognition (SPF) and the peptidyl-tRNA hydrolase reaction (GGQ) are highlighted in gray.
The deduced protein of the frameshifted prfB in Synechocystis exhibits a striking homology of 71.8% with AtprfB and thus appears to be a putative progenitor. The deduced size of 44.7 kD for the mature AtprfB is similar to that calculated for the homologous eubacterial forms (e.g., 41.2 kD for E. coli, 42.0 kD for Bacillus subtilis, and 41.8 kD for Synechocystis) taking the +1 frameshifting site into consideration (this work).
In eubacteria, the N-terminal residues are less conserved, which also was observed in AtprfB. Hydropathy profiles indicate that the N terminus, which is modified in hcf109, is related more closely to the Synechocystis protein than to the E. coli protein (Figure 5B and data not shown). Highly conserved motifs in eubacterial release factor 2 also were identified in conserved positions of the plastid form (Figure 5B). The C-terminal part contains the recently identified tripeptide anticodon Ser-Pro-Phe, which determines release factor 2 spe-cificity in vivo (Ito et al., 2000; Nakamura et al., 2000), and comprises the universal GGQ motif, the Gln of which is N:(5)-methylated in E. coli (Dincbas-Renqvist et al., 2000).
Translation of Plastid mRNAs Containing the UGA Stop Codon Is Impaired in hcf109
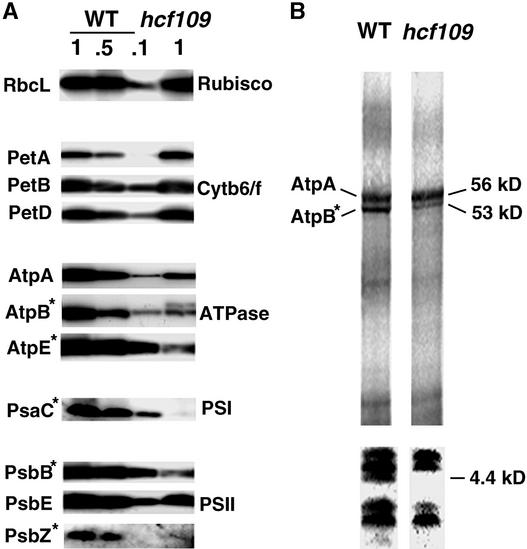
Protein gel blot analysis revealed that plastid proteins that are encoded by genes containing TAA or TAG stop codons (i.e., the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, RbcL, the subunits PetA, PetB, and PetD of the cytochrome b6/f complex, and the photosystem II subunit PsbE) accumulate at normal levels in the mutant. Substantially lower levels were detected for proteins with termination signals stimulated by a UGA stop codon: two subunits of the ATP synthase CF1 subcomplex AtpB and AtpE (<<10%), the photosystem I PsaC protein (<<10%), the photosystem II PsbB (<<10%), and PsbZ (formerly Ycf9) (10%) (Figure 6A). The abundance of the AtpB band of wild-type size was 20% of its normal level in the mutant. An additional overlapping protein of higher molecular mass was detected in the mutant with an antibody raised against AtpB (Figure 6A). The abundance of outer antenna proteins of photosystem II, which are encoded by nuclear genes, is unaffected in the mutant (Meurer et al., 1996a). Protein abundance in the mutant might be limited by translational efficiency. Therefore, plastid translation was quantified by pulse labeling with 35S-Met in the presence of cycloheximide to inhibit cytoplasmic translation. In agreement with previous observations for psbB (Meurer et al., 1996a), this study provides additional evidence for a decrease in translation of the UGA-containing atpB message (20%) (Figure 6B). Translation of the AtpA subunit, which was terminated by a UAA stop codon, was unchanged in the mutant, but only ∼50% of the protein was detectable. This reflects decreased stability of AtpA, probably because of the AtpB deficiency (Figures 6A and 6B). However, we cannot exclude the possibility that some of the labeled AtpA band might arise from the long form of the AtpB detected on the protein gel blot. The translation of a 4.4-kD protein was repressed in hcf109 (Figure 6B). This protein appears to be the photosystem II subunit PsbT, based on the molecular mass and the UGA stop codon in its message.
Figure 6.
Accumulation and Labeling of Plastid Proteins.
(A) Immunoblot analysis of thylakoid membrane proteins from 3-week- old hcf109 and wild-type seedlings. For quantification, 100, 50, and 10% of wild-type membranes were loaded. Asterisks indicate proteins encoded by genes containing a TGA stop codon. Cytb6f, cytochrome b6/f; PSI and PSII, photosystems I and II; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase; WT, wild type.
(B) In vivo labeling of chloroplast membrane proteins separated on 10% (top) and 16% (bottom) polyacrylamide gels. Coseparation of molecular mass standards served to estimate the sizes (in kD) of the labeled proteins.
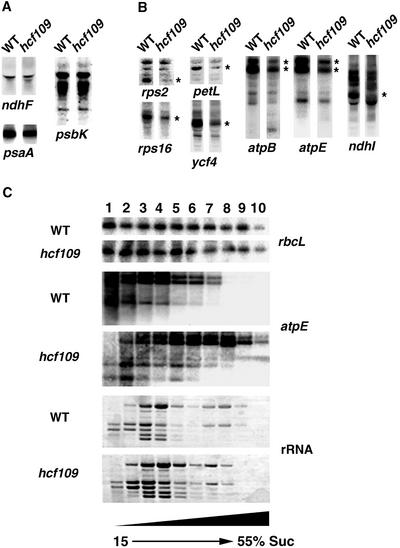
Chloroplast Transcripts Containing Exclusively UGA Stop Codons Are Unstable in hcf109
In hcf109, plastid mRNAs containing the termination signals UAA (28 analyzed out of 47) and UAG (11 analyzed out of 18) and all tested rRNAs and tRNAs were unaffected in size and abundance (Figure 7A and data not shown) (Meurer et al., 1996a). Specific deficiencies were observed for all 12 UGA stop codon–containing plastid transcripts of the photosystem II genes psbB, psbT, and psbZ, the photosystem I gene psaC, the ndhC/K/J (Meurer et al., 1996a) and ndhI genes encoding subunits of the NDH complex, the cytochrome b6/f complex gene petL, the atpB and atpE genes of the ATP synthase, and the rps2 and rps16 genes encoding ribosomal subunits and the ycf4 gene (Figure 7B). We generated macroarrays equipped with probes of all plastid genes encoding proteins, rRNAs, and tRNAs and several controls. These filters were hybridized with end-labeled mutant and wild-type RNA. Specific reductions in UGA-containing transcripts were confirmed by analysis of the plastid expression profile (data not shown). The transcript levels were not reduced uniformly for the genes mentioned and ranged between 5 and 50% of wild-type levels. Run-on analysis demonstrated that transcription of these mRNAs in hcf109 was unaltered (Meurer et al., 1996a). Therefore, the mutant exhibited decreased stability only of UGA stop codon– containing plastid transcripts.
Figure 7.
RNA Gel Blot and Polysome Analysis of Plastid Genes in Mutant and Wild-Type Leaves.
Eight micrograms of total RNA from 3-week-old mutant and wild-type (WT) leaves was analyzed using gene-specific probes.
(A) Transcript levels of plastid genes containing UAA and UAG stop codons.
(B) Transcript levels of plastid genes containing UGA stop codons. Bands of reduced intensity are indicated by asterisks in the mutant lanes.
(C) Intact polysomes fractionated by ultracentrifugation were denatured and subjected to RNA gel blot analysis. The probes used are indicated. Staining of the filter before hybridization resulted in equal distribution of the banding pattern; double amounts of the mutant polysomes were used for the filter hybridized with atpE. The UGA-containing atpE transcript was associated with larger polysomes in hcf109. The experiment was reproduced several times.
Increased Polysome Association of UGA-Containing Transcripts in hcf109 Chloroplasts
An increase in polysome association with the affected transcripts in the mutant is to be expected when elongation is inhibited because of an impaired termination of translation at UGA stop codons in the mutant. Polysomes were separated by Suc density gradient centrifugation. The sizes of polysomes loaded with rbcL transcripts, which contain a UAG stop codon, did not differ in the mutant. rbcL transcripts were distributed equally among the whole gradient (Figure 7C). Transcripts of atpE were shifted dramatically to higher Suc concentrations, indicating enhanced ribosomal association of UGA-containing transcripts in the mutant.
Translational Events Are Responsible for Decreased mRNA Stability in hcf109
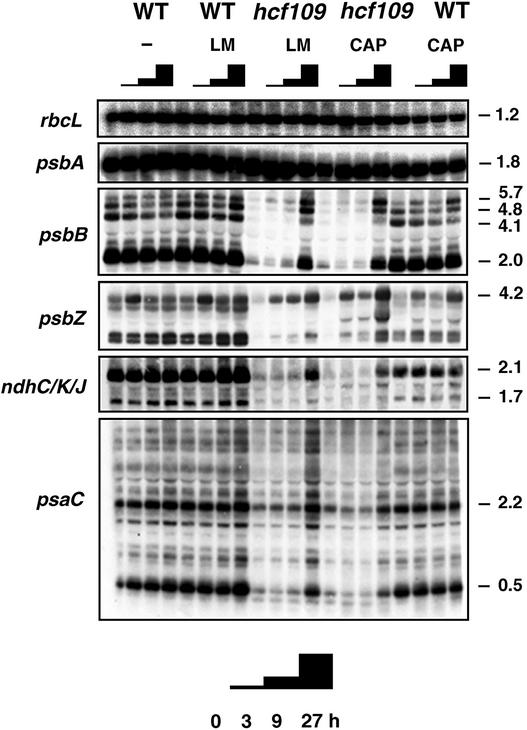
Because AtprfB is involved in ribosomal release, translational or post-translational events at the UGA stop codon could be responsible for the decreased mRNA stability in hcf109. The translation inhibitors lincomycin and chloramphenicol were used to clarify whether translation errors and/or increased polysome association were responsible for decreased mRNA stability in the mutant. Both inhibitors efficiently repressed plastid translation after 15 min of incubation, whereas cytoplasmic translation was unaffected (data not shown). Lincomycin inhibits early peptide bond formation by inducing conformational changes of ribosomes; therefore, ribosomal run-off liberates the transcripts from previously initiated ribosomes (Kallia-Raftopoulos and Kalpaxis, 1999). Chloramphenicol efficiently inhibits the elongation of translation and also is known to interfere with ribosomal release and to force the transcripts into a highly ribosome-associated form (Nierhaus and Wittmann, 1980; Klaff and Gruissem, 1991). Although both drugs are translation inhibitors, mRNA accumulation in wild-type chloroplasts was affected differently (Figure 8).
Figure 8.
Effect of Translation Inhibitors on Plastid Transcript Levels in hcf109.
Three-week-old mutant and wild-type (WT) plants were incubated with the antibiotics chloramphenicol (CAP) and lincomycin (LM). At time 0 and after 3, 9, and 27 h of incubation, transcripts were isolated and analyzed by RNA gel blot hybridization. Incubation without antibiotics (−) served as a control. Probes of the TGA-containing genes psbB, psbZ, ndhJ, and psaC were used in the analysis. The filters were rehybridized with probes of the rbcL and psbA genes, which contain TAG and TAA stop codons, respectively. Within 27 h, UGA-containing transcripts accumulated in the mutant to wild-type levels. Sizes are given in kb.
Generally, endonucleolytic processing of plastid transcripts remained unaffected with both inhibitors: processed forms were mostly unaltered in size and abundance. Chloramphenicol specifically inhibited the splicing of petB and petD introns, which are present in the primary transcript of the Arabidopsis psbB operon (Figure 8) (Meurer et al., 1996a). The unspliced 5.7-kb form accumulated significantly, whereas the completely spliced 4.1-kb form was much weaker after 27 h of inhibition in the wild type. Lincomycin had no effect on the splicing of this operon. An unexpected transient increase of psbZ-containing transcripts was found in all samples at 3 h after incubation. Whereas transcript levels of psbA and rbcL, which do not contain a UGA stop codon, remained constant, both inhibitors clearly gave rise to a substantial increase in the transcript levels of psbB, psbZ, ndhC/K/J, and psaC in the mutant. Complete restoration of mRNA accumulation was finished after 27 h. In spite of the different target sites and effects, both inhibitors used in this study conferred the same stabilizing effect on otherwise completely unstable transcripts in hcf109, indicating that decreased mRNA stability in the mutant is independent of polysome loading but a consequence of translational events in UGA-containing transcripts.
DISCUSSION
+1 Frameshifting of prfB in Synechocystis
An interesting finding of this study was a frameshifting site of a nonsense in-frame UGA codon of prfB in Synechocystis, which raises the following questions: why is there a need to autoregulate prfB expression in eubacteria (Adamski et al., 1993) and which mechanisms, if any, have been adopted by the nucleus for this function? In chloroplasts, +1 frameshifting has not been found. Therefore, regulation by frameshifting is not an essential mechanism for retaining the UGA termination signal in chloroplasts. None of the plastid chromosomes sequenced to date contains release factor–encoding genes, indicating a very early transfer of the gene to the nucleus. Hence, the internal in-frame UGA stop codon of the progenitor gene must have been eliminated to express a functional gene in the nucleus. The conserved amino acids Leu and Asp of the essential eubacterial frameshift site were changed completely in AtprfB (Figure 5B).
Eubacterial Release Factor–Like Genes in Arabidopsis
The comparison of eubacterial prfB mutants and hcf109 implies that the chloroplast protein possesses features for additional regulatory functions in land plants. The two most important characteristics of RF2 (i.e., the SPF anticodon recognition motif and the hydrolytic GGQ center) are conserved in AtprfB, indicating similarities in the mechanism of ribosomal release at UGA and UAA stop codons. Differences from eubacterial features are expected to be present in the nonconserved N-terminal part of the protein, which is thought to interact with ribosomal proteins (Ito et al., 1996) and is mutated in hcf109 (Figure 5B). The evolutionary acquisition of novel nucleus-encoded subunits of chloroplast ribosomes that are not homologous with any known protein in eubacteria further supports the idea of differences in the interaction of release factors with ribosomes and in termination processes (Subramanian, 1993). Inspection of the entire genome of Arabidopsis revealed a eubacterial prfA-like gene, atprfA, that encodes a plastid protein that might terminate translation at plastid UAG and UAA stop codons (data not shown). Because the translation and mRNA stability of UAG- and UAA-containing genes are not affected by the mutation, the function of AtprfA appears to be independent of AtprfB and sufficient for termination at UAA codons.
Expression of atprfB Is Regulated by Light and Tissue
In the dark, the stability and translation of many plastid transcripts generally are reduced (Mullet, 1993), and their expression increases during chloroplast development (Klein et al., 1988; Kim et al., 1993; Bruick and Mayfield, 1999). The low level of atprfB transcripts in the dark might have an impact on the translation and mRNA stability of a set of mRNAs that encode members of the multiprotein complexes photosystem II, photosystem I, NAD(P)H-dependent NDH, ATPase, and ribosomes. The expression of atprfB in Arabidopsis is highly regulated by light in photosynthetic tissue and consequently is connected closely to chloroplast development and function. Otherwise, atprfB expression is low in stems, siliques, and roots but is significantly higher in flowers, suggesting a specific role of polypeptides from messages containing UGA stop codons in this tissue (Figure 2C). Consequently, when constitutive or organ-independent expression of foreign genes in transplastomic approaches is desired, one should avoid equipping the transgene with a TGA stop codon.
AtprfB Affects Plastid mRNA Stability
It has been shown that knockouts in the ribosomal subunits L11 and S17 affect the accumulation of several mRNAs and one mRNA, respectively (Schultes et al., 2000; Pesaresi et al., 2001). However, because run-on experiments have not been performed, it remains to be determined whether changes in mRNA stability are involved in the mentioned knockouts. It is possible that defects in protein synthesis, as in the plastid-encoded RNA polymerase, result in decreased chloroplast mRNA accumulation.
Two transcripts of 2.1 and 1.7 kb that contain sequences of ndhC/K/J are almost absent in hcf109, but only ndhJ is terminated by a UGA stop codon (Figure 8). Thus, messages from genes that are cotranscribed with TGA-containing genes are affected as well. In general, it is difficult to determine the primary effect of a mutation when both RNA stability and translational events are affected (Barkan and Goldschmidt-Clermont, 2000). One way to discriminate between these possibilities is to cross the mutation into another ribosome-deficient mutant background (McCormac and Barkan, 1999). However, RNA polymerase or ribosome-associated proteins involved in mRNA processing might not be active under these conditions. Therefore, we used efficient plastid translation inhibitors to dissect translational events from effects on mRNA stability even in the presence of ribosomes. Application of these inhibitors stabilized psbB, psbZ, ndhC/K/J, and psaC mRNAs, which normally fail to accumulate in the mutant (Figure 8). Because chloramphenicol is assumed to affect release factor function as well, involvement of the termination machinery in the stabilization of plastid mRNAs can be assumed.
Several molecular scenarios are conceivable to illustrate this hypothesis. First, ribosomes traverse into the 3′ untranslated region of the message, which often houses stabilizing elements (Monde et al., 2000). Two processes may be responsible for this: read-through by incorporation of an alternative amino acid or frameshifting. Introduction of genes with an extended reading frame into the genome of Chlamydomonas, for instance, results in a destabilization of transcripts (Lee et al., 1996).
Each kind of read-through or frameshift at UGA stop codons produces larger proteins. In the case of AtpB, we were able to detect an additional larger protein that might result from +1 frameshifting (Figure 6A). Although frameshifting has not been reported previously for chloroplasts, the hcf109 mutant clearly demonstrates potential frameshifting in chloroplasts, which might reveal new or alternative gene products of the organelle in future studies.
Stabilizing elements often are represented by secondary structures (Stern and Gruissem, 1987). However, a pronounced stable 3′ untranslated region structure that might have been perturbed could be predicted only for psbT (Zuker et al., 1999). Numerous stop codons in all reading frames downstream of the UGAs in the Arabidopsis plastome would prevent an extensive movement of ribosomes into the 3′ untranslated region in nearly all affected genes (data not shown). Hence, the instability of UGA-containing transcripts in hcf109 cannot always be explained by traversing ribosomes.
Second, decreased mRNA stability might be caused by translational processes at the UGA stop codon that are involved with the functions of AtprfB and ribosomes. On the other hand, recycling of ribosomes with an unidentified connection between the 3′ and 5′ ends of plastid mRNAs might be impaired, provoking decreased mRNA stability (Zerges, 2000).
Third, we propose a eukaryote-like surveillance mechanism (Culbertson, 1999; Hilleren and Parker, 1999a, 1999b) responsible for the nonsense-mediated degradation of RNA in chloroplasts. In eukaryotes, related mechanisms regulate the stability and expression of mRNAs that fail to signal translational termination (Frischmeyer et al., 2002). The protein Ski7p, which is involved in the exosome-mediated degradation of mRNAs lacking a termination signal, shows homology with the GTPase domain of the translation termination factor eRF3 (van Hoof et al., 2002). Similar surveillance might have been acquired for the regulation of chloroplast transcript stability or for the accuracy of the gene expression machinery. Once acquired, such a cleaning system recognizing and disposing of dysfunctional transcripts might have been established for individual transcripts during coevolution of the plastome and the nuclear genome. Therefore, individual differences in the sequences posing as targets of degradation might explain the discrepancies of the mutational effect on the stability of individual UGA-containing transcripts.
Fourth, in eubacteria, stalled ribosomes on mRNAs that lack a stop codon are released for recycling by a transfer mRNA (Keiler et al., 1996; Watanabe et al., 1998; Karzai et al., 2000). This mechanism targets the protein and probably also the displaced mRNA for degradation (Flynn et al., 2001; Karzai and Sauer, 2001). Interestingly, in eubacteria, transfer mRNA tagging is inversely correlated with RF2 activity (Collier et al., 2002). In chloroplasts, such a cleaning system might be expected, but it has yet to be discovered (Watanabe et al., 1998).
Effect of a Modified AtprfB on Plastid Protein Accumulation and Polysome Association
Several UGA-containing transcripts, such as rps2, rps16, petL, ycf4, ndhI, atpB, and atpE, accumulate somehow in hcf109, although at low levels (Figures 7B and 7C). An increased association of atpE transcripts with polysomes in the mutant indicates that initiation of translation takes place, but elongation of translation is inhibited at later stages, presumably because of the inhibition of translation termination. Although atpE transcripts accumulated to ∼50% in the mutant, the AtpE protein level was reduced by <10%, indicating translational deficiencies. Consequently, the measure of polysome association does not reflect translational activities (Figures 6A and 7C). The important question remains how the translation of UGA-containing transcripts is terminated correctly without a functional release factor in hcf109. It is possible that the mutated AtprfB protein contains some residual activities, albeit at reduced levels. A reduced plastid translation rate might be caused by a nonproductive association of the mutant AtprfB with ribosomes, which is assumed to play a key role in termination (Arkov et al., 1998; Yoshimura et al., 1999). On the other hand, interaction with still unknown factors involved in ribosomal release might be perturbed. A yet unknown prfC (release factor 3)–like protein (Crawford et al., 1999) and/or the recently identified plastid rrf (ribosomal recycling factor) (Rolland et al., 1999) in Arabidopsis are good candidates for involvement in termination and/or post-translational processes. Although functional complementation has been achieved by expressing the wild-type cDNA in the mutant background (Figure 3), it remains possible that the phenotype of hcf109 might be allele specific.
Evolutionary Constraints Keep the Number of Plastid TGA Stop Codons High in Land Plants
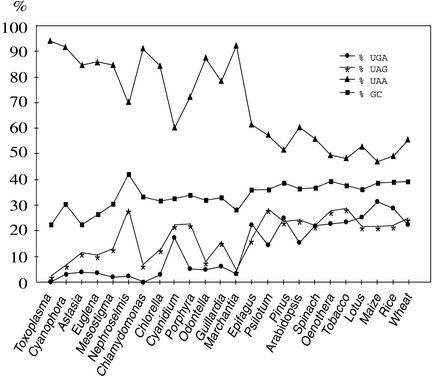
To understand the evolutionary role of the nucleus-encoded AtprfB in chloroplasts of higher plants, we analyzed the distribution of stop codons and the GC content in all 24 plastid chromosomes sequenced to date (Figure 9). Annotation of plastid genomes gives rise to two classes of reading frames, those with known functions or homologs in any other organism, named ycf (hypothetical chloroplast reading frame), and open reading frames (ORFs) of unknown function and without any obvious homology with other known genes. Surprisingly, the number of TGA-containing ORFs in most algae and higher plants is severalfold higher than that among known genes, including ycfs (e.g., 17.0-, 7.4-, and 6.7-fold higher in Nephroselmis olivacea, Odontella sinensis, and Guillardia theta, respectively, and up to 3.3-fold higher in Oenothera elata, spinach, and maize) (data not shown). The only exceptions are in the algae Porphyra purpurea and Cyanidium caldarium, which exhibit an almost identical distribution of TGA elements in both gene classes. Lateral gene transfer into the plastome, which has been proposed only in the case of N. olivacea (Turmel et al., 1999), was responsible for the acquisition of ORFs with a high bias of TGA stop codons.
Figure 9.
Distribution of Stop Codons and GC Content in Plastid Chromosomes.
Completely sequenced plastid chromosomes of the algae Toxoplasma gondii, Cyanophora paradoxa, Astasia longa, Euglena gracilis, Mesostigma viride, Nephroselmis olivacea, Chlamydomonas reinhardtii, Chlorella vulgaris, Cyanidium caldarium, Porphyra purpurea, Odontella sinensis, and Guillardia theta, the moss Marchantia polymorpha, the fern Psilotum nudum, Pinus thunbergii, the dicot plants Epifagus virginiana, Arabidopsis thaliana, Spinacia oleracea, Oenothera elata, Nicotiana tabacum, and Lotus japonicus, and the Liliopsida species Zea mays, Oryza sativa, and Triticum aestivum (http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/plastids_tax.html) were included in our analysis. ORFs were excluded from the analysis (see Discussion).
The two annotated TGA stop codons in the Chlamydomonas plastome (J. Maul, J.W. Lilly, and D.B. Stern, unpublished results; http://www.biology.duke.edu/chlamy_genome/chloro.html) both are present in ORFs. Therefore, it is reasonable to assume that Chlamydomonas has completely lost the release factor 2 function. Assuming this to be the case, caution is advised when performing transplastomic experiments using this alga: reporter genes containing TGA stop codons might not work because of the failure of translation termination. However, two reports have shown that UGA stop codons can be functional in Chlamydomonas chloroplasts (Lee et al., 1996; Sizova et al., 1996), although in these cases, the possibility of frameshifting and read-through was not investigated. These statistical considerations led us to the general conclusion that many annotated TGA-containing ORFs do not seem to represent genes, or, at least, are not always representative of cyanobacteria-derived genes in plastid chromosomes. Therefore, we excluded the ORFs from our analyses with respect to the distribution of stop codons (Figure 9).
The distribution of TAG codons in algae and higher plants often directly correlates with GC content (Figure 9). Generally, changes in the number of TAA-containing genes are mostly accompanied by inverse changes in the use of TAG and in GC content.
TGA-containing genes in plastid chromosomes are not represented by gene groups that are connected functionally, structurally, or biochemically to each other. They are distributed randomly on the chromosomes and are embedded into different nucleotide surroundings of the TGA element (data not shown). Plastids in Toxoplasma gondii have lost the TGA as a stop codon but are assumed to use this codon to decode Trp (Denny et al., 1998). The amount of TGA in algae most often is far <5% (Figure 9). The moss Marchantia polymorpha, at the border to land plants, contains as few TGA stop codons as algae. The only exception is represented by C. caldarium, with a GC content comparable to that of other algae but with a significant increase in TGA use of 18%. Despite a higher GC content of 42.1% in N. olivacea compared with land plants (<40%), UGA stop codon use is not increased in this species. These data clearly indicate discrimination of the two guanidine-containing stop codons in algae and the independence of TGA stop codon use from GC content. Compared with algae, the TGA content is increased significantly in land plant plastomes. It ranges from 15.4% in Arabidopsis to 31.3% in Liliopsida. An ∼10- to 15-fold increase in UGA stop codon use in plastid messages of land plants with respect to that in algae is not reflected by a comparable increase in GC content. Therefore, we assume that important functions other than solely ribosomal release are associated with the plastid RF2, which might explain the significant bias for keeping the number of TGA stop codons high in land plants.
Plastid genomes of algae tend to lose TGA stop codons in correlation with the complete loss in Toxoplasma plastids (Denny et al., 1998) and all nonplant and several algal mitochondria (Inagaki et al., 1998; Kück et al., 2000). We found that in human, yeast, Drosophila, and Caenorhabditis elegans, this reduction was accompanied by a loss of eubacterial prfB-like genes in the entire genomes (http://www.ncbi.nlm.nih.gov). Comparatively, algal mitochondria have none or few TGAs, whereas there is a significant accumulation of this stop codon in land plant mitochondria (e.g., 38.6% in the Arabidopsis mitochondria genome; data not shown) (Unseld et al., 1997). Thus, we propose evolutionary constraints for the bias to accumulate TGA stop codons that became operative almost exclusively in land plant organelles. The differences in algae might arise from the acquisition of additional functions involved with ribosomal release that enabled tissue specificity and the adaptation to adverse and changing environmental conditions for sessile life, which is feasible only with the annexation of regulatory mechanisms acting at the molecular level. Our results demonstrate the close evolutionary relationship between the regulation of gene expression in eubacteria and chloroplasts but also suggest differences reflecting the impact of the intracellular compartmentalization of genomes.
METHODS
Growth Conditions
Sterile growth conditions of mutant and wild-type Arabidopsis thaliana plants were as described previously (Meurer et al., 1996a). Light-dependent expression was performed on seedlings grown either in continuous light for 12 days and subsequently for 2, 8, and 24 h in darkness or in complete darkness for 12 days and subsequently for 2, 8, and 24 h in light. Three-week-old plants grown under greenhouse conditions were used for expression studies of roots and rosette leaves. Six-week-old plants grown under the same conditions were used for expression analyses of stems, green siliques, and young flowers.
Map-Based Cloning of hcf109 and cDNA Isolation
The identification of dinucleotide repeat motifs in the Arabidopsis genome located close to the mutation resulted in the molecular simple sequence length polymorphism markers ms1 (5′-ATGGTAAACAATTATGTCACGTATG-3′ and 5′-CATTGGTTCTCCCATCGTCTCTCT-AG-3′) on BAC T30G6 and ms2 (5′-CATCGAGAAGAGGAATTG-ATCC-3′ and 5′-ACCTAAACACCCTCAACTAATG-3′) and ms3 (5′-TAAGGTAAAGATAATTCAGATGTC-3′ and 5′-TGGTACAAGACACAC-TGATAAGTG-3′) on BAC MAB16 (At5 g36170) (Figure 1A). The additional cleaved amplified polymorphic sequence markers of the same BAC, C-Tru9 (5′-GACCTCGTTAGCTCCGAAGTTC-3′ and 5′-GGACAGCAGATCAGGAACTACGTG-3′), C-MspI (5′-TACTCTCCTGTGCAGCGAATACTA-3′ and 5′-CTCCAGTTAGCCAGAGAATAAGAG-3′), and C-HaeIII (5′-CTCTTATTCTCTGGCTAACTGGAG-3′ and 5′-GGC-ATAAGTCCTTGTGTATCTGCCAAGC-3′), were found to detect DNA polymorphisms between the accessions Landsberg and Columbia using the restriction enzymes Tru9, MspI, and HaeIII, respectively (Figure 1A). atprfB cDNA was isolated from a cDNA library of Arabidopsis (accession Columbia λZAPII Cloning Kit; Stratagene, La Jolla, CA [provided by the European Union Stock Center, Max-Planck-Institut für Züch-tungsforschung, Cologne, Germany]). Nucleotide sequences were determined using the ABI377 system (Applied Biosystems, Foster City, CA) and Li-Cor 4200IR2 (MWG Biotech, Ebersberg, Germany). Sequences were evaluated using Sequencher 3.0 software (Gene Codes Corp., Ann Arbor, MI).
Complementation of the hcf109 Mutant
The full-size atprfB cDNA was amplified with the 5′ phosphorylated oligonucleotides 5′-ATGTCCATGGAGCTCACCGTTCTCGGAC-3′ and 5′-CTAATTGGTTACAGCAGAAGCCATGGC-3′ using Pfu DNA Polymerase (Stratagene) and ligated into the SmaI site of the plant binary expression vector pS001-VS under the control of the 35S promoter of Cauliflower mosaic virus (Meurer et al., 1998a). The plasmid obtained, pbinatprfB, was introduced into Agrobacterium tumefaciens strain GV3101(pMP90RK); subsequently, mutant segregants were transformed by vacuum infiltration (Bechtold and Pelletier, 1998). The point mutation in hcf109 induced a restriction site that could be scored easily by amplifying the genomic region with the intron 2 and exon 4 primers in2-f (5′-TTTCCCAGTGAATCTGAATTC-TTA-3′) and ex4-r (5′-CAAGTAAACTAACATCAGTAGAGTCCAT-3′), respectively, and subsequent digestion with Tru9I. Separation of the obtained restriction fragments resulted in polymorphic and nonpolymorphic control bands, leading to the identification of the genotype of the mutant locus. Mutant plants in the progeny of transformed lines were selected by testing for homozygosity, antibiotic resistance conferred by the T-DNA, and the presence of the expressed cDNA using the exon 1 and exon 3 primers ex1-f (5′-GCCGGTCGGAGT-TTCGCAATCGC-3′) and ex3-r (5′-CTTGAGCTTTAGTACGATCAT-CCC-3′), respectively.
Fluorescence Analyses
Chlorophyll a fluorescence analyses were performed using plants of the same age grown under identical conditions. A pulse amplitude–modulated fluorometer (PAM 101; Walz, Effeltrich, Germany) interphased with the data acquisition system (PDA-100; Walz) and a personal computer using Wincontrol version 1.72 software (Walz) for data collection were used to measure and analyze in vivo chlorophyll a fluorescence. After induction, saturating pulses of 4.0 μE light intensity and 1 s duration were applied in 30-s intervals to estimate quenching parameters (Meurer et al., 1996b).
Expression Studies of Nuclear and Plastid Genes
RNA gel blot analysis and the hybridization probes were as described by Meurer et al. (1996a). Additional plastid probes were generated by amplifying specific regions of the Arabidopsis plastome (Sato et al., 1999). Reverse transcriptase–mediated PCR analysis of atprfB was performed with the oligonucleotides ex1-f and ex3-r generated from exon regions. To avoid embolisms, inhibition of plastid translation was achieved by clipping the hypocotyls of 3-week-old plants in a solution containing 450 mg/L chloramphenicol or lincomycin. Leaves were not immersed to ensure transpiration, which allowed increased uptake of the antibiotics.
Polysome Analysis
Polysome isolation from leaf tissue was performed as described previously (Barkan, 1998). Aliquots (0.5 mL) of polysomes were layered onto 4.4-mL 15 to 55% Suc gradients and centrifuged for 65 min at 45,000 rpm at 4°C in a SW60.1 rotor (Beckman, Munich, Germany). Fractions (0.4 mL) were collected, and the RNA obtained was purified, glyoxylized, and subjected to RNA gel blot analysis.
Protein Analysis
Soluble and membrane proteins of 3-week-old plants were isolated and analyzed on SDS-polyacrylamide gels as described (Meurer et al., 1996b). Separated proteins were transferred to polyvinylidene difluoride membranes (Amersham Buchler, Braunschweig, Germany) according to the manufacturer's instructions, incubated with specific antibodies (Meurer et al., 1996a), and visualized using the enhanced chemiluminescence technique (Amersham Buchler). In vivo labeling of leaf proteins was performed as described (Meurer et al., 1998b), with the exception that the hypocotyls of plants were first cut in the antibiotic-containing solution and subsequently immersed in the appropriate medium supplemented with 35S-Met.
Chloroplast Isolation and Protein Import Experiments
In vitro transcripts of the full-length cDNA of atprfB were generated using T3 RNA polymerase. They were translated in cell-free wheat germ extracts in the presence of 35S-Met according to the manufacturer's instructions (Promega, Mannheim, Germany). Chloroplasts were isolated from young spinach leaves and used for in organello import experiments as described (Michl et al., 1999). Chloroplasts were pelleted, resuspended in 500 μL of 50 mM Hepes and 0.33 M sorbitol, pH 8.0, and treated with thermolysin (100 μg/mL). Unbroken chloroplasts were reisolated by centrifugation through 45% Percoll and divided into membrane and soluble proteins. The fractions were subjected to SDS gel electrophoresis followed by fluorography essentially as described by Clausmeyer et al. (1993).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession number for the atprfB gene described in this article is AJ298098. Accession numbers for other genes, proteins, and genomes mentioned in this article are AB018112 (BAC MAB16), NC_000911 (Synechocystis sp PCC6803), NC_000913 (Escherichia coli), Z99122 AL009126 (Bacillus subtilis), Y08501 (Arabidopsis thaliana), and Y08502 (Arabidopsis thaliana).
Acknowledgments
The authors are grateful to Claudia Nickel for excellent technical assistance. Peter Westhoff (Institut für Entwicklungs und Molekularbiologie der Pflanzen, Düsseldorf, Germany) and Masood Z. Hadi (Bio-systems Research Department, Sandia National Laboratories, Livermore, CA) are acknowledged for critical reading of the manuscript. We also thank Ralf Bernd Klösgen for help with the import experiments. This research was supported by a grant from the German Science Foundation (SFB-TR1) to J.M.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006809.
References
- Adamski, F.M., Donly, B.C., and Tate, W.P. (1993). Competition between frameshifting, termination and suppression at the frameshift site in the Escherichia coli release factor-2 mRNA. Nucleic Acids Res. 21, 5074–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkov, A.L., Freistroffer, D.V., Ehrenberg, M., and Murgola, E.J. (1998). Mutations in RNAs of both ribosomal subunits cause defects in translation termination. EMBO J. 17, 1507–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297, 38–57. [Google Scholar]
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Bruick, R.K., and Mayfield, S.P. (1999). Light-activated translation of chloroplast mRNAs. Trends Plant Sci. 4, 190–195. [DOI] [PubMed] [Google Scholar]
- Clausmeyer, S., Klösgen, R.B., and Herrmann, R.G. (1993). Protein import into chloroplasts: The hydrophilic lumenal proteins exhibit unexpected import and sorting specificities in spite of structurally conserved transit peptides. J. Biol. Chem. 268, 13869–13876. [PubMed] [Google Scholar]
- Collier, J., Binet, E., and Bouloc, P. (2002). Competition between SsrA tagging and translational termination at weak stop codons in Escherichia coli. Mol. Microbiol. 45, 745–754. [DOI] [PubMed] [Google Scholar]
- Craigen, W.J., and Caskey, C.T. (1986). Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature 322, 273–275. [DOI] [PubMed] [Google Scholar]
- Craigen, W.J., Cook, R.G., Tate, W.P., and Caskey, C.T. (1985). Bacterial peptide chain release factors: Conserved primary structure and possible frameshift regulation of release factor 2. Proc. Natl. Acad. Sci. USA 82, 3616–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, D.J., Ito, K., Nakamura, Y., and Tate, W.P. (1999). Indirect regulation of translational termination efficiency at highly expressed genes and recoding sites by the factor recycling function of Escherichia coli release factor RF3. EMBO J. 18, 727–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson, M.R. (1999). RNA surveillance: Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 15, 74–80. [DOI] [PubMed] [Google Scholar]
- Denny, P., Preiser, P., Williamson, D., and Wilson, I. (1998). Evidence for a single origin of the 35 kb plastid DNA in Apicomplexans. Protist 149, 51–59. [DOI] [PubMed] [Google Scholar]
- Dincbas-Renqvist, V., Engstrom, A., Mora, L., Heurgue-Hamard, V., Buckingham, R., and Ehrenberg, M. (2000). A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 19, 6900–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggertsson, G., and Söll, D. (1988). Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol. Rev. 52, 354–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Flynn, J.M., Levchenko, I., Seidel, M., Wickner, S.H., Sauer, R.T., and Baker, T.A. (2001). Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. USA 98, 10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer, P.A., van Hoof, A., O'Donnell, K., Guerrerio, A.L., Parker, R., and Dietz, H.C. (2002). An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295, 2258–2261. [DOI] [PubMed] [Google Scholar]
- Frolova, L.Y., Merkulova, T.I., and Kisselev, L.L. (2000). Translation termination in eukaryotes: Polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA 6, 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova, L.Y., Tsivkovskii, R.Y., Sivolobova, G.F., Oparina, N.Y., Serpinsky, O.I., Blinov, V.M., Tatkov, S.I., and Kisselev, L.L. (1999). Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5, 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P., and Parker, R. (1999. a). Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet. 33, 229–260. [DOI] [PubMed] [Google Scholar]
- Hilleren, P., and Parker, R. (1999. b). mRNA surveillance in eukaryotes: Kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA 5, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki, Y., Ehara, M., Watanabe, K.I., Hayashi-Ishimaru, Y., and Ohama, T. (1998). Directionally evolving genetic code: The UGA codon from stop to tryptophan in mitochondria. J. Mol. Evol. 47, 378–384. [DOI] [PubMed] [Google Scholar]
- Ito, K., Ebihara, K., Uno, M., and Nakamura, Y. (1996). Conserved motifs in prokaryotic and eukaryotic polypeptide release factors: tRNA-protein mimicry hypothesis. Proc. Natl. Acad. Sci. USA 93, 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K., Uno, M., and Nakamura, Y. (2000). A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature 403, 680–684. [DOI] [PubMed] [Google Scholar]
- Kallia-Raftopoulos, S., and Kalpaxis, D.L. (1999). Slow sequential conformational changes in Escherichia coli ribosomes induced by lincomycin: Kinetic evidence. Mol. Pharmacol. 56, 1042–1046. [DOI] [PubMed] [Google Scholar]
- Karzai, A.W., Roche, E.D., and Sauer, R.T. (2000). The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7, 449–455. [DOI] [PubMed] [Google Scholar]
- Karzai, A.W., and Sauer, R.T. (2001). Protein factors associated with the SsrA-SmpB tagging and ribosome rescue complex. Proc. Natl. Acad. Sci. USA 98, 3040–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler, K.C., Waller, P.R., and Sauer, R.T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993. [DOI] [PubMed] [Google Scholar]
- Kim, M., Christopher, D.A., and Mullet, J.E. (1993). Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol. Biol. 22, 447–463. [DOI] [PubMed] [Google Scholar]
- Klaff, P., and Gruissem, W. (1991). Changes in chloroplast mRNA stability during leaf development. Plant Cell 3, 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, R.R., Mason, H.S., and Mullet, J.E. (1988). Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. J. Cell Biol. 106, 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück, U., Jekosch, K., and Holzamer, P. (2000). DNA sequence analysis of the complete mitochondrial genome of the green alga Scenedesmus obliquus: Evidence for UAG being a leucine and UCA being a non-sense codon. Gene 253, 13–18. [DOI] [PubMed] [Google Scholar]
- Lee, H., Bingham, S.E., and Webber, A.N. (1996). Function of 3′ non-coding sequences and stop codon usage in expression of the chloroplast psaB gene in Chlamydomonas reinhardtii. Plant Mol. Biol. 31, 337–354. [DOI] [PubMed] [Google Scholar]
- Lutcke, H.A., Chow, K.C., Mickel, F.S., Moss, K.A., Kern, H.F., and Scheele, G.A. (1987). Selection of AUG initiation codons differs in plants and animals. EMBO J. 6, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield, S.P., Yohn, C.B., Cohen, A., and Danon, A. (1995). Regulation of chloroplast gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 147–166. [Google Scholar]
- McCormac, D.J., and Barkan, A. (1999). A nuclear gene in maize required for the translation of the chloroplast atpB/E mRNA. Plant Cell 11, 1709–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer, J., Berger, A., and Westhoff, P. (1996. a). A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhC operons. Plant Cell 8, 1193–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer, J., Grevelding, C., Westhoff, P., and Reiss, B. (1998. a). The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol. Gen. Genet. 258, 342–351. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Meierhoff, K., and Westhoff, P. (1996. b). Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterization by spectroscopy, immunoblotting and RNA gel hybridization. Planta 198, 385–396. [DOI] [PubMed] [Google Scholar]
- Meurer, J., Plücken, H., Kowallik, K.V., and Westhoff, P. (1998. b). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17, 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl, D., Karnauchov, I., Berghofer, J., Herrmann, R.G., and Klösgen, R.B. (1999). Phylogenetic transfer of organelle genes to the nucleus can lead to new mechanisms of protein integration into membranes. Plant J. 17, 31–40. [DOI] [PubMed] [Google Scholar]
- Mikuni, O., Kawakami, K., and Nakamura, Y. (1991). Sequence and functional analysis of mutations in the gene encoding peptide-chain-release factor 2 of Escherichia coli. Biochimie 73, 1509–1516. [DOI] [PubMed] [Google Scholar]
- Moffat, J.G., and Tate, W.P. (1994). A single proteolytic cleavage in release factor 2 stabilizes ribosome binding and abolishes peptidyl-tRNA hydrolysis activity. J. Biol. Chem. 269, 18899–18903. [PubMed] [Google Scholar]
- Monde, R.A., Schuster, G., and Stern, D.B. (2000). Processing and degradation of chloroplast mRNA. Biochimie 82, 573–582. [DOI] [PubMed] [Google Scholar]
- Mullet, J.E. (1993). Dynamic regulation of chloroplast transcription. Plant Physiol. 103, 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y., and Ito, K. (1998). How protein reads the stop codon and terminates translation. Genes Cells 3, 265–278. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Ito, K., and Ehrenberg, M. (2000). Mimicry grasps reality in translation termination. Cell 101, 349–352. [DOI] [PubMed] [Google Scholar]
- Nierhaus, K.H., and Wittmann, H.G. (1980). Ribosomal function and its inhibition by antibiotics in prokaryotes. Naturwissenschaften 67, 234–250. [DOI] [PubMed] [Google Scholar]
- Ogawara, H., Urabe, H., Ohtaki, R., and Nakamura, Y. (1995). Properties of peptide chain release factor 2 from Streptomyces coelicolor A3(2): Conserved primary structure but no frameshift regulation. J. Bacteriol. 177, 5342–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi, P., Varotto, C., Meurer, J., Jahns, P., Salamini, F., and Leister, D. (2001). Knock-out of the plastid ribosomal protein L11 in Arabidopsis: Effects on mRNA translation and photosynthesis. Plant J. 27, 179–189. [DOI] [PubMed] [Google Scholar]
- Rolland, N., Janosi, L., Block, M.A., Shuda, M., Teyssier, E., Miege, C., Cheniclet, C., Carde, J.P., Kaji, A., and Joyard, J. (1999). Plant ribosome recycling factor homologue is a chloroplastic protein and is bactericidal in Escherichia coli carrying temperature-sensitive ribosome recycling factor. Proc. Natl. Acad. Sci. USA 96, 5464–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., and Tabata, S. (1999). Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 6, 283–290. [DOI] [PubMed] [Google Scholar]
- Schultes, N.P., Sawers, R.J., Brutnell, T.P., and Krueger, R.W. (2000). Maize high chlorophyll fluorescent 60 mutation is caused by an Ac disruption of the gene encoding the chloroplast ribosomal small subunit protein 17. Plant J. 21, 317–327. [DOI] [PubMed] [Google Scholar]
- Scolnick, E., Tompkins, R., Caskey, T., and Nirenberg, M. (1968). Release factors differing in specificity for terminator codons. Proc. Natl. Acad. Sci. USA 61, 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova, I.A., Lapina, T.V., Frolova, O.N., Alexandrova, N.N., Akopiants, K.E., and Danilenko, V.N. (1996). Stable nuclear transformation of Chlamydomonas reinhardtii with a Streptomyces rimosus gene as the selective marker. Gene 181, 13–18. [DOI] [PubMed] [Google Scholar]
- Song, H., Mugnier, P., Das, A.K., Webb, H.M., Evans, D.R., Tuite, M.F., Hemmings, B.A., and Barford, D. (2000). The crystal structure of human eukaryotic release factor eRF1: Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100, 311–321. [DOI] [PubMed] [Google Scholar]
- Stern, D.B., and Gruissem, W. (1987). Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell 51, 1145–1157. [DOI] [PubMed] [Google Scholar]
- Subramanian, A.R. (1993). Molecular genetics of chloroplast ribosomal proteins. Trends Biochem. Sci. 18, 177–181. [DOI] [PubMed] [Google Scholar]
- Tate, W.P., Poole, E.S., Dalphin, M.E., Major, L.L., Crawford, D.J., and Mannering, S.A. (1996). The translational stop signal: Codon with a context, or extended factor recognition element? Biochimie 78, 945–952. [DOI] [PubMed] [Google Scholar]
- Turmel, M., Otis, C., and Lemieux, C. (1999). The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: Insights into the architecture of ancestral chloroplast genomes. Proc. Natl. Acad. Sci. USA 96, 10248–10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unseld, M., Marienfeld, J.R., Brandt, P., and Brennicke, A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15, 57–61. [DOI] [PubMed] [Google Scholar]
- van Hoof, A., Frischmeyer, P.A., Dietz, H.C., and Parker, R. (2002). Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295, 2262–2264. [DOI] [PubMed] [Google Scholar]
- Vestergaard, B., Van, L.B., Andersen, G.R., Nyborg, J., Buckingham, R.H., and Kjeldgaard, M. (2001). Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell 8, 1375–1382. [DOI] [PubMed] [Google Scholar]
- von Heijne, G. (1990). Protein targeting signals. Curr. Opin. Cell Biol. 2, 604–608. [DOI] [PubMed] [Google Scholar]
- Watanabe, T., Sugita, M., and Sugiura, M. (1998). Identification of 10Sa RNA (tmRNA) homologues from the cyanobacterium Synechococcus sp. strain PCC6301 and related organisms. Biochim. Biophys. Acta 1396, 97–104. [DOI] [PubMed] [Google Scholar]
- Yoshimura, K., Ito, K., and Nakamura, Y. (1999). Amber (UAG) suppressors affected in UGA/UAA-specific polypeptide release factor 2 of bacteria: Genetic prediction of initial binding to ribosome preceding stop codon recognition. Genes Cells 4, 253–266. [DOI] [PubMed] [Google Scholar]
- Zerges, W. (2000). Translation in chloroplasts. Biochimie 82, 583–601. [DOI] [PubMed] [Google Scholar]
- Zuker, M., Mathews, D.H., and Turner, D.H. (1999). Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide. In RNA Biochemistry and Biotechnology, J. Barciszewski and B.F.C. Clark, eds (New York: Kluwer Academic Publishers), pp. 11–43.