Abstract
Screening for seed pigmentation phenotypes in Arabidopsis led to the isolation of three allelic yellow-seeded mutants, which defined the novel TRANSPARENT TESTA16 (TT16) locus. Cloning of TT16 was performed by T-DNA tagging and confirmed by genetic complementation and sequencing of two mutant alleles. TT16 encodes the ARABIDOPSIS BSISTER (ABS) MADS domain protein. ABS belongs to the recently identified “B-sister” (BS) clade, which contains genes of unknown function that are expressed mainly in female organs. Phylogenetic analyses using a maximum parsimony approach confirmed that TT16/ABS and related proteins form a monophyletic group. TT16/ABS was expressed mainly in the ovule, as are the other members of the BS clade. TT16/ABS is necessary for BANYULS expression and proanthocyanidin accumulation in the endothelium of the seed coat, with the exception of the chalazal-micropylar area. In addition, mutant phenotype and ectopic expression analyses suggested that TT16/ABS also is involved in the specification of endothelial cells. Nevertheless, TT16/ABS apparently is not required for proper ovule function. We report the functional characterization of a member of the BS MADS box gene subfamily, demonstrating its involvement in endothelial cell specification as well as in the increasingly complex genetic control of flavonoid biosynthesis in the Arabidopsis seed coat.
INTRODUCTION
In angiosperm seeds, the embryo and the endosperm are surrounded by the seed coat, which develops from the ovular integuments and therefore is of maternal origin. Important functions of the seed coat include protecting the embryo from biotic and abiotic stresses, providing nutrients for the embryo during its development as well as water and oxygen during germination, and delaying germination by controlling the strength of dormancy (reviewed by Boesewinkel and Bouman, 1995).
Recent studies have used histological analyses and/or mutant characterization to focus on the development of the Arabidopsis seed coat (Léon-Kloosterziel et al., 1994; Beeckman et al., 2000). The Arabidopsis seed coat consists of five cell layers: two of them form the outer integument and three form the inner integument (Léon-Kloosterziel et al., 1994); it also is the location of pigment deposition (Albert et al., 1997). Seed coat pigments in Arabidopsis are predominantly flavonoids—more precisely, proanthocyanidins (or condensed tannins) of the cyanidin type and flavonols of the quercetin type (Chapple et al., 1994) (Figure 1). Proanthocyanidins (PAs) are located specifically in the innermost cell layer of the seed coat, which also is called the endothelium (Devic et al., 1999). They accumulate as colorless compounds during the early steps of embryogenesis and subsequently give the seed its brown pigmentation after oxidative reactions during seed maturation and desiccation processes (Debeaujon et al., 2000). The pigmentation of Arabidopsis seeds is not uniform, showing a darker coloration around both the chalaza and the micropyle. Interestingly, the pigmentation of the seed body and the chalazal-micropylar area can be lost independently, suggesting that different genetic controls occur (Debeaujon et al., 2001). PAs have been shown to participate in the maintenance of seed coat–imposed dormancy as well as in seed longevity during storage (Winkel-Shirley, 1998; Debeaujon et al., 2000).
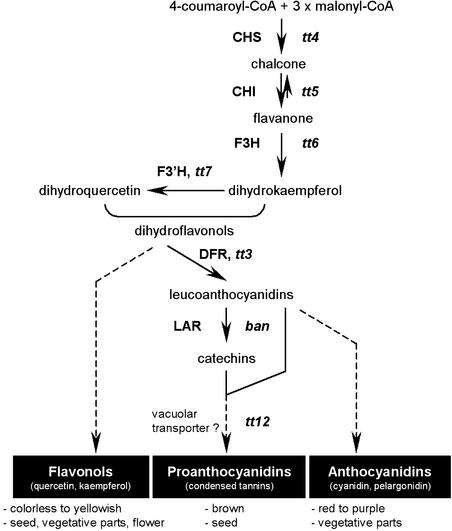
Figure 1.
Main Routes for Flavonoid Biosynthesis in Arabidopsis.
The pathway is initiated by the condensation of 4-coumaroyl-CoA (derived from phenylpropanoid metabolism) with malonyl-CoA. The three major end products of flavonoid metabolism in Arabidopsis are presented with indications of their respective color and localization. Enzymes are designated in capital letters. Mutants corresponding to genes of known function are indicated in lowercase italic letters. Dotted lines indicate different unidentified steps. ban, banyuls; CHI, chalcone isomerase; CHS, chalcone synthase; DFR, dihydroflavonol 4-reductase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; LAR, leucoanthocyanidin reductase; tt, transparent testa.
Numerous flavonoid mutants have been isolated in Arabidopsis on the basis of altered seed color. Collectively, these mutants are named transparent testa (tt), because null mutations in TT genes result in reduced, abolished, or modified pigmentation of the seed coat, depending on the intermediate flavonoid pigments (Koornneef, 1990). To date, ∼20 TT loci have been identified, and several of the corresponding genes have been cloned (Figure 1). In particular, mutations in tt3, tt4, tt5, tt6, tt7, tt12, and banyuls (ban) affect the following structural genes: dihydroflavonol 4-reductase (Shirley et al., 1992), chalcone synthase (Feinbaum and Ausubel, 1988; Shirley et al., 1995), chalcone isomerase (Shirley et al., 1992), flavanone 3-hydroxylase (Pelletier and Shirley, 1996; Wisman et al., 1998), flavonoid 3′-hydroxylase (Koornneef et al., 1982; Schoenbohm et al., 2000), a multidrug secondary transporter–like protein (Debeaujon et al., 2001), and a putative leucoanthocyanidin reductase (Devic et al., 1999), respectively. The TT1, TT2, TT8, TRANSPARENT TESTA GLABROUS1 (TTG1), and TTG2 loci have been shown to encode regulatory proteins with similarities to zinc finger motif–containing proteins (Sagasser et al., 2002), R2R3-MYB DNA binding domain proteins (Nesi et al., 2001), basic helix-loop-helix (bHLH) DNA binding domain transcription factors (Nesi et al., 2000), WD repeat–containing proteins (Walker et al., 1999), and WRKY domain proteins (Eulgem et al., 2000), respectively. Among these genes, TT1, TT2, TT12, and BAN are expressed specifically in seeds. Finally, six other TT loci remain to be characterized at the molecular level. The effects of the tt9, tt10, tt13, and tt15 mutations are restricted to seed pigmentation (Shirley et al., 1995; Focks et al., 1999; Debeaujon et al., 2000), whereas tt11 and tt14 mutations also affect anthocyanin accumulation in vegetative parts (Debeaujon et al., 2000).
It is known that pigmentation patterns are established by the cell-specific accumulation of different flavonoids. The activation of the flavonoid biosynthetic genes is regulated largely at the transcriptional level (reviewed by Weisshaar and Jenkins, 1998). As a consequence, regulatory genes that activate these structural genes are major determinants of pigmentation pattern. Such regulatory genes have been identified in several plant species (Mol et al., 1998; Winkel-Shirley, 2002; reviewed by Weisshaar and Jenkins, 1998). A conserved model for the tissue-specific regulation of flavonoid biosynthesis in monocots and dicots involves MYB- and bHLH-related proteins (Mol et al., 1998). For example, in Arabidopsis, TT2 and TT8 act in concert with TTG1 to regulate BAN expression in the endothelium (Nesi et al., 2000, 2001), and TT2 is a major determinant for BAN accumulation during seed development (Nesi et al., 2001). This scheme closely resembles the situation in maize, in which R- and C1-like genes encode members of the bHLH and MYB transcription factor families (Cone et al., 1986; Paz-Ares et al., 1987; Ludwig et al., 1989), and also the situation in petunia with ANTHOCYANIN2 (AN2), AN1, and AN11, which encode proteins of the MYB, bHLH, and WD repeat families, respectively (de Vetten et al., 1997; Quattrocchio et al., 1999; Spelt et al., 2000). Recent characterization of the Arabidopsis TT1 and TTG2 genes revealed the existence of additional flavonoid regulators in seeds, although their target genes have not been identified.
Diversity in plant organ pigmentation also can be influenced by factors such as vacuolar pH, copigmentation, and cell shape (Mol et al., 1998). In particular, the structure of the pigmented cells modifies their optical properties and thus the intensity of the refracted color. For instance, petals of mixta mutants from Antirrhinum majus exhibit a paler color than their wild-type counterparts as a result of a flattening of the epidermal cells (Noda et al., 1994). In Arabidopsis, Sagasser and co-workers (2002) reported that the tt1 mutation caused the absence of PAs and also the alteration of endothelial cell morphology. In the same way, ANTHOCYANINLESSL2 (ANL2), which encodes a homeodomain protein belonging to the HD-GLABRA2 group, is required for both anthocyanin biosynthesis in vegetative parts and the normal cellular organization of the primary root (Kubo et al., 1999). However, in both cases, it is not clear whether cellular organization and pigment deposition are regulated as two independent pathways or if altered cellular differentiation prevents normal pigment biosynthesis and/or accumulation.
Here, we report the identification and characterization of a novel tt mutant named tt16. T-DNA tagging allowed molecular cloning of the TT16 gene, which was shown to define a MADS domain protein. MADS box genes represent a large group of regulatory genes found in plants and animals (see Discussion; for reviews, see Shore and Sharrocks, 1995; Riechmann and Meyerowitz, 1997; Theissen et al., 2000) that have not been associated previously with flavonoid metabolism. Sequence comparison led to the conclusion that TT16 is identical to the ARABIDOPSIS BSISTER (ABS) gene, which was identified recently on the basis of phylogenetic analyses (Becker et al., 2002) and shown to belong to a new clade of genes of unknown function that are expressed preferentially in female reproductive organs. Histological observations of immature seeds revealed that endothelial cells had a distorted shape in tt16 compared with the wild type, suggesting that TT16/ABS is involved in the control of endothelium development. In addition, our results demonstrated that a tt16 null mutation prevented the activation of the BAN promoter in the endothelium layer, thus corroborating the fact that tt16 did not accumulate PAs.
RESULTS
Identification and Genetic Analysis of a Novel Seed Color Mutant
The dxt32 mutant line was isolated during a screen of 20,000 independent T-DNA Arabidopsis transformants for altered seed coat pigmentation phenotypes (Nesi et al., 2000), as described by Bechtold et al. (1993). Seeds from dxt32 homozygotes were straw colored with a characteristic dark brown pigmentation around the chalaza-micropyle area (Figure 2B). In addition, dxt32 seeds were larger than their wild-type counterparts and occasionally displayed preharvest sprouting (Figures 2A and 2B). Whole dxt32 plants did not show any additional phenotypes under our growth conditions. In particular, vegetative parts appeared to synthesize wild-type pigments (data not shown).
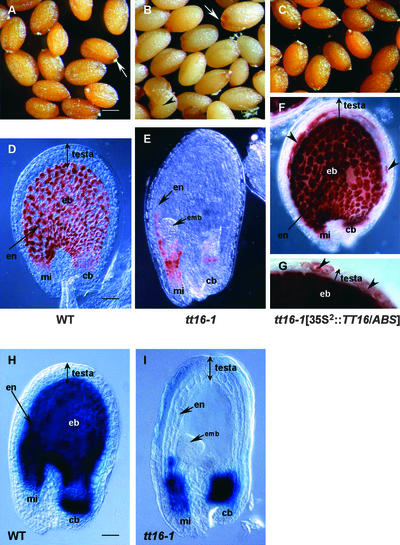
Figure 2.
Seed Pigmentation Phenotypes.
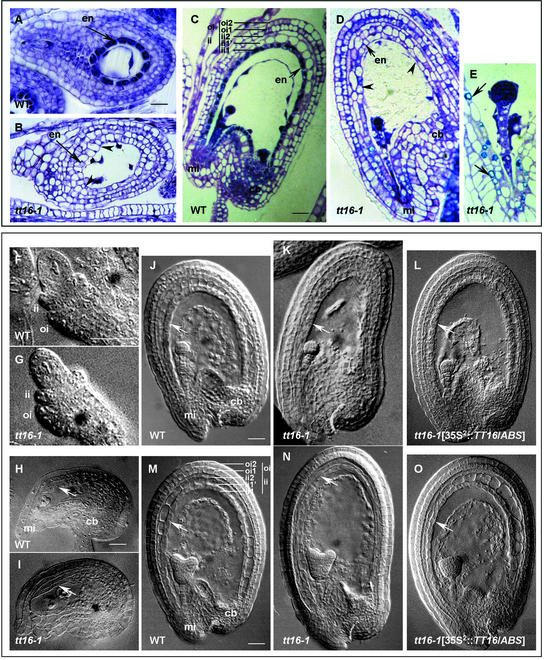
(A) to (C) Mature seeds from a wild-type plant (A), a tt16-1 homozygous plant (B), and the T2 progeny of a tt16-1 homozygote transformed with the TT16/ABS gene under the control of the double-enhanced 35S promoter (35S2::TT16/ABS) (C). White arrows in (A) and (B) point to the chalaza-micropyle area in mature seeds. tt16-1 seeds are larger than wild-type seeds and occasionally display premature germination (black arrowhead in [B]). All genotypes are from the Wassilewskija-2 ecotype. Bar in (A) = 200 μm for (A) to (C).
(D) to (G) PA deposition pattern in immature seeds was visualized with vanillin staining. Genotypes shown in (D), (E), and (F) and (G) correspond to the genotypes shown in (A), (B), and (C), respectively. Seeds were approximately at the globular stage of embryo development. The endothelium layer of tt16-1 seed does not exhibit PA accumulation, except in the chalazal-micropylar area (E). Ectopic vanillin staining is shown in seeds carrying the 35S2::TT16/ABS construct (black arrowheads in [F] and [G]). Bar in (D) = 30 μm for (D) to (F) and ∼15 μm for (G).
(H) and (I) Histochemical analysis of GUS reporter gene expression driven by the BAN promoter in seeds from the wild type (H) and the tt16-1 mutant (I). GUS expression patterns in (H) and (I) fully mimic the PA deposition profiles shown in (D) and (E), respectively. Seeds were at the early heart stage of embryo development. Photographs were made with phase-contrast optics. Bar in (H) = 35 μm for (H) and (I).
cb, chalazal bulb; eb, endothelium body; emb, embryo; en, endothelium; mi, micropyle; WT, wild type.
To investigate the inheritance of the seed phenotype, we backcrossed dxt32 plants with the wild type and then observed the segregation of the phenotype in the progeny. All F1 seeds displayed the phenotype conferred by the maternal genotype, which is consistent with a mutation affecting the seed coat, a sporophytic tissue. In addition, all F2 seeds exhibited the wild-type phenotype (brown testa), demonstrating that the mutation was recessive. Finally, F2 plants segregated as 18 homozygous dxt32 plants (giving yellow seeds) among 75 plants, in agreement with a 1:3 ratio observed for a monogenic nuclear trait (χ2 = 0.04, Pc > 0.05). Together, these results demonstrated conclusively that a recessive, monogenic, and nuclear mutation caused the maternally inherited seed phenotype in dxt32.
Because the seed coat phenotype of the dxt32 transformant strongly resembles that of transparent testa (tt) mutations, we performed allelism tests between the dxt32 mutant and each of the 18 tt mutants that were available at the time (tt1 to tt15, ttg1, ttg2, and ban) (Debeaujon et al., 2000). F2 seeds resulting from all of the crosses were dark brown, indicating that genetic complementation had occurred. This finding demonstrated that the mutation in the dxt32 line specified a novel genetic locus involved in the coloration of the seed coat, which was designated TT16. The dxt32 line harbors the tt16-1 mutation, and two additional tt16 mutant alleles, tt16-2 and tt16-3, were isolated subsequently from the Versailles T-DNA–mutagenized Arabidopsis population. The three tt16 alleles showed similar seed phenotypes under our growth conditions (data not shown).
Pigmentation Pattern in tt16 Seeds
The phenotype of tt16 mature seeds prompted us to examine pigment levels and localization in mutant seeds compared with wild-type seeds. Cyanidins, which result from the hydrolysis of PAs under acidic conditions, were barely detectable in tt16 mature seeds (Table 1). On the contrary, tt16 seeds exhibited wild-type amounts of flavonol compounds (Table 1). Quercetin rather than kaempferol accumulated in tt16 seeds (Table 1), as reported previously for wild-type seeds (Chapple et al., 1994) (Figure 1). Together, these results suggested that all of the biosynthetic steps upstream of the flavonol branch were fully functional in tt16 seeds, whereas the PA subpathway was downregulated.
Table 1.
Quantification of Cyanidins and Flavonols in Mature Seeds of Wild-Type and tt16-1 Plants by HPLC
| Sample | Wild Type | tt16-1 |
|---|---|---|
| Cyanidins | 1.32 ± 0.04 | 0.08 ± 0.00 |
| 1.54 ± 0.17 | 0.07 ± 0.01 | |
| Quercetin | 5.56 ± 0.01 | 4.97 ± 0.11 |
| 4.13 ± 0.24 | 4.01 ± 0.05 | |
| Kaempferol | 0.38 ± 0.01 | 0.33 ± 0.01 |
| 0.36 ± 0.02 | 0.34 ± 0.01 |
Wild-type seeds were from the Wassilewskija-2 ecotype. Cyanidins are released after acid hydrolysis of proanthocyanidins. Quercetin and kaempferol are the major flavonols in Arabidopsis. Analyses were performed on two independent batches of seeds for each sample. The values shown indicate ± se. Results are given in mg/g.
To gain insight into flavonoid biosynthesis during tt16 seed development, we analyzed the presence of the PA precursors, catechins and leucoanthocyanidins, in immature seeds of tt16. Because these compounds remain colorless until oxidative browning occurs during seed maturation, we used a colorimetric test to determine their presence during early seed development (Jende-Strid, 1993). In wild-type seed coats, uncolored PAs and their precursors were detected by their dark red staining with vanillin in the endothelium covering the whole seed body, hereafter called the endothelium body, as well as in the endothelial cells around the chalazal bulb and the micropylar area (Figure 2D). By contrast, only the chalazal bulb and the micropylar end retained red stain in tt16 seeds (Figure 2E), demonstrating that PA accumulation was restricted to these two regions in the mutant seed coat. The presence of PA-related compounds in the chalaza-micropyle area should account for the remaining dark brown pigmentation observed in the corresponding zone in tt16 mature seeds (Figure 2B) and for the detection of a small quantity of cyanidins in tt16 seeds as well (Table 1). The PA deposition pattern in tt16 seeds strongly resembles the pattern observed in tt1 seeds (Debeaujon et al., 2000; Sagasser et al., 2002).
Cloning and Structural Analysis of the TT16 Gene
To determine whether the tt16-1 mutation was caused by the insertion of the T-DNA, we analyzed the segregation of both kanamycin resistance, which is conferred by the T-DNA, and seed color phenotype in the F2 progeny from a cross between the mutant and the wild type. The tt16-1 line segregated for a single functional kanamycin locus (3:1 ratio of kanamycin-resistant to kanamycin-sensitive plants). In addition, all homozygous tt16-1/tt16-1 plants were kanamycin resistant. Therefore, genetic analyses strongly supported the conclusion that the mutation was linked closely to a single kanamycin resistance locus.
Plant genomic sequences flanking the right and left T-DNA insertions in tt16-1 were isolated using PCR walking (Devic et al., 1997). The two products were nearly identical to adjacent portions of a genomic clone, MKD15, found in the P1 Arabidopsis genomic library from the Kazusa group (Liu et al., 1995). The MKD15 clone was located at the top of chromosome 5, close to the KG-10 physical marker (http://www.kazusa.or.jp/kaos/kazusa/). Full sequence and gene annotations of MKD15 are available in the databases (Nakamura et al., 1997). The T-DNA insertion in tt16-1 was located in a coding region of MKD15, the product of which (protein BAB11181) showed sequence similarity to MADS domain factors. Recently, Becker and co-workers (2002) reported the identification of the BAB11181 putative MADS domain protein on the basis of database searches for angiosperm orthologs of the Gnetum gnemon MADS13 (GGM13) gene, a member of a novel clade of MADS box genes related to the B-class genes, named the B-sister (BS) clade. Therefore, the corresponding gene was named ARABIDOPSIS BSISTER (ABS) (Becker et al., 2002). Because the TT16 locus encodes the ABS gene, it will be referred to herein as TT16/ABS.
TT16/ABS transcripts were amplified by reverse transcription (RT) PCR using RNA from immature siliques and sequenced. Alignment of the genomic and cDNA sequences showed that TT16/ABS consists of six exons (Figure 3A) and that exon 4 was alternatively spliced, leading to two mRNA molecules that differed by 15 bp (Figure 3B). The longer molecule defined a 759-bp cDNA (hereafter referred to as cDNA1) that had been identified previously as an EST (Becker et al., 2002). The shorter molecule was 744 bp long (referred to as cDNA2). As a consequence, two proteins with 252 and 247 amino acid residues, respectively, were encoded. Six independent cDNA clones were isolated from immature siliques and sequenced, three of which corresponded to cDNA1 and three of which corresponded to cDNA2.
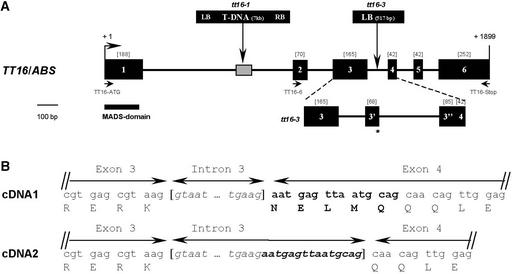
Figure 3.
Characterization of the TT16/ABS Gene.
(A) Structure of the TT16/ABS gene and positions of tt16 mutations. Exons are represented by closed rectangles with their sizes indicated in brackets, and introns are represented by lines. The positions and the nature of mutations in tt16 alleles are shown at top. T-DNA insertion in tt16-1 causes a deletion of plant genomic DNA, as indicated by the crosshatched box. The exon-intron structure of the TT16/ABS gene in tt16-3 is detailed below the scheme. Insertion of a fragment from T-DNA creates two novel exons (3′ and 3′′, respectively) and introduces a premature termination codon (star). Primers used for molecular analyses are indicated. Numbering of the nucleotide positions is given in base pairs from the translational start codon. The conserved MADS domain found in TT16/ABS is shown by the solid bar. LB, left border; RB, right border.
(B) Donor and acceptor splicing sites of intron 3 in the wild-type TT16/ABS gene. Alternative splicing occurs at the 3′ end of intron 3, leading to two cDNA molecules that differ by 15 bp (highlighted in boldface letters in the longer cDNA sequence). The amino acid residues deduced from the translation of each cDNA sequence are given below the corresponding codons.
Predicted Amino Acid Sequence of TT16/ABS
The amino acid sequences predicted for the two TT16/ABS proteins showed the typical structure of plant MADS box–related proteins, with a highly conserved MADS domain (56 amino acids) located at the N terminus (M), followed by a less conserved region, which includes a linker (called the inter [I] domain), a keratin-like (K) domain, and a C-terminal (C) end (Figure 4A). The MADS domain is believed to be involved in DNA contacts and protein dimerization (reviewed by Riechmann and Meyerowitz, 1997). The putative base-contacting residues found in the MADS domain are conserved in TT16/ABS (Figure 4A). The K domain is a coiled-coil segment, which is suggested to promote protein dimerization through interactions with other K boxes. The K box in TT16/ABS is capable of forming two putative amphipathic α-helices (Figure 4A). In addition, the C terminus of TT16/ABS contained numerous Pro and Gln residues (20% of the last 73 residues), a common feature of transcriptional activators (Ptashne, 1988).
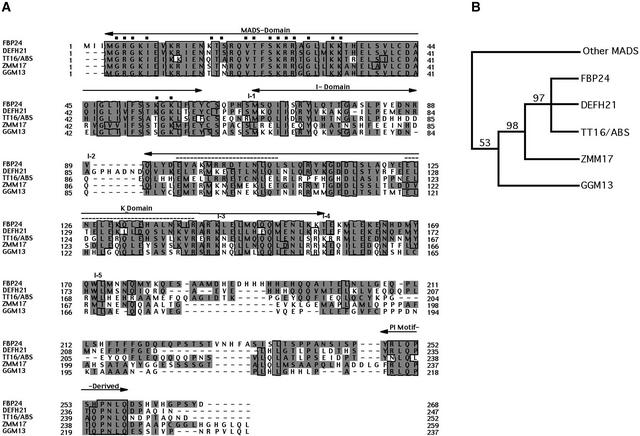
Figure 4.
Comparison of TT16/ABS with Other Related MADS Domain Proteins from the BS Clade.
(A) Alignment of deduced amino acid sequences for Arabidopsis TT16/ABS and related proteins from petunia (FBP24), A. majus (DEFH21), maize (ZMM17), and G. gnemon (GGM13). The MADS, I, K, and PI domains are indicated by arrows. Identical amino acid residues are boxed, and similar residues are shaded in gray. Dashes indicate gaps in the sequences to allow maximal alignment. The intron positions in TT16/ABS are shown above the alignment (I-1 to I-5). Putative base-contacting residues in the MADS domain of TT16/ABS are indicated by closed squares. The two putative amphipathic α-helices in the K domain are highlighted by dotted lines.
(B) Phylogenetic relationships between TT16/ABS and related MADS domain proteins. Shown is a simplified cladogram illustrating the consensus most-parsimonious pattern of relationships obtained using weighted MP analysis (see Methods). The monophyletic origin of the BS clade is presented with that of FBP24 (petunia), DEFH21 (A. majus), TT16/ABS (Arabidopsis), ZMM17 (maize), and GGM13 (G. gnemon). Bootstrap percentages are indicated before nodes, and nodes with a bootstrap score of <50% were discarded.
TT16/ABS-Like Genes Form a Monophyletic Clade
BLAST searches revealed that TT16/ABS was more closely related to MADS domain proteins from G. gnemon (GGM13), maize (ZMM17), A. majus (DEFH21), and petunia (FBP24) than to other plant MADS domain factors identified to date (Figure 4A). In particular, these proteins share a conserved motif, called the PI motif, at the C-terminal end (Becker et al., 2002) (Figure 4A). The corresponding genes were shown to form the new BS monophyletic group, which is related closely to MADS box genes from the B family (Becker et al., 2002). However, because the G. gnemon sequence (GGM13) was not found at the base of the clade in distance trees, as expected for a gymnosperm sequence, Becker and co-workers (2002) could not conclude whether these genes were orthologous. Therefore, to confirm that TT16-like MADS box genes formed a monophyletic group and were orthologous, we performed additional phylogenetic studies based on a maximum parsimony (MP) approach.
Unweighted MP analyses found three most parsimonious trees and their strict consensus, which suggested that the BS clade might be related to the B group (data not shown), as suggested previously (Becker et al., 2002). However, these results had only weak bootstrap support, because <50% of the trees displayed these relationships. PROTPARS analyses based on weighted MP did not improve this result (Figure 4B). However, it strongly supported the monophyletic origin of TT16/ABS-like genes (Figure 4B). In addition, the positioning of the gymnosperm sequence (GGM13) relative to monocot (ZMM17) and dicot (DEFH21, FBP24, and TT16/ABS) sequences strongly suggested that these genes are orthologous (Figure 4B).
Mutations of TT16/ABS Are Responsible for the Phenotype
To provide evidence that the disruption of the TT16/ABS locus was responsible for the mutant phenotype, we performed molecular characterization of two tt16 mutants. In tt16-1, the T-DNA insertion was present in the first intron of the TT16/ABS gene and was accompanied by a 78-bp deletion of plant genomic DNA (Figure 3A). In addition, tt16-1 might be a null allele, because no transcript was detected by RT-PCR (data not shown). Characterization of the TT16/ABS gene in tt16-3 showed that a 517-bp fragment from the T-DNA left border was inserted into the third intron (Figure 3A). Sequencing of the full-length TT16/ABS cDNA in tt16-3 revealed that insertion of the T-DNA fragment creates two novel splice sites within intron 3, resulting in a 912-bp unique transcript (data not shown) that contains a premature termination codon (Figure 3A). The resulting predicted protein contains 22 extra amino acid residues derived from translation of the T-DNA fragment and lacks the last 111 amino acids of the native TT16/ABS protein. Therefore, insertion of the T-DNA fragment in the third intron of the TT16/ABS gene likely confers the tt16-3 mutant phenotype. These results provide strong evidence that mutations in the TT16/ABS gene are responsible for the tt16 seed phenotype.
To confirm this idea, the TT16/ABS gene was expressed ectopically in the tt16-1 background in an effort to restore the wild-type phenotype. Homozygous tt16-1 plants were transformed with the TT16/ABS gene placed under the double-enhanced 35S promoter (henceforth called the 35S2:: TT16/ABS transgene). Five independent primary transformants were recovered, and in all cases, the presence of the transgene was sufficient to restore the ability of the plants to produce brown wild-type seeds (Figure 2C). In addition, a vanillin test conducted on the brown seeds of the primary transformants revealed that PA accumulation was not restricted to the endothelium, as was observed in nontransformed seeds of wild-type plants, but also occurred in the four other layers of the seed coat (Figures 2F and 2G). This finding suggested that ectopic expression of TT16/ABS was sufficient to induce competence for pigment biosynthesis and accumulation in the whole testa, although pigment distribution and quantity in the parenchymal and epidermal layers were disturbed compared with endothelium pigmentation (Figures 2F and 2G). We did not observe ectopic accumulation of PAs in embryos, in silique tissues, or in other vegetative organs of the transgenic plants (data not shown).
Expression of the TT16/ABS Gene
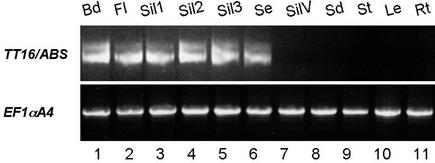
Because the effects of tt16 mutations were restricted to the seed, we wondered whether TT16/ABS transcripts accumulated specifically during seed development. Because TT16/ABS mRNA was barely detectable by RNA gel blot analysis (data not shown) (Becker et al., 2002), the accumulation of transcripts was monitored by RT-PCR. Primers TT16-6 and TT16-Stop, which do not hybridize within the conserved MADS domain (Figure 3A), were used to amplify RT products from several wild-type organs. The specificity of the PCR products was confirmed by direct sequencing. As shown in Figure 5, TT16/ABS was expressed in buds, flowers, and immature seeds throughout seed development (Figure 5, lanes 3 to 5), but no signal was detected in silique valves, seedlings, leaves, stems, and root samples. Our results demonstrated that TT16/ABS expression was restricted to reproductive organs, consistent with the expression data reported recently by Becker et al. (2002). In addition, we noticed that the two cDNA forms were always coexpressed in the tissues investigated (Figure 5).
Figure 5.
RT-PCR Showing TT16/ABS mRNA Accumulation in Organs of Arabidopsis Plants.
TT16/ABS transcripts were detected after 35 cycles of PCR amplification on reverse transcription products using primers TT16-6 and TT16-Stop (see Figure 3A). PCR products were visualized on a 2.5% (w/v) agarose gel stained with ethidium bromide. The two bands correspond to the two cDNA forms. Transcript levels of the Arabidopsis elongation factor EF1αA4 were used as a control (migrated on a 1% [w/v] agarose gel). Abbreviations for the organs examined are as follows: Bd, floral buds; Fl, full-blown flowers; Sil, immature siliques at different ages of embryo development; Se, seeds; SilV, silique valves; Sd, 4-day-old seedlings; St, stems; Le, rosette leaves; Rt, roots. For silique samples, Sil1 comprises siliques from 1 and 2 days after pollination (dap), Sil2 comprises siliques from 3 to 5 dap, and Sil3 comprises siliques from 6 to 10 dap. Seeds in lane 6 were harvested from the globe to late-torpedo stages of embryo development and removed from the silique valves.
Effect of a tt16 Null Mutation on the Activation of the BAN Promoter
PA biosynthesis requires the BAN gene, which most likely encodes leucoanthocyanidin reductase (Devic et al., 1999) (Figure 1). Because TT16/ABS defines a transcription factor involved in the accumulation of PAs in the endothelium, it was of interest to analyze BAN expression in a tt16 genetic background. For that purpose, we compared the expression of the GUS reporter gene driven by the BAN promoter in wild-type plants and a homozygous tt16-1 mutant. We found that the BAN promoter was activated specifically in the endothelium of young immature wild-type seeds (I. Debeaujon and L. Lepiniec, unpublished data) (Figure 2H), a result that faithfully mimics the BAN mRNA accumulation pattern revealed previously by RNA gel blot analysis and in situ hybridization (Devic et al., 1999). When introduced into the tt16-1 null mutant, activation of the BAN promoter was shown to be restricted to the chalaza-micropyle area (Figure 2I), in agreement with the pigmentation pattern and the vanillin profile of mutant seeds (Figures 2B and 2E, respectively). These results demonstrated that TT16/ABS is fully necessary for the normal activation of the BAN promoter in the endothelium body.
The Structure of Endothelial Cells Is Altered in tt16
To determine other possible roles for TT16/ABS in seed coat formation, we performed structural analyses of tt16 seeds. Scanning electron microscopy of the tt16 mature seed surface did not reveal any abnormalities in the seed coat epidermis (data not shown). Lengthwise sections of immature tt16 seeds were observed to investigate the internal anatomy of the five layers that form the seed coat after staining with toluidine blue, which reveals phenolic compounds in bluish green. The testa of tt16 seeds appeared to be more fragile than that of wild-type seeds because it often was damaged by microtome slicing (data not shown). As shown in Figure 6C, endothelial cells in immature wild-type seeds are small, almost rectangular in shape, regularly spaced, and filled with a bluish-green precipitate, so that the endothelium (called the ii1 layer by Beeckman et al. [2000]) clearly differs from the two parenchymal layers of the inner integument (ii1′ and ii2). In tt16 immature seeds, cells of the endothelium seemed to be flatter and more irregularly shaped than those in the wild type, resembled parenchymatic cells, and often seemed to collapse (Figure 6D). In addition, the tt16 endothelium did not accumulate the bluish-green polyphenol granules, except around the chalazal-micropylar region (Figure 6D), in agreement with the pigmentation pattern of the mutant seeds (Figures 2B and 2E). Detailed observation of the subcellular localization of the remaining polyphenols in the micropylar zone revealed that these compounds were deposited around the inside perimeter of the cells in tt16 seeds (Figure 6E), whereas they filled the entire central vacuole in wild-type cells (Beeckman et al., 2000) (Figure 6C).
Figure 6.
Structure of the Seed Coat.
(A) to (E) Sections (2 μm thick) through immature seeds after staining with toluidine blue. Ovules in (A) and (B) were taken at the one-celled stage of embryo development (before pigment deposition). Seeds in (C) to (E) were harvested at the globular embryo stage. The five layers of the seed coat are indicated in (C), according to Beeckman et al. (2000). Arrowheads in (B) and (D) show flattened and irregular cells in tt16-1 endothelium. (E) represents an enlarged view of the micropylar zone in tt16-1 seed, and the remaining pigmentation is indicated by arrows. (C) is reprinted from Nesi et al. (2001). Bars = 5 μm for (A) and (B) and 20 μm for (C) and (D).
(F) to (O) Cleared seeds viewed with Nomarski optics. Comparison of the development of the wild-type and tt16-1 seed coat, with an emphasis on the development of the endothelium.
(F) and (G) Early development of the two ovule integuments.
(H) and (I) Prefertilization stages with mature embryo sacs.
(J) to (L) Globular stage of embryo development.
(M) to (O) Seeds at the heart-shaped embryo stage.
The endothelium is indicated by white arrows ([H] to [O]). (L) and (O) show seeds carrying the 35S2::TT16/ABS construct. Bars = 5 μm for (F) and (G), 20 μm for (H) and (I), 25 μm for (J) to (L), and 30 μm for (M) to (O).
cb, chalazal bulb; en, endothelium; ii, inner integument; mi, micropyle; oi, outer integument; WT, wild type.
Because we did not know whether the altered morphology of endothelial cells in tt16 was caused by the lack of pigments or if these two phenotypes were independent, we examined sections through very young seeds that were harvested just after fertilization but before pigment deposition in the endothelium. In one-celled embryo wild-type seeds, the endothelial cells were regularly shaped and stained dark blue (Figure 6A) as a result of dense cytoplasm (Beeckman et al., 2000). By contrast, cells of tt16 endothelium already displayed a modified morphology (Figure 6B). These observations suggest that tt16 affects the endothelium cell structure in addition to pigment accumulation.
To extend these preliminary data, we examined the development of the tt16 endothelium in detail by analyzing cleared immature ovules and seeds using Nomarski microscopy. The altered morphology of the tt16 endothelium was particularly obvious in immature seeds of the globular and heart-embryo stages (Figures 6K and 6N, respectively), compared with that of the wild-type endothelium (Figures 6J and 6M). In addition, it appeared that the two parenchymal layers of the inner integument (ii1′ and ii2) also were affected in tt16 (Figure 6N), but no alterations were observed in the tt16 outer integument (oi1 and oi2). However, the integrity of the inner integument in tt16 did not appear to be affected in very young developing ovules (Figures 6F and 6G) or in mature ovules (Figures 6H and 6I).
Together, these results suggest that tt16 alters the endothelium structure and to some extent the integrity of the whole inner integument (ii1, ii1′, and ii2 layers), probably after fertilization. Nevertheless, alteration of the endothelium in tt16 apparently did not affect the normal development of the seeds, although they are more oblong than their round wild-type counterparts (Figures 6K and 6N). Finally, brown seeds of the transgenic plants carrying the 35S2::TT16/ABS construct in a tt16-1 genetic background exhibited a wild-type seed coat (Figures 6L and 6O), demonstrating that TT16/ABS was sufficient to fully restore the morphology of the endothelium.
Ectopic Expression of TT16/ABS Induced New Phenotypes
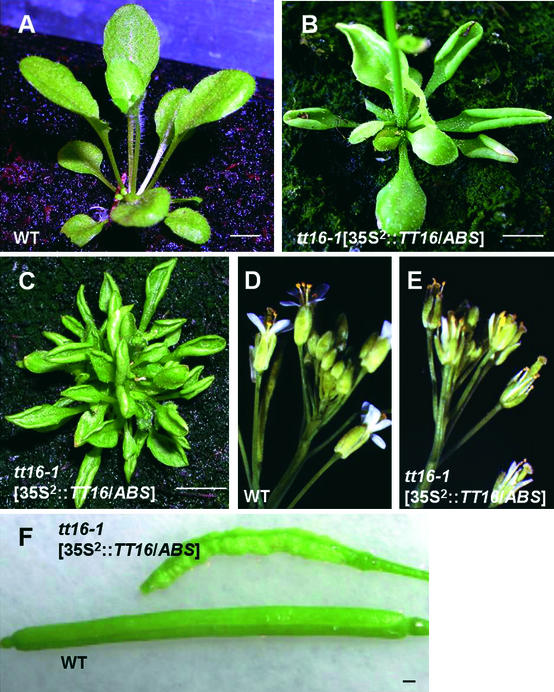
To investigate the putative effects of TT16/ABS when expressed ectopically, we analyzed transgenic plants carrying the 35S2::TT16/ABS construct in the wild-type and tt16 backgrounds. Approximately half of 15 independent hygromycin-resistant 35S2::TT16/ABS plants displayed aberrant characteristics in vegetative parts. In particular, rosette leaves were curled (Figures 7A and 7B), and in some cases, the rosette became bushy and the growth of the primary bolt was delayed (Figure 7C). Plants showed late flowering, and inflorescences had smaller flowers that were partially to completely sterile (Figures 7D and 7E). Siliques that did develop were shrunken (Figure 7F). The presence of the transgene was verified by RT-PCR in rosette leaves of all of the transformants (data not shown). These observations suggest that, when expressed ectopically, TT16/ABS is able to interact with cell differentiation and developmental processes.
Figure 7.
Effects of TT16/ABS Ectopic Expression on Plant Phenotype.
Phenotypes of plants carrying the 35S2::TT16/ABS construct ([B], [C], [E], and [F]) compared with nontransformed wild-type plants ([A], [D], and [F]). Approximately 50% of the 35S2::TT16/ABS transgenic lines showed curled leaves ([B] and [C]), reductions in flower size (E), and shrunken siliques (F). Plants in (A), (B), and (C) are shown at ∼20, 30, and 60 days after sowing, respectively. WT, wild type. Bars = 0.5 cm.
DISCUSSION
TT16 Encodes a MADS Domain Protein of the BS Family and Is Expressed Specifically in Seed
Screening for seed color mutants has led to the identification of a novel TT locus, TT16. The results of this study demonstrate that TT16 codes for the ABS MADS domain protein. The MADS domain is a conserved region of ∼56 amino acids that is involved in DNA binding and dimerization (Pellegrini et al., 1995; Shore and Sharrocks, 1995). MADS domain proteins are regulatory proteins found in all eukaryotic kingdoms (reviewed by Theissen et al., 2000) that acquired their names from the four founding members of the family: MCM1 (from yeast), AGAMOUS (from Arabidopsis), DEFICIENS (from A. majus), and SRF (human serum response factor). Approximately 82 MADS box genes have been identified in Arabidopsis with the completion of genome sequencing (Riechmann et al., 2000).
Previous studies have suggested that an ancestral MADS box gene duplication occurred before the divergence of plants and animals (Alvarez-Buylla et al., 2000), giving rise to two lineages of MADS box genes, types I and II. A monophyletic group containing plant AGL34-like sequences and animal SRF-related genes defines the type-I lineage, whereas other plant sequences and animal MEF2-like genes form the type-II group (Alvarez-Buylla et al., 2000). Plant proteins, including TT16/ABS, belong predominantly to the type-II lineage and share a conserved structure consisting of four domains: a MADS (M), an inter (I), a keratin-like (K), and a C-terminal (C) domain (Theissen et al., 1996; Münster et al., 1997). The DNA binding partner specificity is mediated to a large extent by the I domain, and the K domain likely promotes protein dimerization (Riechmann et al., 1996; Riechmann and Meyerowitz, 1997).
Two TT16/ABS cDNA forms are produced by alternative splicing of exon 4. The expression ratio of the two forms is almost constant in all tissues tested. The two corresponding proteins differ by five amino acid residues at the C-terminal end of the K domain. These changes could interfere with the specificity of protein–protein interactions mediated by the K dimerization domain. Alternative splicing also was observed for different MADS domain proteins in several plant species (Montag et al., 1995; Schmitz et al., 2000), although no biological function has been associated to date.
In plants, MADS box genes regulate the development of different organs, such as flower, ovule, fruit, leaf, and root (reviewed by Riechmann and Meyerowitz, 1997; Riechmann et al., 2000; Smyth, 2000; Ng and Yanofsky, 2001). A well-characterized example is the fate determination of floral organ primordia by genes of the A, B, and C classes, leading to the ABC model (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994). Today, the MADS box gene family is divided into different functional clades, namely A, B, C, D, and E (Becker et al., 2000; Theissen et al., 2000). Recently, a new class of MADS box genes, BS, was described (Becker et al., 2002). It includes genes from G. gnemon (GGM13), maize (ZMM17), petunia (FBP24), A. majus (DEFH21), and Arabidopsis (TT16/ABS), which are related closely to the B-class genes but expressed predominantly in the female reproductive organ. These genes form a monophyletic group that emerged before gymnosperm and angiosperm divergence, ∼300 million years ago (Becker et al., 2002). Our additional phylogenetic analysis demonstrates that GGM13 is positioned at the base of the group, as expected for a gymnosperm sequence, allowing the conclusion that the identified BS genes are orthologous.
Expression analyses showed that TT16/ABS mRNA accumulated in buds, flowers, and seeds but not in vegetative parts, including silique valves. Because of the very weak expression of TT16/ABS, it was difficult to localize transcript accumulation within the seed. However, TT16/ABS ectopic expression led to the accumulation of PAs in the different layers of the ovule integuments (Figures 2F and 2G), suggesting that TT16/ABS expression is confined to the endothelium layer in the wild-type seed coat, where PAs accumulate. The TT16/ABS mRNA accumulation profile matches the expression pattern of the other known BS genes (Becker et al., 2002), in particular DEFH21 transcripts, which were detected specifically in the inner integument layers of A. majus ovules (Becker et al., 2002).
TT16/ABS Is a New Regulator of Seed Flavonoid Pigmentation
Three independent tt16 allelic mutants were isolated from the Versailles T-DNA transformant collection, and two of them were sequenced during the course of this work. The mutation in tt16-1 induced a complete loss of function (i.e., no transcript was produced). In tt16-3, insertion of a T-DNA fragment in the third intron probably leads to the production of a severely truncated protein (lack of the C terminus). Because the tt16-3 phenotype was similar to the tt16-1 phenotype, the mutated protein is unlikely to be functional at all.
tt16 seeds displayed a characteristic pigmentation pattern, with a straw-colored seed body but a dark chalazal-micropylar area, similar to wild-type seeds. This suggested that the pigmentation of the chalaza-micropyle region is not under the control of TT16/ABS, as opposed to pigmentation of the seed body. In this way, TT16/ABS clearly differed from three other seed flavonoid regulators identified previously, TT2, TT8, and TTG1 (Walker et al., 1999; Nesi et al., 2000, 2001), but closely resembled a fourth one, TT1, which encodes a zinc finger protein required for pigmentation of the endothelium body (Sagasser et al., 2002). The existence of different genetic controls for pigmentation regulation in the chalaza-micropyle zone and the seed body was described previously in Arabidopsis (Debeaujon et al., 2001; Sagasser et al., 2002). A very similar situation also was reported for soybean and Brassica campestris seeds, in which one gene controlled pigment deposition in the hilum region specifically (Schwetka, 1982; Todd and Vodkin, 1993). The occurrence of genetic controls specific to the development of the chalazal-micropylar region may account for its critical roles, especially in preventing early radicle protrusion. Regulators involved in pigment accumulation specifically in the chalaza-micropyle area remain to be identified in Arabidopsis.
Histological analyses of immature tt16 seeds showed that endothelial cells completely lacked phenolic compounds, with the exception of the bluish-green polyphenol granules around the chalazal-micropylar pole, in agreement with the pigmentation profile of mature seeds. These data suggested that tt16 is affected in PA biosynthesis or accumulation, because these flavonoids are the major pigments found in the endothelium of Arabidopsis seeds (Albert et al., 1997). A vanillin test confirmed that tt16 endothelium was deprived of PAs, except in the chalaza-micropyle region. Accumulation of PAs in the chalaza-micropyle area of tt16 seeds suggests that downstream of leucoanthocyanidins at least two regulatory pathways control PA biosynthesis. One would be dependent on TT16/ABS and activated in the endothelium body, and the other would be TT16/ABS independent and occur in the chalaza-micropyle end. Further biochemical analyses revealed normal levels of flavonols in tt16 mature seeds, suggesting that the flavonoid biosynthetic pathway was blocked at the level of dihydroflavonol 4-reductase or downstream (Figure 1). In addition, flavonoid biosynthesis in vegetative parts of the tt16 plant was not altered, indicating that tt16 mutations affect the pigmentation of the seed coat only.
The effect of tt16 mutations on germination behavior could be correlated with the reduction in pigment accumulation in the mutant seed coat, as reported for some other tt mutants (Debeaujon et al., 2000). Indeed, seed coat pigments have been shown to participate in the hardness, and therefore in the permeability, of the testa. Histological observations of immature seeds revealed that the tt16 seed coat was damaged easily by microtome sectioning, confirming that tt16 testa was weaker than its wild-type counterpart, which could account for preharvest sprouting.
Finally, the BAN promoter was shown to be inactive in the endothelium body of tt16 seeds, demonstrating that the lack of PAs was correlated with the absence of BAN mRNA accumulation. Together, these results strongly support the conclusion that TT16/ABS is required for the normal activation of BAN expression in the endothelium body, and, as a consequence, for PA accumulation. Recent works have reported that TT1, TT2, TT8, and TTG1 also are involved in BAN regulation (Nesi et al., 2000, 2001; Sagasser et al., 2002). It is possible that some or all of these regulators act in concert with TT16/ABS. In particular, the pigmentation pattern of tt16 seeds closely resembles the tt1 seed phenotype, suggesting that TT1 and TT16/ABS could act together to regulate BAN expression. A comparison of temporal gene expression patterns revealed that TT16/ABS transcripts were maintained throughout seed development, as shown for TT8 (Nesi et al., 2000), whereas the expression of TT2 and BAN decreased rapidly from day 5 after pollination onward (Nesi et al., 2000, 2001). This finding suggests that although TT2, TT8, and TT16/ABS all are required for BAN activation, only TT2 determines the specificity of the BAN expression profile.
TT16/ABS Controls Cell Shape
Another important aspect of the phenotype that is invoked by TT16/ABS gene disruption is the abnormal structure of tt16 endothelial cells. Indeed, endothelial cells in tt16 usually resembled parenchymal cells and occasionally showed flattening. Our results suggest a role for TT16/ABS in the control of endothelium cell identity. The abnormal phenotypes of transgenic plants that ectopically express TT16/ABS support its involvement in the developmental regulation of the endothelium.
The effect of tt16 mutations on the shape of endothelial cells is similar to the situation for the FLORAL BINDING PROTEIN7 (FBP7) and FBP11 MADS box genes in petunia (Colombo et al., 1997). Indeed, simultaneous downregulation of FBP7 and FBP11 gene expression causes aberrant seed development attributable to premature degeneration of the endothelium layer. However, two lines of evidence suggest that TT16/ABS may not be the Arabidopsis ortholog of petunia genes. First, phylogenetic analyses demonstrate that FBP7 and FBP11 do not belong to the BS clade. Second, the putative Arabidopsis ortholog of FBP7/FBP11, the AGL11 gene (Rounsley et al., 1995), is expressed predominantly in the chalazal region of the fertilized ovule, whereas analyses of tt16 seed phenotypes suggest that TT16/ABS is expressed at least in the seed body. In addition, the seedstick mutation, which affects the AGL11 gene, induces abnormal development of the seed integuments without modifying seed pigmentation (A. Pinyopich and M.F. Yanofsky, personal communication).
During the past decade, several reports have underscored the relationships between cell morphology and pigmentation. For instance, maize defective aleurone pigmentation mutations affect aleurone development, giving patches of yellow sectors. Aleurone cells corresponding to the colorless areas are abnormal in shape, and anthocyanin biosynthesis is blocked in these cells, suggesting a possible connection between flavonoid biosynthesis and cell shape (Gavazzi et al., 1997). Another example is provided by the mixta mutant from A. majus, which exhibits pale petals with abnormal epidermal cells. However, in sharp contrast to tt16 endothelium, pigments are produced normally in mixta petals; therefore, the effect of the mutation on pigmentation is more likely indirect, by enhancing light absorption as a result of altered cell structure (Noda et al., 1994).
Other studies indicate that a lack of phenolic compounds could directly induce alterations of cell structure. For example, leaves of tobacco plants overexpressing the A. majus AmMYB308 factor lack phenolic intermediates, resulting in abnormal leaf palisade development and the induction of premature cell death in mature leaves (Tamagnone et al., 1998). Therefore, the authors postulated a role for phenolic intermediates as signaling molecules in tissue development and senescence. However, two lines of evidence clearly indicate that the abnormal structure of tt16 endothelial cells does not result from the absence of PA. (1) We first noticed aberrant endothelium morphology during early seed development, even before pigment deposition. (2) The absence of PAs in other tt mutants is not correlated with a defect in cell specification; for instance, endothelium develops normally in tt2 seeds (Nesi et al., 2001).
One attractive possibility is that TT16/ABS determines the structural features of endothelial cells and/or controls cell differentiation, which subsequently interferes with flavonoid accumulation in the endothelium. Analyses of the ectopic expression of pigmentation-specific factors in a tt16 genetic background may help test this hypothesis. Indeed, preliminary results obtained by ectopic expression of the Arabidopsis MYB factor TT2 (Nesi et al., 2001) suggested complementation of the tt16 pigmentation phenotype (N. Nesi, I. Debeaujon, C. Jond, and L. Lepiniec, unpublished results), indicating that TT2 may act downstream of TT16/ABS. Therefore, TT16/ABS would specify endothelium cell identity, and may regulate BAN expression and flavonoid biosynthesis indirectly, by controlling factors such as TT2. Remarkably, Sagasser and co-workers (2002) reported that tt1 mutations affected endothelium development in addition to PA accumulation in a manner similar to that of tt16. Thus, investigating possible interactions between TT1 and TT16/ABS will be an important approach in elucidating the complex regulatory network that controls endothelium formation and flavonoid biosynthesis in Arabidopsis seeds.
To conclude, we report the functional characterization of a MADS box gene of the BS family. TT16/ABS is required for normal endothelium development as well as for seed pigmentation in the endothelium body. This reinforces the conclusion that mutants that resemble typical flavonoid mutants may provide the access to the genes required for the development of specific tissues, as in anl2 or tt1 (Kubo et al., 1999; Sagasser et al., 2002). Furthermore, it has been hypothesized that BS genes may function either at the level of homeotic genes, by regulating the development and specification of female reproductive organs, or downstream of homeotic genes (Becker et al., 2002). However, analysis of tt16 knockout mutants demonstrates that TT16/ABS is not necessary for normal ovule function. Therefore, our results favor the second hypothesis, placing the BS genes downstream of homeotic genes, at least in Arabidopsis.
METHODS
Plant Strains
The tt16-1, tt16-2, and tt16-3 mutant alleles were obtained from the Versailles T-DNA–mutagenized Arabidopsis thaliana population (ecotype Wassilewskija-2) (Bechtold et al., 1993). The mutant lines were originally designated dxt32, bsf7, and dzr22, respectively. The dzr22 line was screened simultaneously by our team, on the basis of an altered seed coat pigmentation, and by J.C. Palauqui and colleagues (Institut National de la Recherche Agronomique, Versailles, France), on the basis of specific β-glucuronidase (GUS) expression (J.C. Palauqui, V. Lefèbvre, and S. Dinant, unpublished results), as a result of the structure of the T-DNA insert (Bouchez et al., 1993). In the initial dzr22 transformant, the mutations leading to the tt16 phenotype and GUS expression were shown to segregate as two independent traits. All methods and conditions used for plant growth, plant transformation, and selection of transgenic plants were as reported previously (Nesi et al., 2000). Plant materials used in this study are available at the Nottingham Arabidopsis Stock Center (Nottingham, UK) with the seed stock numbers given in parentheses: tt16-1 (N57500), tt16-2 (N57501), and tt16-3 (N57502).
Molecular Characterization of the TT16/ABS Gene
To identify the T-DNA integration site in tt16-1, a PCR walking strategy, as described by Devic et al. (1997), was used. Briefly, genomic DNA from mutant plants was extracted using a cetyl-trimethyl-ammonium bromide method (Doyle and Doyle, 1990) and used to construct PCR walking libraries. A 773-bp fragment of plant genomic DNA was amplified from a SspI library using a pair of nested primers specific for the T-DNA left border, LBBAR1 (5′-CAACCCTCAACT-GGAAACGGGCCGGA-3′) and LBBAR2 (5′-CGTGTGCCAGGTGCCCACGGAATAGT-3′). At the right border, a 91-bp fragment of plant genomic DNA was amplified from a DraI library using the nested primers RBGUS1 (5′-CCAGACTGAATGCCCACAGGCCGTC-3′) and RBGUS2 (5′-TCACGGGTTGGGGTTCTACAGGAC-3′) derived from the T-DNA sequence. In tt16-1, a unique full-length T-DNA copy was found to be inserted in the first intron of the TT16 gene, between nucleotides 620 and 697, creating a 78-bp deletion of genomic DNA. In tt16-3, a 517-bp T-DNA fragment was inserted at nucleotide 1314.
Detection and amplification of transcripts by reverse transcription PCR were performed as described elsewhere (Nesi et al., 2000). For cDNA cloning, we used primers TT16-ATG (5′-CATGGGTAGAGGGAAGATAG-3′) and TT16-Stop (5′-TTAATCATTCTGGGCCGTTG-3′) to amplify the predicted coding sequence. Analysis of TT16/ABS expression was conducted with primers TT16-6 (5′-AGGATGCCTCAACTCATTGACCG-3′) and TT16-Stop, which amplify a 573-bp fragment of cDNA1 and a 558-bp fragment of cDNA2. Control experiments were performed with primers EF1αA4-UP and EF1αA4-RP, designed by Nesi et al. (2000), and showed that EF1αA4 cDNA was amplified with similar efficiency from each reverse transcription reaction.
Constructs
All PCR products used for the construction of transgenic plants were generated with a high-fidelity DNA polymerase (pfu polymerase; Stratagene) and sequenced to ensure their integrity. Constructs for transformation were based on the pBIB-Hyg binary vector (Becker, 1990), which carries a hygromycin resistance gene, for the selection of transgenic plants. The resulting constructs were introduced into Agrobacterium tumefaciens C58C1Rif(pmp90) by electroporation and used to transform mutant or wild-type Arabidopsis (accession Wassilewskija-2). Transgenic plants were identified by their resistance to hygromycin and by the presence of transgene-specific sequences, as verified by PCR.
For complementation and ectopic expression assays, the 35S2::TT16/ABS construct was generated as follows. The TT16/ABS gene was amplified by PCR with primers TT16-ATG and TT16-Stop and cloned into the pMagic vector. The pMagic vector was constructed by cloning the expression cassette 35S2::Term (i.e., double-enhanced 35S promoter of Cauliflower mosaic virus [CaMV]–CaMV polyadenylation signals) from pLBR19 (Guerineau et al., 1992) as a KpnI-XhoI fragment into the KpnI-SalI–digested pBIB-Hyg binary vector. The TT16/ABS gene was directly blunt-end ligated in the SmaI-digested pMagic vector downstream of the 35S2 promoter. Orientation of the clones was double checked by restriction enzyme and sequence analyses.
The detailed construction of the pBAN::GUS transgene used in this study will be reported elsewhere (I. Debeaujon and L. Lepiniec, unpublished data). Briefly, the pBIB-Hyg binary vector, containing a 2.3-kb BAN promoter fragment fused translationally to a GUS::CaMV terminator cassette, was used to transform Arabidopsis wild-type or tt16-1 plants. For the analysis of pBAN::GUS expression in the tt16 genetic background, three independent transformants were recovered that expressed the GUS transgene in a similar pattern.
Biochemical Analyses
Preliminary experiments were performed to ascertain the most effective acid hydrolysis conditions for the tissues involved in this study. To analyze flavonols, Arabidopsis seeds (20 mg) were hydrolyzed at 90°C for 2 h in a 3-mL glass vial containing 2 mL of 1.2 M HCl in 50% aqueous methanol and 20 mM sodium diethyldithiocarbamate as an antioxidant. A Teflon-coated magnetic stir was placed in the vial, which was sealed tightly with a polytetrafluoroethylene-faced septum before heating in a Reacti-Therm Heating/Stirring Module (Pierce, Rockford, IL). For cyanidins, the hydrolysis time was optimized at 3 h. Extract aliquots of 100 μL, taken both before and after acid hydrolysis, were made up to 250 μL with distilled water containing 0.5% trifluoroacetic acid (flavonols) or 2.5% formic acid (cyanidins) before the analysis of 10-μL volumes by gradient elution reversed-phase HPLC. The flavonol analysis was performed as described by Stewart et al. (2001) except that UV light absorbance was measured with a Waters 996 photodiode array (Milford, MA) scanning from 250 to 600 nm and the chromatographs were analyzed using Waters Millennium Chromatography Manager software. For cyanidins, HPLC separations were performed at 40°C using a 150 × 3.0-mm (i.d., 4 μm) Nemesis Phenomenex C18 column (Phenomenex, Torrance, CA). The mobile phase was a 45-min, 5 to 25% gradient of acetonitrile in 2.5% formic acid, eluted at a flow rate of 1 mL/min. Column eluent was directed to the Waters 996 photodiode array with absorbance monitored at 520 nm. The analyses were performed three times on two independent batches of seeds.
Microscopy
Methods used for toluidine blue staining of immature seed sections and for the vanillin HCl test were as described by Nesi et al. (2001). Observations and photographs were made with an Axioplan 2 light microscope (Zeiss, Jena, Germany).
For analyses using Nomarski optics, immature seeds at different developmental stages were removed from siliques and cleared overnight in a chloral hydrate solution (chloral hydrate:water:glycerol, 8:2:1 [w/v/v]) before examination with the Axioplan 2 microscope equipped with Nomarski optics.
GUS histochemical analysis was performed according to Jefferson et al. (1987) in the presence of 2.5 mM each potassium ferricyanide and ferrocyanide. Samples were vacuum infiltrated for 1 h with GUS solution, incubated at 37°C for 12 to 16 h, and depleted of chlorophyll in 70% ethanol before mounting in a chloral hydrate solution and being observed with the Axioplan 2 microscope equipped with Nomarski optics.
Phylogenetic Analyses
Selection of sequences was designed to sample all major groups of plant MADS domain proteins listed in recent studies (Alvarez-Buylla et al., 2000; Becker et al., 2000, 2002). Sequence alignment was performed using ClustalX (version 1.8; Thompson et al., 1997) with gap penalties of 10 for opening and 0.10 for extension, and PAM350 as protein weight matrix for pairwise alignment, with gap penalties of 10 for opening and 0.20 for extension. The final alignment is available on request. Phylogenetic studies were conducted using heuristic maximum parsimony (MP) analyses with the Phylogenetic Analysis Using Parsimony (PAUP) program (version 3.1.1) (developed by D.L. Swofford, Laboratory of Molecular Systematics, Smithsonian Institution, Washington, DC). A weighted MP analysis was performed with the PROTPARS matrix given in the PAUP package and described in PHYLIP (Phylogeny Inference Package, version 3.5c; Joseph Felsenstein, University of Washington, Seattle). For weighted analyses, 100 replicates were used and gaps were treated as additional character states. For unweighted MP analyses, bootstrap measurements were conducted with 1000 iterations of random sequences, keeping all optimal trees, tree-bisection-reconnection branch swapping, and the no maxtrees limit for each replicate. Gaps were treated as additional data.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The accession numbers for the sequences mentioned in this article are as follows: MKD15 clone (AB007648), ABS (AJ318098), cDNA1 (AV556586), Arabidopsis TT16/ABS (AJ318098), petunia FBP24 (AF335242), A. majus DEFH21 (AJ307056), maize ZMM17 (AJ271208), and G. gnemon GGM13 (AJ132219).
Acknowledgments
We are especially grateful to Brenda Winkel-Shirley, Jérôme Giraudat, Francis Quétier, and Dao Zhou for enthusiastic discussions during the progress of this work. Brenda Winkel-Shirley also is acknowledged for critically reading the manuscript. We thank Nicole Bechtold and Georges Pelletier for providing access to the Versailles T-DNA transformant collection, Jocelyne Kronenberger for helpful advice with microscopy analysis, Jean-Christophe Palauqui for characterization of the dzr22 mutant, and Alan Crozier for access to facilities for biochemical analysis. This work was supported by a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche (Grant 97-5-11772 to N.N.). A.J.S. was employed on a grant (17/RSP07833) from the U.K. Biotechnology and Biological Sciences Research Council.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.004127.
References
- Albert, S., Delseny, M., and Devic, M. (1997). BANYULS, a novel negative regulator of flavonoid biosynthesis in the Arabidopsis seed coat. Plant J. 11, 289–299. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla, E.R., Pelaz, S., Liljegren, S.J., Gold, S.E., Burgeff, C., Ditta, G.S., Ribas de Pouplana, L., Martinez-Castilla, L., and Yanofsky, M.F. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97, 5328–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Becker, A., Kaufmann, K., Freialdenhoven, A., Vincent, C., Li, M.-A., Saedler, H., and Theissen, G. (2002). A novel MADS-box gene subfamily with a sister-group relationship to class B floral homeotic genes. Mol. Genet. Genomics 266, 942–950. [DOI] [PubMed] [Google Scholar]
- Becker, A., Winter, K.-U., Meyer, B., Saedler, H., and Theissen, G. (2000). MADS-box gene diversity in seed plants 300 million years ago. Mol. Biol. Evol. 17, 1425–1434. [DOI] [PubMed] [Google Scholar]
- Becker, D. (1990). Binary vectors which allow the exchange of plant se-lectable markers and reporter genes. Nucleic Acids Res. 18, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman, T., De Rycke, R., Viane, R., and Inzé, D. (2000). Histological study of seed coat development in Arabidopsis thaliana. Plant Res. 113, 139–148. [Google Scholar]
- Boesewinkel, F.D., and Bouman, F. (1995). The seed: structure and function. In Seed Development and Germination, J. Kigel and G. Galili, eds (Washington, DC: Library of Congress), pp. 1–24.
- Bouchez, D., Camilleri, C., and Caboche, M. (1993). A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C. R. Acad. Sci. Paris 316, 1188–1193. [Google Scholar]
- Chapple, C.C.S., Shirley, B.W., Zook, M., Hammerschmidt, R., and Somerville, S.C. (1994). Secondary metabolism in Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Sommerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 989–1030.
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Colombo, L., Franken, J., Van der Krol, A.R., Wittich, P.E., Dons, H.J., and Angenent, G.C. (1997). Downregulation of ovule-specific MADS box genes from petunia results in maternally controlled defects in seed development. Plant Cell 9, 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K.C., Burr, F.A., and Burr, B. (1986). Molecular analysis of the maize anthocyanin regulatory locus C1. Proc. Natl. Acad. Sci. USA 83, 9631–9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon, I., Léon-Kloosterziel, K.M., and Koornneef, M. (2000). Influence of the testa on seed dormancy, germination and longevity in Arabidopsis. Plant Physiol. 122, 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon, I., Peeters, A.J.M., Léon-Kloosterziel, K.M., and Koornneef, M. (2001). The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13, 853–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten, N., Quattrocchio, F., Mol, J., and Koes, R. (1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 11, 1422–1434. [DOI] [PubMed] [Google Scholar]
- Devic, M., Albert, S., Delseny, M., and Roscoe, T.J. (1997). Efficient PCR walking on plant genomic DNA. Plant Physiol. Biochem. 35, 331–339. [Google Scholar]
- Devic, M., Guilleminot, J., Debeaujon, I., Bechtold, N., Bensaude, E., Koornneef, M., Pelletier, G., and Delseny, M. (1999). The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J. 19, 387–398. [DOI] [PubMed] [Google Scholar]
- Doyle, J.J., and Doyle, J.L. (1990). Isolation of plant DNA from fresh tissues. Focus 12, 13–15. [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Feinbaum, R.L., and Ausubel, F.M. (1988). Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol. Cell. Biol. 8, 1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks, N., Sagasser, M., Weisshaar, B., and Benning, C. (1999). Characterization of tt15, a novel transparent testa mutant of Arabidopsis thaliana (L.) Heynh. Planta 208, 352–357. [DOI] [PubMed] [Google Scholar]
- Gavazzi, G., Dolfini, S., Allegra, D., Castiglioni, P., Todesco, G., and Hoxha, M. (1997). Dap (Defective aleurone pigmentation) mutations affect maize aleurone development. Mol. Gen. Genet. 256, 223–230. [DOI] [PubMed] [Google Scholar]
- Guerineau, F., Lucy, A., and Mullineaux, P. (1992). Effect of two consensus sequences preceding the translation initiator codon on gene expression in plant protoplasts. Plant Mol. Biol. 18, 815–818. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jende-Strid, B. (1993). Genetic control of flavonoid biosynthesis in barley. Hereditas 119, 187–204. [Google Scholar]
- Koornneef, M. (1990). Mutations affecting the testa colour in Arabidopsis. Arabidopsis Inf. Serv. 27, 1–4. [Google Scholar]
- Koornneef, M., Luiten, W., de Vlaming, P., and Schram, A.W. (1982). A gene controlling flavonoid 3′ hydroxylation in Arabidopsis. Arabidopsis Inf. Serv. 19, 113–115. [Google Scholar]
- Kubo, H., Peeters, A.J.M., Aarts, M.G.M., Pereira, A., and Koornneef, M. (1999). ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 11, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon-Kloosterziel, K.M., van de Bunt, G.A., Zeevaart, J.A.D., and Koornneef, M. (1994). Arabidopsis mutants with a reduced seed dormancy. Plant Physiol. 110, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.-G., Mitsukawa, N., Vazquez-Tello, A., and Whittier, R.F. (1995). Generation of a high-quality P1 library of Arabidopsis suitable for chromosome walking. Plant J. 7, 351–358. [Google Scholar]
- Ludwig, S.R., Habera, L.F., Dellaporta, S.L., and Wessler, S.R. (1989). Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. USA 86, 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol, J., Grotewold, E., and Koes, R. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3, 212–217. [Google Scholar]
- Montag, K., Salamini, F., and Thompson, R.D. (1995). ZEMa, a member of a novel group of MADS box genes, is alternatively spliced in maize endosperm. Nucleic Acids Res. 25, 2168–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münster, T., Pahnke, J., DiRosa, A., Kim, J., Martin, W., Saedler, H., and Theissen, G. (1997). Floral homeotic genes were re-cruited from homologous MADS-box genes preexisting in the common ancestor of ferns and seed plants. Proc. Natl. Acad. Sci. USA 94, 2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y., Sato, S., Kaneko, T., Kotani, H., Asamizu, E., Miyajima, N., and Tabata, S. (1997). Structural analysis of Arabidopsis thaliana chromosome 5. III. Sequence features of the regions of 1,191,918 bp covered by seventeen physically assigned P1 clones. DNA Res. 4, 401–414. [DOI] [PubMed] [Google Scholar]
- Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for the proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, M., and Yanofsky, M.F. (2001). Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2, 186–195. [DOI] [PubMed] [Google Scholar]
- Noda, K., Glover, B.J., Linstead, P., and Martin, C. (1994). Flower colour intensity depends on specialized cell shape controlled by a MYB-related transcription factor. Nature 369, 661–664. [DOI] [PubMed] [Google Scholar]
- Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P.A., and Saedler, H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6, 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, L., Tan, S., and Richmond, T.J. (1995). Structure of serum response factor core bound to DNA. Nature 376, 490–498. [DOI] [PubMed] [Google Scholar]
- Pelletier, M.K., and Shirley, B.W. (1996). Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings: Coordinate regulation with chalcone synthase and chalcone isomerase. Plant Physiol. 111, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne, M. (1988). How eukaryotic transcriptional activators work. Nature 335, 683–689. [DOI] [PubMed] [Google Scholar]
- Quattrocchio, F., Wing, J., van der Woude, K., Souer, E., de Vetten, N., Mol, J., and Koes, R. (1999). Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., Krizek, B.A., and Meyerowitz, E.M. (1996). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93, 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1997). MADS domain proteins in plant development. Biol. Chem. 378, 1079–1101. [PubMed] [Google Scholar]
- Rounsley, S.D., Ditta, G.S., and Yanofsky, M.F. (1995). Diverse roles for MADS-box genes in Arabidopsis development. Plant Cell 7, 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagasser, M., Lu, G.-H., Hahlbrock, K., and Weisshaar, B. (2002). A. thaliana TRANSPARENT TESTA1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 16, 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, J., Franzen, R., Ngyuen, T.H., Garcia-Maroto, F., Pozzi, C., Salamini, F., and Rohde, W. (2000). Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol. Biol. 42, 899–913. [DOI] [PubMed] [Google Scholar]
- Schoenbohm, C., Martens, S., Eder, C., Forkmann, G., and Weisshaar, B. (2000). Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol. Chem. 381, 749–753. [DOI] [PubMed] [Google Scholar]
- Schwetka, A. (1982). Inheritance of seed colour in turnip rape (Brassica campestris L.). Theor. Appl. Genet. 62, 161–169. [DOI] [PubMed] [Google Scholar]
- Shirley, B.W., Hanley, S., and Goodman, H.M. (1992). Effects of ionizing radiation on a plant genome: Analysis of two Arabidopsis transparent testa mutations. Plant Cell 4, 333–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley, B.W., Kubasek, W.L., Storz, G., Bruggemann, E., Koornneef, M., Ausubel, F.M., and Goodman, H.M. (1995). Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8, 659–671. [DOI] [PubMed] [Google Scholar]
- Shore, P., and Sharrocks, A.D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229, 1–13. [DOI] [PubMed] [Google Scholar]
- Smyth, D. (2000). A reverse trend: MADS functions revealed. Trends Plant Sci. 5, 315–317. [DOI] [PubMed] [Google Scholar]
- Spelt, C., Quattrocchio, F., Mol, J.N.M., and Koes, R. (2000). anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12, 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, A.J., Chapman, W., Jenkins, G.I., Graham, I., Martin, T., and Crozier, A. (2001). The effect of nitrogen and phosphorus deficiency on flavonol accumulation in plant tissues. Plant Cell Environ. 24, 1189–1197. [Google Scholar]
- Tamagnone, L., Merida, A., Stacey, N., Plaskitt, K., Parr, A., Chang, C.F., Lynn, D., Dow, J.M., Roberts, K., and Martin, C. (1998). Inhibition of phenolic acid metabolism results in precocious cell death and altered cell morphology in leaves of transgenic tobacco plants. Plant Cell 10, 1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J.T., Munster, T., Winter, K.U., and Saedler, H. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149. [PubMed] [Google Scholar]
- Theissen, G., Kim, J.T., and Saedler, H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43, 484–516. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTALX Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, J.J., and Vodkin, L.O. (1993). Pigmented soybean (Glycine max) seed coats accumulate proanthocyanidins during development. Plant Physiol. 102, 663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, A.R., Davison, P.A., Bolognesi-Winfield, A.C., James, C.M., Srinivasan, N., Blundell, T.L., Esch, J.J., Marks, M.D., and Gray, J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Meyerowitz, E.M. (1994). The ABCs of floral homeotic genes. Cell 78, 203–209. [DOI] [PubMed] [Google Scholar]
- Weisshaar, B., and Jenkins, G.I. (1998). Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1, 251–257. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley, B. (1998). Flavonoids in seeds and grains: Physiological function, agronomic importance and the genetics of biosynthesis. Seed Sci. Res. 8, 415–422. [Google Scholar]
- Winkel-Shirley, B. (2002). A mutational approach to dissection of flavonoid biosynthesis in Arabidopsis. In Recent Advances in Phytochemistry: Proceedings of the Annual Meeting of the Phytochemical Society of North America, Vol. 36, J.T. Romeo, ed (New York: Elsevier), in press.
- Wisman, E., Hartmann, U., Sagasser, M., Baumann, E., Palme, K., Hahlbrock, K., Saedler, H., and Weisshaar, B. (1998). Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc. Natl. Acad. Sci. USA 95, 12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]