Abstract
Human severe combined immunodeficiency (SCID) can be caused by defects in Janus kinase 3 (JAK3)-dependent cytokine signaling pathways. As a result, patients are at high risk of life-threatening infection. A JAK3 −/− SCID mouse model for the human disease has been used to test whether transplant with retrovirally transduced bone marrow (BM) cells (JAK3 BMT) could restore immunity to an influenza A virus. The immune responses also were compared directly with those for mice transplanted with wild-type BM (+/+ BMT). After infection, approximately 90% of the JAK3 BMT or +/+ BMT mice survived, whereas all of the JAK3 −/− mice died within 29 days. Normal levels of influenza-specific IgG were present in plasma from JAK3 BMT mice at 14 days after respiratory challenge, indicating restoration of B cell function. Influenza-specific CD4+ and CD8+ T cells were detected in the spleen and lymph nodes, and virus-specific CD8+ effectors localized to the lungs of the JAK3 BMT mice. The kinetics of the specific host response correlated with complete clearance of the virus within 2 weeks of the initial exposure. By contrast, the JAK3 −/− mice did not show any evidence of viral immunity and were unable to control this viral pneumonia. Retroviral-mediated JAK3 gene transfer thus restores diverse aspects of cellular and humoral immunity and has obvious potential for human autologous BMT.
The Janus kinases comprise a family of protein tyrosine kinases that play critical roles in cytokine signal transduction. Janus kinase-3 (JAK3) is unique in that it is expressed almost exclusively in hematopoietic cells (1). JAK3 associates with the common γ chain for the receptors to interleukins 2, 4, 7, 9, and 15 (2) and is phosphorylated on tyrosine residues after ligand binding (3, 4). Activated JAK3 then phosphorylates members of the signal transducers and activators of transcription (STAT) family of latent transcription factors known to be involved in a wide range of cellular processes (5). Recently, it has been demonstrated that cases of SCID can result from either the complete absence of the JAK3 protein (6, 7) or expression of a nonfunctional mutant form (8). JAK3 deficiency is responsible for roughly 7% of human severe combined immunodeficiency (SCID) cases, presenting with normal or elevated levels of B cells but very low T cell counts (9). The consequence of the resultant deficiencies in both humoral and cellular immunity is that patients are highly susceptible to opportunistic infections, which commonly cause mortality at an early age (10).
Bone marrow transplantation (BMT) is currently the best available treatment option for SCID patients. However, because most of these patients lack an HLA-matched sibling donor, haploidentical BMT often is performed (10). Although this procedure results in clinical improvement in most patients (10, 11), there can be incomplete reconstitution of B lymphocyte function (12), graft failure (13), and dysfunctional cytokine production by host-reactive T cell clones (14). As a result, many of these patients must remain on i.v. gamma globulin therapy (10). Transplant of genetically corrected autologous cells has the potential to circumvent most of these problems.
Gene therapy approaches for treating JAK3-deficient SCID patients with autologous BM currently are being tested in preclinical experiments. A study by Candotti et al. (15) demonstrated biochemical correction of JAK3-deficient Epstein–Barr virus-immortalized patient B cell lines in vitro after retroviral-mediated transfer of the human JAK3 gene. In addition, recently developed mouse models for JAK3-deficient SCID (16–18) have allowed studies aimed at correction of the immune deficient phenotype by targeting repopulating hematopoietic stem cells. Previously, we have demonstrated that retroviral-mediated gene transfer of murine JAK3 into BM from JAK3 −/− mice (JAK3 BMT) was sufficient to restore peripheral blood T and B lymphocyte numbers and increase plasma Ig levels in transplanted mice (19). However, it was still unclear whether the levels of lymphocyte reconstitution were physiologically significant. Because patients commonly succumb to viral infections, we have used this JAK3 BMT model to test whether gene correction would protect against a potentially lethal viral pathogen. These studies provide strong support for the potential efficacy of gene therapy for treating JAK3-deficient SCID patients.
MATERIALS AND METHODS
Retroviral Transduction and BMT.
BM was harvested from the hind limbs of JAK3 −/− mice (B6 × 129; F2), cultured with hematopoietic growth factors on GP + E86/MPSV-JAK3 ecotropic producer cells, and injected into irradiated (900 rad) mice as described (19). Beginning at 4–6 weeks after BMT, mice were analyzed by flow cytometry for restoration of absolute B and T lymphocyte numbers in the peripheral blood (19).
Influenza Virus Infection.
JAK3 −/− mice were infected intranasally with the Hong Kong influenza x31 (HKx31) virus, which is a laboratory-generated reassortant between A/Aichi/68 (H3N2) and A/PR/8/34 (PR8, H1N1) that contains the H3N2 surface hemagglutinin (H) and neuraminidase (N) molecules and the internal components of the PR8 virus (20). The mice were anesthetized with avertin (2,2,2-tribromoethanol), then infected with 106.8 EID50 (egg infectious doses). Those used in survival studies were monitored daily and euthanized if found severely moribund.
ELISA for Influenza-Specific IgG.
Blood was obtained from anesthetized mice by bleeding from the retro-orbital sinus (21). Purified HKx31 was dissociated and 96-well ELISA plates (Nunc-maxisorp, Fisher Scientific) were coated with 0.5 μg of disrupted virus per well. Wells were washed with PBS/0.05% Tween-20, blocked with PBS/3% BSA, and then washed with PBS/0.05% Tween-20. Plasma samples were diluted into PBS/0.5% BSA/0.05% Tween-20 and added to the 96-well plates. The plates then were washed and developed by using an established protocol (19).
Tissue Sampling and Treatment.
Inflammatory cells were obtained from anesthetized, infected mice by bronchoalveolar lavage (BAL). The BAL cells first were adhered onto plastic Petri dishes (Falcon) for 1 hr at 37°C to remove macrophages. Single cell suspensions were made from the cervical lymph nodes, mediastinal lymph nodes, and spleens, followed by lysis of red blood cells. The lungs were frozen (−70°C) and later homogenized in 1 ml of PBS. Virus was assayed by allantoic inoculation of 100 μl in embryonated hen’s eggs (22) and virus titers were expressed as log10 EID50.
Staining Virus-Specific CD8+ T Cells.
Tetramers of major histocompatibility complex class I glycoprotein plus viral peptide (NPP), H-2Db complexed with influenza nucleoprotein NP366–374, ASNENMETM, have been described (23). Before staining, Fc receptors were blocked by using purified anti-mouse CD16/CD32 (Fc-γRIII/II receptor, PharMingen), and the lymphocytes were incubated with the NPP for 1 hr at room temperature, followed by staining with fluorescein isothiocyanate-conjugated anti-CD8α (53–6.7, PharMingen) for 30 min on ice. The lymphocytes then were washed and analyzed on a FACScan using Cell Quest software (Becton Dickinson).
Assaying Functional CD8+ T Cells.
For cytotoxicity assays, spleen, mediastinal lymph nodes, and cervical lymph nodes cells were pooled and incubated under bulk culture conditions as described (24). Briefly, cells were cultured for 5 days in 12-well tissue culture plates (Costar) at a responder-to-stimulator ratio of 2:1 and at a density of 3 × 106 cells/ml. The stimulators were irradiated (2,000 Rad) HKx31-infected or uninfected T-depleted splenocytes from wild-type (B6 × 129) F2 mice. Syngeneic MC57G target cells were infected with HKx31, pulsed with the NP366–374 peptide, or left untreated. The % specific lysis was determined as: [(experimental release − spontaneous release)/(maximum release − spontaneous release)] × 100. The enzyme-linked immunospot (ELISpot) assay for viral peptide-specific CD8+ T cells (25) started with 4 × 105 pooled lymph node and spleen cells that were serially diluted 2-fold. NP-pulsed, irradiated splenocytes from (B6 × 129) F2 mice were added at 2 × 105 cells per well as antigen-presenting cells, and numbers of spot-forming cells (SFC) were determined after 40 hr of incubation.
Quantitating Functional CD4+ T Cells.
The CD4+ set was enriched by incubating lymph node and spleen populations with anti-CD8α (53–6.72; ATCC) and anti-major histocompatibility complex class II (TIB 120; ATCC), followed by anti-rat-Ig and anti-mouse-Ig-coated Dynabeads and depletion with a magnet. The final population was examined by flow cytometry and contained >85% CD4+ T cells and less than 0.03% CD8+ T cells. The ELISpot assay was started with 4 × 105 enriched CD4+ T cells, using 2-fold serial dilutions. The CD4+ T cells were stimulated with either uninfected or HKx31-infected, irradiated syngeneic splenocytes, and SFC numbers were determined after 72 hr of incubation. The CD4+ T cell ELISpot assay has been described (26) and does not detect virus-specific CD8+ T cells, which must be stimulated with peptide.
Statistical Analyses.
Average values ± SD were compared by two-tailed Student’s t tests. Analyses were performed by using statistical analysis software for the Macintosh (instat 2.03 shareware).
RESULTS
To test whether gene transfer into JAK3-deficient mice is sufficient to restore immune function, mice were transplanted with either wild-type BM expressing the endogenous JAK3 gene (+/+ BMT) or BM from JAK3 knockout mice (JAK3 −/−) that had been transduced (19) with a JAK3-expressing retroviral vector (JAK3 BMT). These chimeric mice were tested for the presence of reconstituted B and T lymphocytes in the peripheral blood, beginning 4–6 weeks later. In all cases, +/+ BMT (n = 10) and JAK3 BMT (n = 16) mice showed a highly significant increase in the absolute number of B and T lymphocytes relative to JAK3 −/− mice, of the same magnitude as those reported previously (19). Mice transplanted with either unmanipulated JAK3 −/− BM or JAK3 −/− BM cultured on naive GP+E86 cells (mock BMT) had lymphocyte counts identical to untransplanted mice and displayed an activated T cell phenotype as described (16–18). By contrast, the numbers of activated T cells in JAK3 BMT mice were significantly lower in spleen and lymph nodes, similar to previous results (19) analyzing peripheral blood T lymphocytes (data not shown). However, because the magnitude of B and T lymphocyte reconstitution in JAK3 BMT mice was typically lower (data not shown) than in normal wild-type mice, the extent of functional reconstitution of the immune system remained questionable. The JAK3 BMT mice thus were tested for immune competence by challenge with 106.8 EID50 of the HKx31 influenza virus at times ranging from 2 to 5 months after BMT.
Virus-Specific Antibody Response in JAK3 BMT Mice.
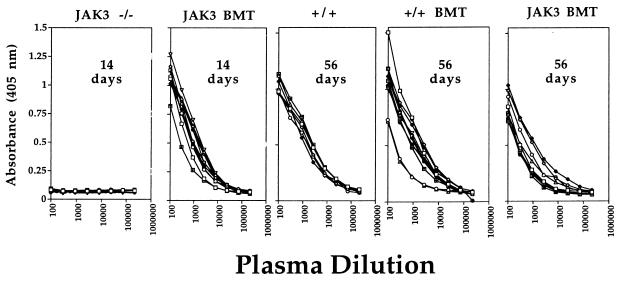
Plasma samples were obtained at intervals after infection and influenza-specific IgG levels were determined by ELISA (Fig. 1). The negative controls, which included untransplanted JAK3 −/− or mock BMT mice, showed no detectable antibody response at 14 days after infection. However, all JAK3 BMT mice developed a normal virus-specific response within 14 days of virus challenge, with the levels in plasma remaining high until day 56 (Fig. 1). The virus-specific Ig titers on day 56 in JAK3 BMT mice were also roughly equivalent to those found for mice transplanted with wild-type BM or untransplanted, wild-type mice (Fig. 1). The humoral immune response to this respiratory pathogen was thus fully restored by JAK3 gene transfer into the JAK3 −/− recipients.
Figure 1.
Levels of influenza-specific IgG in plasma from JAK3 BMT mice 14 and 56 days after challenge. Mice were infected intranasally with 106.8 EID50 of the HKx31 influenza virus, and plasma was collected from the peripheral blood at various time points. Antibody titers were determined by ELISA using the absorbance at 405 nm. No antibody response was detectable for the JAK3 −/− mice 14 days after infection. By contrast, all JAK3 BMT mice showed normal titers of specific antibody at this early time point. Anti-influenza IgG levels also were determined in plasma from JAK3 BMT mice 56 days after challenge. The +/+ BMT and JAK3 BMT mice showed normal levels of circulating virus-specific Ig when compared with untransplanted wild-type mice (+/+). The titers in JAK3 BMT and +/+ BMT mice did not differ significantly between days 14 and 56. Each line represents the antibody titer from an individual mouse.
Reconstitution of Cell-Mediated Immunity.
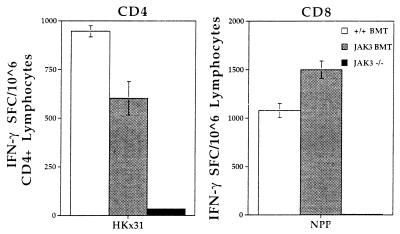
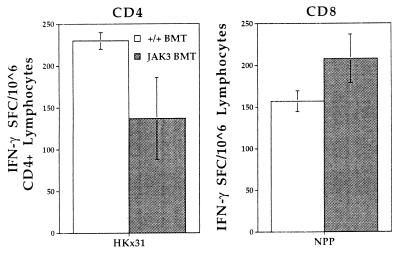
Virus-specific CD4+ and CD8+ T cell responses were analyzed in the lymphoid compartment and the site of pathology in the lung for mice challenged intranasally with the HKx31 virus. Lymph nodes and spleens from three mice were pooled for ELISpot analysis to measure influenza-specific, interferon (IFN) γ SFC numbers 13 days after infection (Fig. 2). The lack of SFC established that the JAK3 −/− mice did not generate a virus-specific T cell response. By contrast, mice transplanted with either wild-type BM or JAK3-transduced BM showed similar frequencies for both the CD4+IFN-γ+and CD8+IFN-γ+ SFC, which were significantly above those for untreated controls (P < 0.0001). Comparable results were obtained at 7 days after infection (data not shown). The lymphoid organs of the JAK3 BMT mice thus contain virus-specific T cells capable of proliferation and function in response to viral challenge.
Figure 2.
ELISpot assay to quantitate the cellular immune response in spleen and lymph nodes of JAK3 BMT mice 13 days after influenza challenge. Spleen and lymph nodes were collected from infected mice and pooled for analysis. The cells were cocultured with antigen-presenting cells that were either HKx31-infected (CD4) or NP366-pulsed (CD8). Uninfected or nonpulsed samples served as negative controls and the background level of IFN-γ production was less than 38 SFC/106 cells. Numbers of IFN-γ producing cells were determined by ELISpot assay in 96-well plates. The JAK3 BMT (gray bars) and +/+ BMT (white bars) mice showed a similar response for both CD4+ and CD8+ T cells. However, no significant response was detectable in the JAK3 −/− mice (black bars). Results are expressed as the number of SFCs and represent data from three mice assayed as a pool.
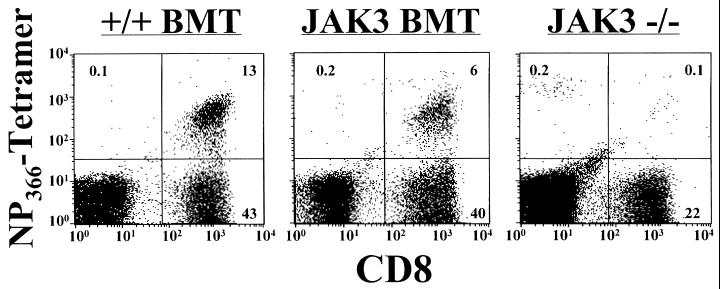
Virus-specific CD8+ T cells then were quantitated for the BAL population from the lung, as the primary site of influenza virus infection is the respiratory epithelium. The virus-specific CD8+ T cell response was measured by direct flow cytometric analysis using the NPP (27), which binds to T cells bearing receptors for immunodominant Db-NP366–374 epitope (Fig. 3) (28, 29). Mice transplanted with wild-type BM (+/+ BMT) showed a large percentage of CD8+NPP+ cells (24%, calculated as the percentage of CD8+ T cells that costained with the tetramer) infiltrating the lung. The JAK3 BMT mice also showed evidence of a significant CD8+NPP+ response (14%). However, the CD8+NPP+ population was virtually absent from the JAK3 −/− mice (0.6%). These data indicate that influenza-specific CD8+ effector T cells clearly had localized to the site of virus-induced pathology in the JAK3 BMT mice.
Figure 3.
Influenza-specific CD8+ T cells in the lungs of JAK3 BMT mice 13 days after influenza challenge. The BAL population was stained for influenza virus-specific CD8+ T cells by using the NPP tetramer (NP366 + H-2Db)-phycoerythrin and CD8- fluorescein isothiocyanate, followed by flow cytometric analysis. Significant populations of CD8+NPP+ cells were found by flow cytometric analysis of BAL cells from the +/+ BMT and JAK3 BMT mice, demonstrating the presence of infiltrating CD8+ T cells specific for influenza virus. No significant numbers could be detected in JAK3 −/− mice.
Virus Clearance and Survival.
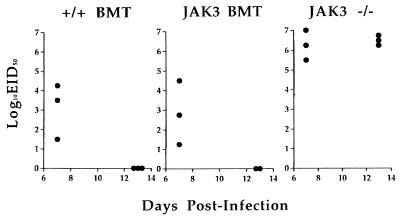
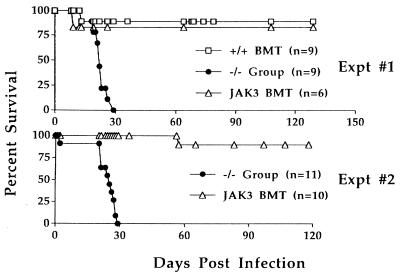
Lung virus titers were determined at 7 and 13 days after challenge with the HKx31 virus (Fig. 4). Evidence of clearance was already apparent for the +/+ BMT and JAK3 BMT mice by day 7, as the lung titers were obviously lower than those in the JAK3 −/− mice (P < 0.0270). No evidence of residual infection was apparent on day 13 in +/+ BMT and JAK3 BMT, when lung virus titers were high in the JAK3 −/− group. The virus-infected JAK3 −/− mice became progressively ill, lost weight, and succumbed to the infection (Fig. 5) within 3–4 weeks of challenge (n = 25 from three separate infections). The +/+ BMT or JAK3 BMT mice were protected, showed survival rates of 90%, and remained healthy for 11 months, the latest time point examined.
Figure 4.
Clearance of influenza virus from the JAK3 BMT mice. Lungs were collected from infected mice 7 and 13 days after intranasal challenge with 106.8 EID50 of the HKx31 influenza A virus. Virus was detectable after inoculation in embryonated hen’s eggs for all three groups on day 7, but the titers in the +/+ BMT and JAK3 BMT mice were much lower than those in JAK3 −/− mice. By day 13, the +/+ BMT and JAK3 BMT mice had cleared the infection from the lungs, but titers remained high in the JAK3 −/− mice. Each symbol represents a single mouse.
Figure 5.
Protection against influenza virus in JAK3 BMT mice. The HKx31-infected mice were followed long term to determine the effects of JAK3 gene transfer on survival. All mice from the JAK3 −/− group (includes mock BMT mice and untransplanted JAK3 −/− mice) became progressively ill and died by day 29 after infection. By contrast, roughly 90% of JAK3 BMT mice survived the challenge and lived long term. The relative survival of JAK3 BMT mice was the same as that seen after transplant of normal wild-type BM (+/+ BMT, Expt #1).
T Cell Memory.
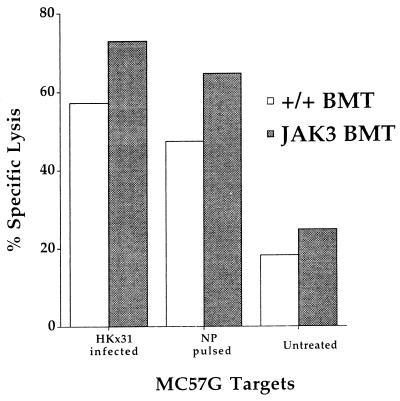
Spleen and lymph nodes were harvested from surviving mice on day 130 after infection and analyzed by ELISpot (Fig. 6). The frequencies of CD4+IFN-γ+ or CD8+IFN-γ+ SFCs were similar for the +/+ BMT and JAK3 BMT mice, and significantly above background levels (P < 0.0117) (Fig. 6). This in vitro result, which in naive T cells involves JAK3-dependent signal transduction pathways, was obtained 9 months after transplant. In addition, a virus-specific cytotoxic T cell response was demonstrated for in vitro cultured T cells by Cr51 release assay at this late time point (Fig. 7). The retroviral gene transfer protocol thus effectively reconstituted long-term T cell memory.
Figure 6.
Memory T cells in JAK3 BMT mice. Spleen and lymph nodes were collected from HKx31-infected surviving mice at 130 days after challenge. ELISpot assay of pooled lymphocyte populations for IFN-γ SFC demonstrated the presence of substantial numbers of memory CD4+ and CD8+ T cells from both +/+ BMT (open bars) and JAK3 BMT (filled bars) mice. The background level of IFN-γ production after incubation with untreated stimulators was <33 SFC/106 cells.
Figure 7.
Influenza-specific cell-mediated cytotoxicity after in vitro stimulation of memory CD8+ T cells. Spleen and lymph nodes were collected and pooled from +/+ BMT (open bars) or JAK3 BMT (filled bars) mice that had been infected 130 days previously with the HKx31 virus. After 5 days of in vitro stimulation in the presence of irradiated stimulator cells that were HKx31-infected or untreated, effectors were harvested and cytotoxic T cell activity measured in 51Cr release assay against HKx31-infected, NP-pulsed, or untreated MC57G (H-2b) target cells. This result shows lysis of targets at an effector-to-target cell ratio of 3.8:1. The background level of specific lysis was <2% for lymphocytes cultured with uninfected stimulator cells.
DISCUSSION
Earlier experiments (19) established the feasibility of targeting hematopoietic stem cells from JAK3-deficient mice with a retroviral vector expressing murine JAK3. The key question that remained, however, was the extent of the correction under conditions of immune challenge. To fully test the reconstituted immune system of the transplanted mice, we chose to infect them with a potentially lethal influenza A virus. The capacity of the JAK3 BMT mice to deal with such challenge is clearly an important test for this gene therapy model, establishing that the repertoire of reconstituted T cells is sufficiently diverse to allow a specific host response to a complex foreign antigen. This test is relevant for patients receiving JAK3 gene therapy, because virus infections are a major cause of morbidity and mortality. In a retrospective study of 108 SCID infants, common problems at presentation were respiratory syncytial virus and adenovirus infections (9). Uncontrolled growth of cytomegalovirus and Epstein–Barr virus infections is also a major cause of death in SCID patients (10).
The choice of influenza virus for this study was based on several considerations. The immune response to influenza infection has been extensively characterized in wild-type mice (22, 28–30). Lung viral titers peak 3–4 days after infection and decline to undetectable levels by day 10 (31). Although mice lacking either CD8+ T cells (32) or B cells (33–35) have been shown to survive infection, both T and B lymphocytes appear to work in synergy to protect against influenza virus and play important roles in recovery (34, 36, 37). The consequences of influenza infection in SCID mice are clearly much more severe than for normal immuno-competent controls. In a study by Albert et al. (38), infected C.B-17 SCID mice showed dehydration, weight loss, and lung disease consistent with proliferative pneumonia. Similar findings were noted here in JAK3 −/− mice after infection with the HKx31 influenza A virus. The influenza model is thus an appropriate experimental system for determining whether restoration of JAK3 expression can confer resistance to infection.
The results presented here demonstrate that not only were the JAK3 BMT mice able to respond to virus challenge, but the magnitude of the immune response was not significantly different from mice transplanted with wild-type BM. The JAK3 BMT mice developed virus-specific Ig and CD4+ and CD8+ T cell response profiles comparable to those found in controls receiving +/+ BMT. Both humoral and cellular immune responses correlated with clearance of virus from the lungs and 90% of JAK3 gene-corrected mice survived in the long term.
Even moderate levels of JAK3 gene transfer appear to be sufficient to restore a functional immune system, because gene transfer levels into JAK3 −/− BM-derived myeloid progenitors were typically in the range of only 15 to 25% with the MPSV/JAK3 vector. Thus far, of 48 mice we have transplanted with JAK3-transduced BM, all have shown highly significant levels of lymphocyte correction, which was likely the result of the natural in vivo selective advantage for the gene-modified lymphocytes (19). The presence of an in vivo selective advantage for JAK3-corrected cells is an important consideration for gene therapy, given that gene transfer into hematopoietic stem cells remains relatively inefficient in patients (39). A strong selective advantage also has been observed in humans containing mutations in the common γ chain, a cytokine receptor component that associates with JAK3 and is necessary for its activation. Female carriers bearing mutations in the X-linked common γ chain gene show nonrandom X chromosome inactivation in both circulating T and B cells (40, 41), indicating a selective advantage for lymphocytes that express the normal gene on the active X chromosome. Recent evidence also suggests that a spontaneous reversion of a common γ chain mutation, presumably occurring in a single T cell precursor, was sufficient to partially restore T cell function in a patient with X-linked SCID (42). In addition, gene therapy clinical trials for adenosine deaminase deficiency also have shown a selective advantage for lymphocytes containing the ADA vector (43, 44). Altogether, these data suggest that in the context of a gene therapy trial, a small number of JAK3-corrected cells may be sufficient for conferring significant benefit.
To date, two other examples have been reported where gene therapy has been used to correct animal models of immunodeficiency. These studies used models for chronic granulomatous disease (CGD) resulting from a deficiency in phagocyte oxidase (phox) p47phox (45) or gp91phox (46) proteins. After retroviral transduction of corrective genes into murine BM, mice were transplanted and subsequently challenged with either Burkholderia cepacia or Aspergillus fumigatus, respectively. In both studies, significant enhancement in survival of mice after infection was obtained when compared with the controls. However, the degrees of protection were substantially lower than that for normal wild-type mice. Also in those studies, the percentage of corrected cells obtained was found to be the rate-limiting step for providing resistance. The lack of an in vivo selective advantage for corrected cells in the CGD disease model is because the defect occurs in a terminally differentiated function of the granulocyte, rather than any step affecting granulocyte development or survival. In this case, as in most other disorders of the myeloid system, other approaches may be necessary to enhance levels of transduced cells (47).
The JAK3-deficient mouse (16–18) model differs from the human disease in the lack of phenotypically mature B cells. However, deficiencies in both humoral and cellular immunity are apparent for JAK3-deficient patients, and the resulting functional phenotypes are very similar with those found in the JAK3 −/− mice. Thus, correction of the defect in the mouse model represents an important step toward the goal of gene therapy for this disease. Clinical translation of this approach may require some modification of the protocol described here, however. The ideal setting for BMT in SCID patients is transplantation without prior irradiation. The necessity for myeloablation currently is being tested in JAK3 −/− mice to model a clinical approach for umbilical cord blood stem cell transduction and transplant into neonatal recipients. In summary, these studies demonstrate the proof of principle that retroviral-mediated JAK3 gene transfer into repopulating hematopoietic cells can be sufficient to restore specific immunity and provide a high degree of protection against challenge with a virus pathogen.
Acknowledgments
We thank Mark Sangster for technical advice on ELISA assays for detecting influenza-specific IgG and for providing purified HKx31 virus. We also thank John Altman (Department of Microbiology and Immunology, Emory University, Atlanta, GA) in whose laboratory the NP366-tetramer was made. This work was supported in part by National Heart, Lung, and Blood Institute Program Project Grant P01 HL 53749, James S. McDonnell Foundation Grant 94-50, Public Health Service Grants P01 CA 31922, AI29579, and AI38359, Cancer Center Support Grant P30 CA 21765, and the American Lebanese Syrian Associated Charities (ALSAC).
ABBREVIATIONS
- JAK3
Janus kinase 3
- NP
influenza virus nucleoprotein peptide
- NPP
tetramer of NP + H-2Db
- BM(T)
bone marrow (transplantation)
- HKx31
Hong Kong influenza x31
- SFC
spot-forming cell
- BAL
bronchoalveolar lavage
- H
influenza virus hemagglutinin molecule
- N
influenza virus neuraminidase glycoprotein
- SCID
severe combined immunodeficiency
- EID
egg infectious doses
- ELISpot
enzyme-linked immunospot
- IFN
interferon
References
- 1.Gurniak C B, Berg L J. Blood. 1996;87:3151–3160. [PubMed] [Google Scholar]
- 2.Chen M, Cheng A, Chen Y Q, Hymel A, Hanson E P, Kimmel L, Minami Y, Taniguchi T, Changelian P S, O’Shea J J. Proc Nat Acad Sci USA. 1997;94:6910–6915. doi: 10.1073/pnas.94.13.6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ihle J N. Philos Trans R Soc London B. 1996;351:159–166. doi: 10.1098/rstb.1996.0012. [DOI] [PubMed] [Google Scholar]
- 4.Oakes S A, Candotti F, Johnston J A, Chen Y Q, Ryan J J, Taylor N, Liu X, Hennighausen L, Notarangelo L D, Paul W E, et al. Immunity. 1996;5:605–615. doi: 10.1016/s1074-7613(00)80274-5. [DOI] [PubMed] [Google Scholar]
- 5.Teglund S, McKay C, Schuetz E, van Deursen J M, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle J N. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 6.Macchi P, Villa A, Gillani S, Sacco M G, Frattini A, Porta F, Ugazio A G, Johnston J A, Candotti F, O’Shea J J. Nature (London) 1995;377:65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- 7.Russell S M, Tayebi N, Nakajima H, Riedy M C, Roberts J L, Aman M J, Migone T S, Noguchi M, Markert M L, Buckley R H, et al. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- 8.Candotti F, Oakes S A, Johnston J A, Giliani S, Schumacher R F, Mella P, Fiorini M, Ugazio A G, Badolato R, Notarangelo L D, et al. Blood. 1997;90:3996–4003. [PubMed] [Google Scholar]
- 9.Buckley R H, Schiff R I, Schiff S E, Markert M L, Williams L W, Harville T O, Roberts J L, Puck J M. J Pediatr. 1997;130:378–387. doi: 10.1016/s0022-3476(97)70199-9. [DOI] [PubMed] [Google Scholar]
- 10.Buckley R H, Schiff S E, Schiff R I, Roberts J L, Markert M L, Peters W, Williams L W, Ward F E. Semin Hematol. 1993;30:92–101. [PubMed] [Google Scholar]
- 11.Fischer A, Landais P, Friedrich W, Morgan G, Gerritsen B, Fasth A, Porta F, Griscelli C, Goldman S F, Levinsky R. Lancet. 1990;336:850–854. doi: 10.1016/0140-6736(90)92348-l. [DOI] [PubMed] [Google Scholar]
- 12.Minegishi Y, Ishii N, Tsuchida M, Okawa H, Sugamura K, Yata J. Bone Marrow Transplantation. 1995;16:801–806. [PubMed] [Google Scholar]
- 13.Bacchetta R, Parkman R, McMahon M, Weinberg K, Bigler M, de Vries J E, Roncarolo M G. Blood. 1995;85:1944–1953. [PubMed] [Google Scholar]
- 14.van Leeuwen J E, van Tol M J, Joosten A M, Schellekens P T, van den Bergh R L, Waaijer J L, Oudeman-Gruber N J, van der Weijden-Ragas C P, Roos M T, Gerritsen E J. Blood. 1994;84:3936–3947. [PubMed] [Google Scholar]
- 15.Candotti F, Oakes S A, Johnston J A, Notarangelo L D, O’Shea J J, Blaese R M. J Exp Med. 1996;183:2687–2692. doi: 10.1084/jem.183.6.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nosaka T, van Deursen J M, Tripp R A, Thierfelder W E, Witthuhn B A, McMickle A P, Doherty P C, Grosveld G C, Ihle J N. Science. 1995;270:800–802. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]
- 17.Thomis D C, Gurniak C B, Tivol E, Sharpe A H, Berg L J. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- 18.Park S Y, Saijo K, Takahashi T, Osawa M, Arase H, Hirayama N, Miyake K, Nakauchi H, Shirasawa T, Saito T. Immunity. 1995;3:771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 19.Bunting K D, Sangster M Y, Ihle J N, Sorrentino B P. Nat Med. 1998;4:58–64. doi: 10.1038/nm0198-058. [DOI] [PubMed] [Google Scholar]
- 20.Kilbourne E D. Bull W H O. 1969;41:643–645. [PMC free article] [PubMed] [Google Scholar]
- 21.Spencer H T, Sleep S E, Rehg J E, Blakley R L, Sorrentino B P. Blood. 1996;87:2579–2587. [PubMed] [Google Scholar]
- 22.Allan W, Tabi Z, Cleary A, Doherty P C. J Immunol. 1990;144:3980–3986. [PubMed] [Google Scholar]
- 23.Flynn K J, Belz G T, Altman J D, Ahmed R, Woodland D L, Doherty P C. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- 24.Flynn K, Mullbacher A. Immunol Cell Biol. 1996;74:413–420. doi: 10.1038/icb.1996.71. [DOI] [PubMed] [Google Scholar]
- 25.Murali-Krishna K, Altman J D, Suresh M, Sourdive D J, Zajac A J, Millder J D, Slansky J, Ahmed R. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 26.Riberdy, J. M., Flynn, K. J., Stech, J., Webster, R., Altman, J. D. Altman & Doherty, P. C. (1999) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 27.Altman J D, Moss P H, Goulder P R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 28.Townsend A R, Rothbard J, Gotch F M, Bahadur G, Wraith D, McMichael A J. Cell. 1986;44:959–968. doi: 10.1016/0092-8674(86)90019-x. [DOI] [PubMed] [Google Scholar]
- 29.Vitiello A, Yuan L, Chesnut R W, Sidney J, Southwood S, Farness P, Jackson M R, Peterson P A, Sette A. J Immunol. 1996;157:5555–5562. [PubMed] [Google Scholar]
- 30.Ada G L, Jones P D. Curr Top Microbiol Immunol. 1986;128:1–54. doi: 10.1007/978-3-642-71272-2_1. [DOI] [PubMed] [Google Scholar]
- 31.Astry C L, Yolken R H, Jakab G J. J Med Virol. 1984;14:81–90. doi: 10.1002/jmv.1890140202. [DOI] [PubMed] [Google Scholar]
- 32.Eichelberger M, Allan W, Zijlstra M, Jaenisch R, Doherty P C. J Exp Med. 1991;174:875–880. doi: 10.1084/jem.174.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epstein M M, Di Rosa F, Jankovic D, Sher A, Matzinger P. J Exp Med. 1995;182:915–922. doi: 10.1084/jem.182.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucas S J, Barry D W, Kind P. Infect Immun. 1978;20:115–119. doi: 10.1128/iai.20.1.115-119.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Topham D J, Tripp R A, Hamilton-Easton A M, Sarawar S R, Doherty P C. J Immunol. 1996;157:2947–2952. [PubMed] [Google Scholar]
- 36.Mackenzie C D, Taylor P M, Askonas B A. Immunology. 1989;67:375–381. [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor P M, Askonas B A. Immunology. 1986;58:417–420. [PMC free article] [PubMed] [Google Scholar]
- 38.Albert S E, McKerlie C, Pester A, Edgell B J, Carlyle J, Petric M, Chamberlain J W. J Immunol. 1997;159:1393–1403. [PubMed] [Google Scholar]
- 39.Dunbar C E, Cottler-Fox M, O’Shaughnessy J A, Doren S, Carter C, Berenson R, Brown S, Moen R C, Greenblatt J, Stewart F M. Blood. 1995;85:3048–3057. [PubMed] [Google Scholar]
- 40.Conley M E, Lavoie A, Briggs C, Brown P, Guerra C, Puck J M. Proc Natl Acad Sci USA. 1988;85:3090–3094. doi: 10.1073/pnas.85.9.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puck J M, Stewart C C, Nussbaum R L. Am J Hum Genet. 1992;50:742–748. [PMC free article] [PubMed] [Google Scholar]
- 42.Stephan V, Wahn V, le Deist F, Dirksen U, Broker B, Muller-Fleckenstein I, Horneff G, Schroten H, Fischer A, de Saint B. N Engl J Med. 1996;335:1563–1567. doi: 10.1056/NEJM199611213352104. [DOI] [PubMed] [Google Scholar]
- 43.Bordignon C, Notarangelo L D, Nobili N, Ferrari G, Casorati G, Panina P, Mazzolari E, Maggioni D, Rossi C, Servida P. Science. 1995;270:470–475. doi: 10.1126/science.270.5235.470. [DOI] [PubMed] [Google Scholar]
- 44.Kohn, D. B., Hershfield, M. S., Carbonaro, D., Shigeoka, A., Brooks, J., Smogorzewska, E. M., Barsky, L. W., Chan, R., Burotto, F., Annett, G., et al. (1998) Nat. Med. 775–780. [DOI] [PMC free article] [PubMed]
- 45.Mardiney M, Jackson S H, Spratt S K, Li F, Holland S M, Malech H L. Blood. 1997;89:2268–2275. [PubMed] [Google Scholar]
- 46.Bjorgvinsdottir H, Ding C, Pech N, Gifford M A, Li L L, Dinauer M C. Blood. 1997;89:41–48. [PubMed] [Google Scholar]
- 47.Allay J A, Persons D A, Galipeau J, Riberdy J M, Ashmun R A, Blakley R L, Sorrentino B P. Nat Med. 1998;4:1136–1143. doi: 10.1038/2632. [DOI] [PubMed] [Google Scholar]