Abstract
We used DNA microarray technology to identify genes involved in the low-oxygen response of Arabidopsis root cultures. A microarray containing 3500 cDNA clones was screened with cDNA samples taken at various times (0.5, 2, 4, and 20 h) after transfer to low-oxygen conditions. A package of statistical tools identified 210 differentially expressed genes over the four time points. Principal component analysis showed the 0.5-h response to contain a substantially different set of genes from those regulated differentially at the other three time points. The differentially expressed genes included the known anaerobic proteins as well as transcription factors, signal transduction components, and genes that encode enzymes of pathways not known previously to be involved in low-oxygen metabolism. We found that the regulatory regions of genes with a similar expression profile contained similar sequence motifs, suggesting the coordinated transcriptional control of groups of genes by common sets of regulatory factors.
INTRODUCTION
The plant kingdom displays a wide variation in the extent to which low-oxygen conditions can be tolerated, but the morphological adaptations and metabolic processes responsible for flooding tolerance/sensitivity remain poorly understood. The diffusion of oxygen in water is 10,000 times slower than that in air (Armstrong, 1979), drastically reducing the supply of oxygen, which is vital to the roots of the plant, when the roots are waterlogged. Morphological adaptations to low-oxygen stress include the formation of aerenchyma, root cortical air spaces that promote air transport from shoot to root, as well as the formation of adventitious roots and leaf and shoot elongation (Vartapetian and Jackson, 1997). Metabolic adaptation to anaerobiosis includes the induction of fermentation pathway enzymes (ethanol, lactic acid, and Ala fermentation) (Kennedy et al., 1992). A dramatic change in protein synthesis occurs in roots during anaerobiosis (Sachs et al., 1980; Dolferus et al., 1985). In maize roots, a set of ∼20 anaerobic proteins (ANPs) are synthesized selectively; most of these proteins have been identified as enzymes of glycolysis or sugar-phosphate metabolism (Sachs et al., 1996). ANPs that are part of other metabolic processes also have been reported (Chang et al., 2000), indicating that the low-oxygen response is complex and involves more than a simple adaptation in energy metabolism (Saab and Sachs, 1996; Trevaskis et al., 1997).
The expression of low-oxygen-induced genes is controlled predominantly at the transcriptional level, although post-transcriptional regulation mechanisms also have been demonstrated (Fennoy and Bailey-Serres, 1995). An anaerobic response element (ARE) (Walker et al., 1987) was identified in the promoters of the maize and Arabidopsis alcohol dehydrogenase genes (ADH1) and in the promoters of other anaerobically induced genes, suggesting that the ARE participates in the coordinated control of those genes in response to low-oxygen stress. This element consists of GC and GT motifs (Olive et al., 1991; Dolferus et al., 1994). The transcription factor AtMYB2 binds to the GT motif and is induced by low-oxygen conditions (Hoeren et al., 1998), suggesting that it may be an important regulatory factor. Some components of the signal transduction pathway leading to low-oxygen-induced gene expression are known (Dolferus et al., 1994), but not all of the steps have been elucidated; the components include changes in cytosolic Ca2+ levels, which play a role as a signal for gene expression under hypoxia (Subbaiah et al., 1994a, 1994b), and the phytohormone ethylene, which has been suggested to play a role in the formation of aerenchyma (Drew et al., 2000).
We used microarray technology (Richmond and Somerville, 2000) to further characterize the anaerobic response using root cultures as the experimental material. Sampling points spread over a 20-h time course detected 210 genes whose expression is affected by low-oxygen stress. We used a package of statistical tools that was developed for the analysis of DNA microarray data. The differentially expressed genes include those previously identified as encoding ANPs. In addition, we found genes encoding transcription factors and signal transduction components. We also found genes involved in metabolic processes not known previously to be involved in the low-oxygen stress response. The differentially expressed genes clustered into six groups according to their expression profiles. Analysis of the 5′ regulatory regions of genes within each cluster revealed common sequence motifs, suggesting that expression of the grouped genes may be regulated by common regulatory factors.
RESULTS
Our experimental results are based on a microarray containing 3500 cDNA clones. A total of 1000 clones of this array were selected randomly from a cDNA library prepared from Arabidopsis hairy root cultures; these root cultures were treated for 4 h under low-oxygen conditions (0.5%) in the presence of cycloheximide (10 μM). The cycloheximide treatment of the roots was used to enrich for mRNA with rapid turnover rates, such as signal transduction components. Previous results showed that cycloheximide treatment, although reducing the anaerobic induction of ADH1, strongly induced the transcription factor AtMYB2 (Hoeren et al., 1998). A total of 2500 sequenced Arabidopsis EST cDNA clones were added to the array. These clones encompass a broad array of developmental and metabolic processes in different organs and at different developmental stages; they were assembled by Schenk et al. (2000) and Ruan et al. (1998).
We used cultured roots as the primary experimental system, having found that the response in this system was similar to the response measured in the roots of normal plants (Figure 1A) (Dolferus et al., 1994). In a microarray experiment, in which the two systems were compared using a specialized array containing 10,000 unidentified clones from the low-oxygen-stressed root library, there was a strong correlation between the results from cultured roots and normal plant roots (correlation coefficient [r] = 0.72). Also, all seven anaerobic genes printed on the microarray slides as controls showed parallel induction (Table 1).
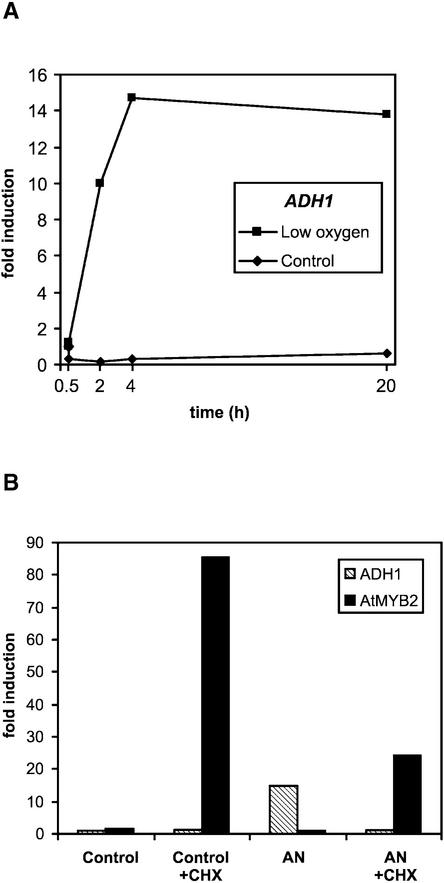
Figure 1.
Gene Expression of ADH1 and AtMYB2 under Low-Oxygen Stress.
(A) ADH1 mRNA profiles in low-oxygen-treated and control Arabidopsis root cultures.
(B) Effect of cycloheximide (CHX) on ADH1 and AtMYB2 mRNA levels in root cultures after 4 h of low-oxygen (5%) treatment (AN), as indicated (10 μM cycloheximide, 2 h before and during low-oxygen treatment). RNA gel blot analyses and quantitation of hybridization signals were performed as described previously (Dolferus et al., 1994). A ubiquitin probe was used to correct for gel-loading differences.
Table 1.
Comparison of Gene Expression in Root Culture and Normal Plant Roots
| Open Reading Frame Identifier |
Function | Plant Roots | Root Culture |
|---|---|---|---|
| At1g77120 | ADH1 | 4.13 | 7.66 |
| At1g77120 | ADH1 | 4.06 | 6.11 |
| At1g17290 | AlaAT1 | 3.85 | 4.53 |
| At1g17290 | AlaAT1 | 4.11 | 4.63 |
| At1g72330 | AlaAT2 | 4.80 | 4.94 |
| At1g72330 | AlaAT2 | 4.86 | 4.16 |
| At3g43190 | ASUS1 | 5.64 | 9.44 |
| At4g01900 | GLB1 | 5.49 | 10.26 |
| At4g01900 | GLB1 | 5.33 | 9.66 |
| At4g17260 | LDH1 | 2.43 | 3.26 |
| At4g17260 | LDH1 | 2.03 | 2.66 |
| At4g33070 | PDC1 | 7.20 | 14.15 |
| At4g33070 | PDC1 | 6.75 | 13.11 |
Shown are the microarray results of seven low-oxygen-induced genes that were printed onefold to twofold onto the slides as controls. Values are ratios (medians from three or four replicate experiments) of low-oxygen-treated compared with aerated roots for 4 h and are log base 2 transformed and normalized (see Methods).
At 0.5, 2, 4, and 20 h of low-oxygen stress (time points based on expression kinetics of Arabidopsis ADH1 and AtMYB2) (Hoeren et al., 1998), gene expression levels were compared with that of the 0-h low-oxygen stress control. At each assay point, we performed three to four microarray hybridizations using cDNA prepared from different samples of root material grown and stress treated under identical conditions (i.e., biological repeats) rather than cDNA samples made from the same RNA (technical repeats).
The fluorescence data derived from the microarray images were normalized using tools for R Microarray Analysis (tRMA [see Methods]; for manual and software, see www.pi.csiro.au/gena/trma). To test the reproducibility of the replicated stress treatments, we calculated the correlations between the data (i.e., the normalized expression ratios of all genes) of the different biological replicates within each time point. We found a strong correlation between the replicates (r > 0.6 for the 0.5-h time point; r > 0.4 for the 2-h time point; r > 0.5 for the 4-h time point; and r > 0.6 for the 20-h time point).
The Low-Oxygen Response Consists of Two Stages
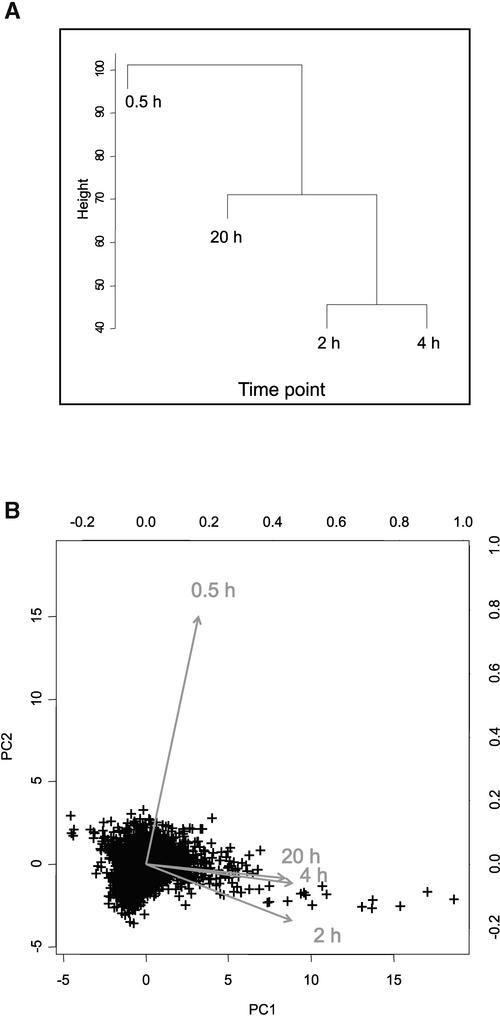
The correlations between the time points were calculated (from the median value of the biological replicates for each of the 3500 clones in each time point); this showed that the 0.5-h time point had a low correlation (r < 0.2) with the later three time points (2, 4, and 20 h). By contrast, the three later time points were highly correlated (r > 0.7). Clustering (Figure 2A) and principal component analyses confirmed that the data from the 0.5-h time point were different from the data from the later time points (Figure 2B) and that the data from the 2-, 4-, and 20-h time points were related. These results indicate that gene expression during low-oxygen treatment can be separated into at least two response stages (the 0.5-h time point as the first stage, and the three later time points combined as the second stage). Different sets of genes were activated or repressed at the two stages.
Figure 2.
The Low-Oxygen Response Consists of Different Stages.
Data from the first time point (0.5 h) are not correlated with data from the other three time points.
(A) Dendrogram of relationships between data from the four time points (averaged for each gene) as determined by cluster analysis.
(B) Principal component analysis (PC1 and PC2) of the data from the four time points (averaged for each gene). The data are presented as a biplot, incorporating the gene effects (scores) as points and the treatments (loadings) as vectors (Gabriel, 1971; Chapman et al., 2002). Vectors that are close together are highly correlated in terms of the gene effects observed for each treatment, whereas vectors that are orthogonal are poorly correlated. Points (genes) that are near the origin of the biplot are either not expressed differentially in all treatments or are explained poorly by the principal component analysis. Points (genes) that are close to the head of a vector have high positive expression values in that treatment, whereas genes on the opposite side of the origin, relative to the head of the vector, have negative expression values for that treatment. The relative expression level of any combination of gene and treatment can be determined by a perpendicular projection of a point onto a vector.
Expression Profiling of Differentially Expressed Genes
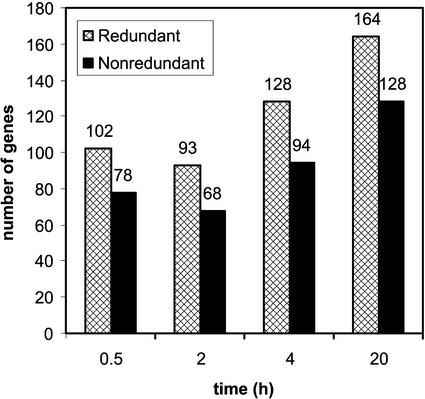
Using tRMA, genes expressed differentially at any of the four time points were identified among the total population, which numbered 274 clones (8% of the total number of clones; see Methods for criteria used to select differentially expressed genes). These 274 clones represent a nonredundant set of 210 genes, of which 78, 68, 94, and 128 genes were expressed at the 0.5-, 2-, 4-, and 20-h time points, respectively (Figure 3). In the list of differentially expressed genes, there was a bias toward the low-oxygen library; 13% of the 1000 clones from the low-oxygen library were expressed differentially compared with 6% of the set of 2500 genes on the 3500-gene array. This implies that a specific subset of genes is activated under low-oxygen stress.
Figure 3.
Differential Gene Expression under Low-Oxygen Stress.
The total number of differentially expressed genes in both the redundant and nonredundant sets in each time point of the low-oxygen time course is shown. Note that genes can be expressed differentially in more than one time point.
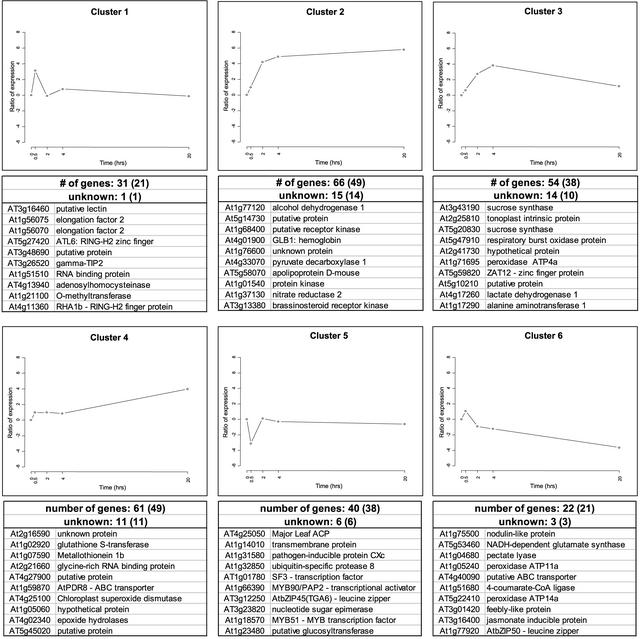
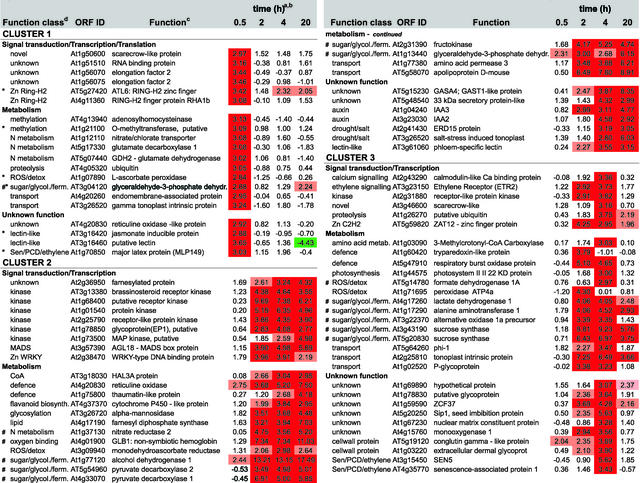
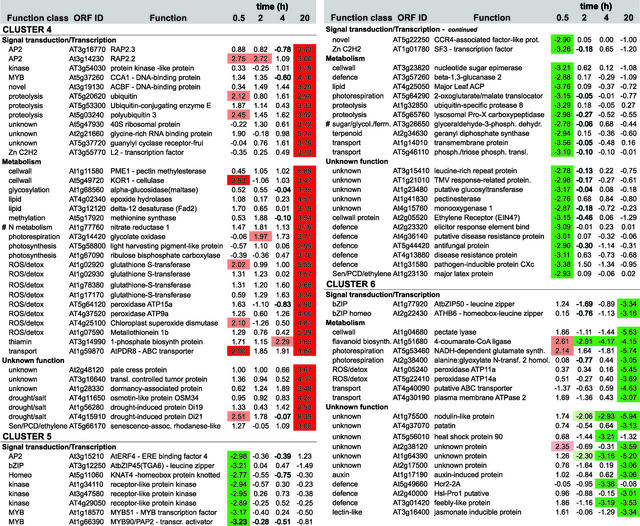
We were able to cluster the 210 differentially expressed genes into six groups according to their expression profiles during the time course of the anaerobic treatment (Figures 4 and 5; a comprehensive version of Figure 5 is available with the online version of this article). Cluster 1 showed a rapid increase of gene expression from 0 to 0.5 h of low-oxygen stress, with reduced expression during the remainder of the time period. Transcription factors, but also transporters, metabolic enzymes, and two unknown proteins, were identified in this cluster.
Figure 4.
Expression Profiles of Genes Expressed Differentially under Low-Oxygen Stress.
The graphs represent the means of expression for each cluster (see Figure 5). The y axis is in log base 2 units. The total number of genes and the number of unknown genes is indicated, with the number of nonredundant genes shown in parentheses. For each expression profile, the 10 most highly induced or repressed genes are listed with both open reading frame identifiers and functions. The complete list of differentially expressed genes is available in the supplemental data online.
Figure 5.
Expression Profiling and Functional Clustering of Genes Differentially Expressed by Low-Oxygen Stress.
Genes with differential expression pattern under low oxygen conditions are grouped in 6 clusters according to their induction profile. Values in the table are ratios of low-oxygen treated compared to aerated roots for the time given, and are transformed (log base 2, so ratio = 2n) and normalised. Positive values indicate induction, whereas negative values indicate repression. High induction is shaded in red, high repression in green; levels of differential induction are indicated by pale red (1.96>n>2.75) and dark red (n>2.75), and the same applies for the levels of repression as indicated by the green colours. Clones from unknown, putative, or hypothetical proteins are not shown, nor are redundant clones. The complete list, including unknown and redundant clones, Genbank accession numbers, clone IDs, gene descriptions, and literature references can be found via the online version of this publication. Known ANPs (#) and transition proteins (*) are indicated.
Clusters 2, 3, and 4 showed increased expression during the later three time points (i.e., after 2 or 4 h [clusters 2 and 3] or after 20 h [cluster 4]). The initial expression profile of cluster 3 was similar to that of cluster 2, but expression levels were decreased at later stages of low-oxygen stress. These three clusters contained a number of genes encoding ANPs, including alcohol dehydrogenase (ADH1), Suc, synthase (ASUS1), and pyruvate decarboxylase (PDC1; labeled # in Figure 5). These genes are activated maximally after 2 to 4 h of anaerobiosis (Dolferus et al., 1994). We found 35 unknown genes showing an increase in gene activity similar to the ANP-encoding genes found in profiles 2, 3, and 4. Cluster 5 contained genes that showed a strong decrease in expression during early anaerobiosis (i.e., 0.5 h), and genes in cluster 6 exhibited a slight increase at the 0.5-h time point, with decreasing gene activity at the three later time points.
Verification of Differentially Expressed Genes
The differential expression of the 210 genes was validated in two different ways. A set of control clones printed among the ∼3500 clones was derived from genes known to increase their expression under low-oxygen stress (e.g., Ala aminotransferase [AlaAT1], ADH1, PDC1, PDC2, lactate dehydrogenase [LDH1], globin1 [GLB1], and AtMYB2). Our results (Figure 5) confirm that each of these genes was induced under low-oxygen stress. AtMYB2 was induced, but its expression ratio remained just below the cutoff, which was estimated to be ∼2.7; hence, it is not represented in Figure 5 (note that the cutoff value was computed from rescaled and normalized log base 2 data; see Methods). The ADH1 control gene displayed a gene expression profile similar to that shown previously (Dolferus et al., 1994). Multiple copies of ADH1 and various other control genes were present on our array, and all were expressed differentially, as expected.
We also used real-time PCR to verify the differential expression of genes. In total, 17 genes (7% of the differentially expressed genes) were selected from each of the expression profiles and functional categories. We chose several hypothetical and unknown genes, based on their expression profiles being similar to that of ADH1 (Figure 5). Expression of these 17 genes was tested in each case using the same cDNA samples that were used for the microarray hybridization experiment. Table 2 shows that the real-time PCR data were similar to the microarray data: in most cases, genes that showed high expression in the microarray experiment showed high expression in the real-time PCR experiment. In a few cases (Zat12, ADH1, unknown protein At3g11930, and pectin methylesterase), the expression ratios were different, although both approaches indicated strong gene induction. The differences might be explained by cross-hybridization among related genes that can occur in the microarray approach but not in real-time PCR. BLAST searches indicated the existence of related genes in some cases.
Table 2.
Confirmation of Microarray Data by Real-Time PCR
| Time Point | cDNA Templates | Open Reading Frame Identifier | Function | Microarray | Real-Time PCR |
|---|---|---|---|---|---|
| 0.5 h | At1g59870 | ABC transporter | 2.09 | 2.14 | |
| At1g21100 | Similarity to O-methyltransferase1 | 1.39 | 1.50 | ||
| At5g10210 | Putative protein | 2.41 | 7.07 | ||
| 2 h | 1 | At2g41730 | Hypothetical protein | 2.01 | 5.20 |
| 2 | At2g41730 | Hypothetical protein | 6.56 | 3.10 | |
| At5g59820 | Zinc finger protein Zat12 | 3.56 | 11.30 | ||
| At4g15760 | Hypothetical protein | 2.72 | 3.47 | ||
| 4 h | At4g01900 | Nonsymbiotic hemoglobin (GLB1) | 3.34 | 3.90 | |
| 1 | At5g48540 | 33-kD secretory protein-like | 2.82 | 3.76 | |
| 2 | At5g48540 | 33-kD secretory protein-like | 2.09 | 2.28 | |
| 1 | At3g46600 | Hypothetical protein | 1.96 | 2.05 | |
| 2 | At3g46600 | Hypothetical protein | 1.48 | 3.16 | |
| At1g77120 | Alcohol dehydrogenase | 13.08 | 4.60 | ||
| 1 | At1g76600 | Unknown protein | 2.86 | 2.98 | |
| 2 | At1g76600 | Unknown protein | 6.25 | 6.67 | |
| 3 | At1g76600 | Unknown protein | 2.66 | 2.57 | |
| 20 h | 1 | At3g11930 | Unknown protein | 2.08 | 2.66 |
| 2 | At3g11930 | Unknown protein | 4.34 | 21.17 | |
| At2g41730 | Hypothetical protein | 7.25 | 5.01 | ||
| 1 | At1g11580 | Pectin methylesterase | 3.69 | 12.31 | |
| 2 | At1g11580 | Pectin methylesterase | 2.59 | 2.46 | |
| At4g17190 | Farnesyl diphosphate synthase2 | 3.51 | 3.49 | ||
| At4g20830 | Reticuline oxidase | 5.27 | 3.64 |
Differentially expressed genes were chosen across the four time points and across functional categories. Some genes were tested with two or three cDNA templates, as indicated. Nontransformed ratios are shown for both the microarray and real-time PCR approaches.
Signal Transduction Components Involved in Low-Oxygen Gene Expression
The high sensitivity of microarray analysis allowed us to identify families of transcription factors and signal transduction components that were affected by low-oxygen stress (Figures 4 and 5). These factors were distributed among the different expression profiles, indicating that they may be involved in the control of different processes throughout the low-oxygen response. Two C2H2 zinc finger factors that contain a membrane-spanning domain (ATL6 and RHA1b) were induced after 0.5 h, a WRKY-type factor and ZAT12 were induced after 2 h, and the AP2-domain factors RAP2.2 and RAP2.3 were induced after 20 h. AtERF4 was reduced after 0.5 h, and AtbZIP50 was reduced after 20 h. Associated with the changes in transcription factors were changes in the expression of other signal transduction pathway genes. One Ser/Thr kinase, a putative mitogen-activated protein kinase, and the ethylene receptor ETR2 were among the factors that were induced in the early stage of the response. On the other hand, expression of a number of protein kinases was reduced in the early stage and increased in the later stage of the response.
The Low-Oxygen Response Consists of a Complex Set of Metabolic Adaptations
The 3500-gene microarray identified many of the metabolic genes that had been characterized previously as ANPs (Figure 5). In addition, the expression of many other metabolic genes was affected by low-oxygen stress (Figure 5). One predominant class of genes is involved in the detoxification of reactive oxygen species (peroxidase, ascorbate peroxidase, monodehydroascorbate reductase, glutathione reductase, and superoxide dismutase); other genes are associated with cell wall biosynthesis, flavonoid biosynthesis, or defense-related processes. A number of these genes also are induced by other biotic and abiotic stress responses (Borsani et al., 2001).
Expression was increased in genes involved in nitrogen metabolism (e.g., glutamate dehydrogenase, glutamate decarboxylase, and nitrate reductase) and genes involved in photorespiration (e.g., peroxisomal glycolate oxidase and 2-oxoglutarate/malate translocator) (Figure 5). These pathways provide substrates for lipid biosynthesis (Grace and Logan, 2000). A set of genes involved directly in lipid metabolism also is induced by low-oxygen stress. We also detected genes involved in methyl group metabolism (Met synthase and O-methyltransferases), ethylene signaling (ETR2), and senescence and in programmed cell death responses, ubiquitination, protein glycosylation, and transport (Figure 5).
Clustered Genes Share 5′ Motifs
Clustered genes might share common regulatory elements. To find DNA motifs co 5mmon to the 5′ regions of clustered genes, we retrieved up to 2000 bp immediately upstream of the ATG of each differentially expressed gene. The resulting upstream fragments were analyzed for overrepresented 6- to 10-bp motifs (see Methods).
Cluster 2 contains the ADH1 gene, which has been well characterized at the promoter level (Dolferus et al., 1994; de Bruxelles et al., 1996). We subjected a subset of 22 genes from cluster 2 with expression profiles similar to that of ADH1 to a search for shared 5′ motifs. Common motifs found in this cluster often could be matched with known regulatory elements of the ADH1 gene, as shown in Table 3. Apart from two motifs that also are present in cluster 3 (see below), the motifs in cluster 2 were not present in any other cluster, confirming the specificity of these motifs for this set of genes (Table 4).
Table 3.
Motif Recognition in 5′ Regions
| −45 | GC (ARE) −146 |
GT (ARE) −155 |
Footprint −195 |
Gbox-1 −216 |
ARE −360 |
|
|---|---|---|---|---|---|---|
| ADH1 motif | CAATTACC | GCCCCTAG | GCAAAACC | GCCAAG | CCACGTGGAC | CCGAAACC |
| Cluster consensus | CmCTTnCC | GCCCATTG | GCAAAACC | GCCAAG | nCACGTGGCC | CCGAmACn |
Shown are motifs found in cluster 2 that are similar to regulatory elements identified in the ADH1 promoter. Motifs are ordered according to their positions in the ADH1 promoter. The ADH1 motif sequences are displayed, as is the consensus sequence for each motif.
Table 4.
Presence and Position of Motifs in Genes of Cluster 2
| Motifs and Their Positions
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Open Reading Frame Identifier |
Function | GC (ARE) | GT (ARE) | Footprint | Gbox-1 | ARE | Length of DNA Investigated |
|
| At1g77120 | Alcohol dehydrogenase | −45 | −146† | −155/−256† | −195† | −218† | −362† | 1045 |
| At3g13380 | Brassinosteroid receptor kinase | −1722 | 2003 | |||||
| At4g37370 | Cytochrome P450-like | −260† | 365 | |||||
| At1g68400 | Putative receptor kinase | −949 | 1349 | |||||
| At2g38470 | WRKY-type DNA-binding protein | −508 | −1821† | 2003 | ||||
| At3g26720 | α-mannosidase, putative | −82* | −66 | −1833 | −1541 | 1979 | ||
| At5g58070 | Outer membrane lipoprotein-like | −159† | −253† | −415* | −143† | −152* | 632 | |
| At1g78850 | Hypothetical protein | −1199* | 2003 | |||||
| At3g18030 | HAL3A | −426* | −537* | −46* | 758 | |||
| At2g41730 | Hypothetical protein | 2003 | ||||||
| At1g05060 | Hypothetical protein | −386† | 2003 | |||||
| At3g11930 | Unknown protein | −317 | −451† | −243† | −228 | −92† | −447* | 1456 |
| At1g37130 | Nitrate reductase | −10* | −367 | −100 | 2003 | |||
| At2g15890 | Unknown protein | −563 | 2003 | |||||
| At3g61060 | Putative protein | −448 | −385† | −143* | −1266* | −636 | −342† | 2003 |
| At1g01540 | Hypothetical protein | −327* | −102* | −893 | −279* | 2003 | ||
| At5g63790 | Putative protein | −261 | −197† | 2003 | ||||
| At5g54960 | Pyruvate decarboxylase | −240 | −406* | −379† | 593 | |||
| At2g25790 | Receptor-like protein kinase | −315 | −25† | −1830 | −400 | 2003 | ||
| At5g15230 | GASA4 | −17 | −270 | −296† | −233 | 2003 | ||
| At1g76600 | Unknown protein | −1073 | 2003 | |||||
| At5g20830 | Suc-UDP glucosyltransferase | −137† | −243 | −487 | 500 | |||
The position for each motif in the ADH1 promoter is shown in the first line. For each gene, the position of the respective motif is shown. A probability score (homology with the consensus sequence, taking the length of the sequence into account) for each motif is indicated: >90% (
and >96% (
. The positions indicated are 5′ of the ATG translation start codon, except for the positions of the ADH1 motifs, which are from the transcription start site (+1; the ADH1 ATG is at position +61).
The GC and GT motifs present in the Arabidopsis ADH1 ARE between positions −142 and −158 (Walker et al., 1987; Dolferus et al., 1994) were found in many genes across the cluster. Although they are adjacent in the ADH1 promoter, in other genes the distance between the two motifs varies from 26 to 240 bp (Table 4). An upstream sequence that has homology with the ARE of the ADH1 promoter (position −360) (Ferl and Laughner, 1989; Dolferus et al., 1994) also is present in many members of the cluster. A G-box-1–like motif (position −218; see Introduction) was found in a substantial number of genes in the cluster. Another motif, referred to as −195 in Table 4, coincides with an area of the ADH1 promoter identified previously by DNA footprinting (Ferl and Laughner, 1989), although deletion analysis did not reveal this site to be functionally important (Dolferus et al., 1994). The −45 motif in the ADH1 promoter (Table 4) has not been found experimentally to be involved in the regulation of the gene; however, the fact that this motif is present in many of the genes within the cluster indicates that this sequence does have a regulatory role.
We also found motifs specific to the other clusters (Table 5), but well-studied promoter sequences that could serve as reference motifs were not available. Some of the motifs found in these clusters resemble binding sites of known transcription factors (Table 5). Because the expression profiles of cluster 2 and 3 differ only slightly, we anticipated that some motifs would be present in both clusters; we identified two motifs present in both cluster 2 and cluster 3 (Table 5).
Table 5.
Shared Motifs in Other Clusters
| Motif | No. | Similar to |
|---|---|---|
| Cluster 1 | (18) | |
| CTCTCTCT | 13 | |
| GGwATGAC | 6 | AP1 |
| CCAAAAAm | 14 | |
| GTwTGAC | 7 | AP1 |
| TCTTACC | 13 | |
| Cluster 3 | (29) | |
| CGTCACAy | 13 | TCF11 |
| *CyTCwCTC | 20 | |
| *TyCTCTs | 16 | |
| AGCTTTT | 14 | DOF/PBF |
| ACCTTAC | 10 | deltaEF1 |
| Cluster 4 | (40) | |
| CTTTyTCT | 26 | DOF/PBF |
| yTCAGCT | 19 | AP-4 |
| TATCTTC | 21 | NIT2 |
| CTyTCTC | 32 | |
| Cluster 5 | (28) | |
| AAAAAGAT | 15 | DOF/PBF |
| mGCGTGyG | 12 | AhR/Arnt |
| TAACGnnC | 12 | GAMYB |
| CTCTksC | 21 | |
| Cluster 6 | (16) | |
| CTTCTTCC | 6 | |
| yCCTyCnC | 12 | |
| CAATmAAA | 12 | |
| ATmyATA | 14 |
For each gene, 500 bp immediately upstream from the ATG was investigated. The number of genes within each cluster containing a motif is indicated; the total number of genes is shown in parentheses. A selection of motifs from each cluster is shown: motifs present in at least 60% of the genes within a cluster, motifs with a high-probability score, and motifs resembling known transcription factor binding sites (sequences shown in boldface). Some motifs in cluster 3 also are present in cluster 2 (
.
DISCUSSION
The aim of this study was to expand our understanding of the plant's response to low-oxygen stress and to identify key regulatory genes that might be used to manipulate the stress response to improve the agronomic performance of crop plants. We used microarray technology, and to optimize this, we used a number of strategies. The quality of the printed DNA was verified by gel electrophoresis. We used biological replication of the treatments to minimize both biological and technical artifacts. We developed a set of statistical tools (tRMA) to assist in the analysis of microarray data. Our microarray results were confirmed through the use of known low-oxygen-responsive genes and by real-time PCR experiments.
One of the driving forces for the design of the tRMA statistics was the need for accurate normalization of microarray data. Normalization is required to adjust for inequalities in the amounts of RNA used for cDNA preparation and to remove possible nonlinear bias in fluorescence as a result of differences in cDNA labeling or in the stability of the fluorescent dyes. Linear normalization (i.e., across the slide) is unreliable because of spatial fluorescence–based biases. Hence, a normalization method was developed that corrects for such biases (D.L. Wilson, M.J. Buckley, C.A. Helliwell, and I.W. Wilson, unpublished data; see Methods for World Wide Web access to software and manual). Another driving force for developing tRMA was the need for empirically based statistical tools, rather than an arbitrary cutoff, to determine which genes are expressed differentially.
Microarrays Confirm Our Current Knowledge of the Low-Oxygen Stress Response
Early data in maize showed that the low-oxygen stress response is evident after 0.5 h. Many proteins disappear from two-dimensional gel patterns in the first hour of low-oxygen stress (Sachs et al., 1980). A similar observation was made in Arabidopsis (Dolferus et al., 1985). We found reduced expression for a relatively large number of genes; however, the change at the transcriptional level seems to be less than the massive disappearance of “aerobic proteins” reported by Sachs et al. (1980) and Dolferus et al. (1985). This could mean that the steady state level of many aerobic mRNAs does not change substantially during low-oxygen treatment and that anaerobic mRNAs are translated preferentially. It has been suggested previously that anaerobic treatment of maize seedlings disrupts polysomes (Bailey-Serres and Freeling, 1990) and leads to a redirection of protein synthesis (Sachs et al., 1980; Russell and Sachs, 1991), which may involve changes in ribosomal proteins and elongation factors (Webster et al., 1991; Perez-Mendez et al., 1993; Manjunath et al., 1999). The strong induction of an RNA binding protein and of elongation factor 2 in our experiments (Figure 5) supports this possibility.
Sachs et al. (1980) detected a group of small (∼33-kD) “transition proteins” induced after 1.5 h of low oxygen. In our experiments, we found seven genes induced after 0.5 h that, according to the length of their coding sequences, would produce 26- to 40-kD proteins (these genes are labeled with asterisks in Figure 5). Three of these genes produce a 33-kD protein: a putative protein (At3g48690), a jasmonate-inducible protein, and a major latex protein (MLP149). Another gene of similar molecular mass codes for a RING-H2 zinc finger factor, which might be involved in ubiquitination (Potuschak et al., 1998; for review, see Tyers and Jorgensen, 2000) and could be implicated in the targeted degradation of aerobic proteins. Functional analysis using sense and antisense transgenic plants will be needed to establish any role of these possible transition proteins.
After the first time point in the low-oxygen response (0.5 h), a different set of genes was induced. After 2 to 4 h of low-oxygen stress, we detected a large increase in the expression level of many ANP-encoding genes. This finding corresponds to the massive induction of ANP-encoding genes described previously (Dennis et al., 2000). At this time point, a number of metabolic changes had occurred, and these persisted for the duration of the analysis. Some genes that encode ANPs were present in the set of 2500 known genes but were not induced under our conditions (e.g., α-amylase and enolase), whereas genes that encode other ANPs, such as Fru bis-phosphate aldolase and xyloglucan endotransglycosylase, were induced weakly and did not meet the cutoff threshold. Note that these genes have been characterized as ANP-encoding genes under different experimental conditions and in different plant species and may not be induced under our conditions. It also is possible that different members of gene families that are induced under different conditions in vivo cross-hybridized in our experiment and caused discrepancies.
Microarrays Show a Range of Genes and Processes Involved in the Low-Oxygen Stress Response
The microarray approach has enabled us to identify a set of transcription factors and signal transduction components that could play a role in the regulation of the anaerobic response. The fact that these factors were induced at different stages of the response suggests that different regulatory events occur during the time course of the response. The use of cycloheximide in the library preparation may have led us to print more signal transduction cDNAs on our array, resulting in their identification.
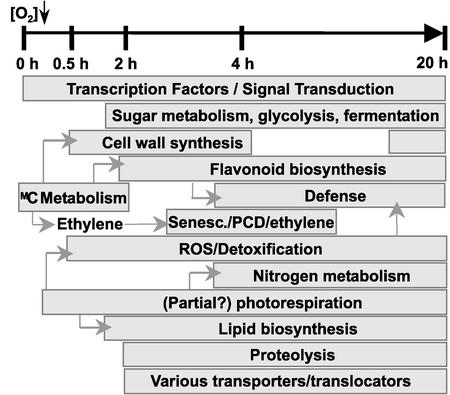
Apart from these regulatory genes and the known ANP-encoding genes, many other genes that encode proteins involved in metabolic processes appear to play a role under low-oxygen stress (Figure 6). Three photorespiratory enzymes are affected by low-oxygen stress (a putative peroxisomal glycolate oxidase is induced at 20 h, and 2-oxoglutarate/malate translocator and Ala:glyoxylate aminotransferase are repressed), and the photorespiratory pathway plays an important role in nitrogen metabolism (Douce and Neuburger, 1999; Wingler et al., 2000). The induction of nitrate reductase and NADH-dependent glutamate synthase instead of the enzymes of the glutamate synthase cycle suggests that a shift occurs in nitrogen metabolism during low-oxygen stress. This also is indicated by the induction of glutamate decarboxylase, which converts glutamate to 4-aminobutyric acid and plays a role in the determination of cytosolic pH (Shelp et al., 1999). The induction of glutamate decarboxylase and glutamate dehydrogenase raises the possibility that the glutamate and 2-oxoglutarate generated may be converted to Ala via Ala aminotransferase, explaining the accumulation of Ala under low-oxygen conditions (Vanlerberghe et al., 1990; Muench and Good, 1994). The production of glyoxylate by glycolate oxidase also establishes a link with lipid biosynthesis (Figure 6). Several lipid biosynthesis genes, as well as the HAL3A protein, which plays a role in acetyl-CoA biosynthesis, were induced by low-oxygen stress.
Figure 6.
Scheme of the Different Biochemical Processes Induced under Low-Oxygen Stress during the 20-h Treatment Period.
PCD, programmed cell death; ROS, reactive oxygen species.
The induction of Met synthase and O-methyltransferases indicates the importance of methylation in the response to low oxygen. This is consistent with the induction of processes that involve methylation as a modification of compounds to accomplish activation or intracellular translocation: flavonoid biosynthesis, cell wall biosynthesis, and defense mechanisms (Figure 6) (Ibrahim et al., 1998; Grace and Logan, 2000). Met also is a precursor in ethylene biosynthesis. We observed the induction of senescence-related genes later in the response (Figure 6). The ethylene receptor ETR2 is induced between 2 and 4 h of low-oxygen stress, and the negative regulator EIN4 is repressed (Hua and Meyerowitz, 1998).
Many genes that we found to be induced by low-oxygen stress also are affected by other stresses, suggesting an overlap in function between low-oxygen stress and other biotic and abiotic stress responses (Chen et al., 2002). The induction of genes involved in free radical scavenging and detoxification of reactive oxygen species (Douce and Neuburger, 1999), peroxidases, and superoxide dismutase is common to a number of abiotic stresses (Reymond et al., 2000; Schenk et al., 2000; Seki et al., 2001).
Gene Expression Profiles and 5′ Motifs
A major objective in the clustering of gene expression profiles is to assist in the identification of unknown genes. Genes grouping in the same expression cluster are expected to be part of the same functional category or biological process. We found 45 genes with unknown functions in our nonredundant set of 210 genes, with the majority (35 genes) showing an increase in gene activity concomitant with ANP-encoding genes (2 to 4 h).
The transcriptional control of a gene is a combinatorial effect of a number of regulatory factors (Singer et al., 2001). Coordinated regulation of groups of genes might occur through the actions of similar regulatory factors. Gene expression profiling enables us to identify common promoter elements, and a well-characterized promoter can serve as a reference. Using the cluster that contains the ADH1 gene, we found that functional regulatory elements of the ADH1 promoter also are present in the 5′ regions of other, often unknown, genes in the same cluster (Table 4). The similarity of expression profiles and the presence of similar 5′ motifs strongly indicate regulation by the same set of transcription factors. The presence of such motifs within gene clusters will assist us in further analysis of the regulatory events involved in the low-oxygen response.
With our microarray experiments, we confirmed existing data and gained a more comprehensive understanding of the low-oxygen stress response. We identified low-oxygen-induced genes, many of them new, and possible DNA sequence elements that may coordinately regulate members of gene clusters. The coupling of microarray data with functional analysis of candidate genes will lead to a more comprehensive understanding of the molecular basis of a plant's response to low oxygen.
METHODS
Preparation of the Microarray Slides
A cDNA library (λZipLox; Life Technologies, Rockville, MD) was prepared using poly(A)+ mRNA isolated from Arabidopsis thaliana hairy root cultures (ecotype C24) induced with Agrobacterium rhizogenes (Shiao et al., 2002). The roots were treated with low oxygen for 4 h in liquid Murashige and Skoog (1962) medium and incubated in a mixture of 5% O2 and 95% N2 (Howard et al., 1987). The root cultures were treated with 10 μM cycloheximide for 2 h before and during the low-oxygen treatment. From this library, 1000 cDNA clones were chosen randomly, and inserts were amplified by PCR directly from bacterial cultures (2 μL of overnight culture per reaction) using primers with a Tm of 72°C (5′-GCCGCCGACTAGTGAGCTCGTCGACCC-GGG-3′ and 5′-GGGAAAGCTGGTACGCCTGCAGGTACCGGT-CCG-3′) and a two-step procedure (35 cycles of 30 s at 94°C and 150 s at 72°C). Also, PCR products were acquired from 2500 sequenced genes known to be involved in developmental and metabolic processes (see Schenk et al., 2000, for the complete list of these genes). The PCR products from both sets of clones were precipitated in ethanol, resuspended in 5 μL of 3 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), and transferred from 96-well to 384-well microtiter plates. PCR fragments were printed onto silylated microscope slides (CEL Associates, Houston, TX) using an Omnigrid Microarrayer (Genemachines, San Carlos, CA) with ChipMaker2 quill pins (TeleChem International, Sunnyvale, CA). Before hybridization, the slides were processed according to Schena et al. (1996).
Several control steps were included to guarantee reproducible results. The quality of PCR products was determined by gel electrophoresis after both PCR and ethanol precipitation. As controls, PCR products from well-characterized genes known to be involved in low-oxygen stress were printed twice randomly across the array.
Slide Hybridization
To minimize experimental artifacts, the procedure described here was performed three to four times for each time point using different lots of plant root material grown and stress treated under identical conditions (biological repeats). Cultured Arabidopsis hairy roots were treated with low oxygen (5% O2 and 95% N2) (Howard et al., 1987) in the dark for the indicated period of time. Control roots were treated similarly, except that they were kept aerobic. RNA was isolated as described (Dolferus et al., 1994). From this RNA, Cy3- and Cy5-labeled (Amersham Pharmacia) cDNA probes were generated using the two-step labeling method described by Schenk et al. (2000). Application of the probe to the microarray slide, hybridization, and subsequent washes of the slide were performed according to Schenk et al. (2000). Slides were scanned with a GenePix 4000A microarray scanner (Axon Instruments, Union, CA), and spots were analyzed using GenePix Pro 3 software. Spots that were poorly segmented by the GenePix Pro software were either adjusted manually or discarded to ensure that only high-quality microarray data were obtained. Among the replicate experiments within each time point, the Cy3 and Cy5 labels were swapped between sample and control DNA to minimize any possible impact of inequalities in DNA incorporation and photobleaching of the fluorescent dyes.
Normalization of Data and Calculation of Median Values
Normalization of the microarray data was performed using a new statistical microarray analysis package, tRMA (tools for R Microarray Analysis; D.L. Wilson, M.J. Buckley, C.A. Helliwell, and I.W. Wilson, unpublished data), a suite of statistical functions written in R code (Ihaka and Gentleman, 1996; for review, see Ellner, 2001). The R software package is freely available (http://www.r-project.org/). A detailed description and manual of tRMA is available online (www.pi.csiro.au/gena/trma). Normalization was performed to allow for differences in the amounts of RNA used for preparation of the cDNA samples. It also removed possible biases in fluorescence as a result of differences in label incorporation during cDNA preparation and in the stability of the dyes. Also, tRMA allows for spatial fluorescence–based biases through a spatial normalization algorithm.
The data were normalized using the “pin-normalization” protocol from the tRMA package developed by Yang et al. (2001). Median values were calculated for each gene from the different replicate experiments within each time point.
Correlation of Data
To calculate the correlation between the four time points, median values for each gene within a time point were ordered into a table (matrix) in which the four time points were represented as columns and the genes were represented as rows (genes from which no average ratio could be calculated were excluded). This table was used for the calculation of correlations between the individual experiments and cluster/principal component analysis (Chapman et al., 2002) using additional R and S-Plus code (Insightful, Seattle, WA). For a detailed description of the methodology, see the supplemental data online.
Detection of Differentially Expressed Genes
From the median values for each gene in each time point and using the relevant function in tRMA, the differentially expressed genes were extracted from the complete list of ∼3500 clones. More specifically, detection of the differentially expressed genes was computed by selecting genes that were considered “outliers” in a standard Gaussian distribution. Under this assumption, a ratio cutoff threshold was computed empirically from the normalized data and estimated to be ∼2.7 (note that this value was computed from rescaled and normalized log base 2 data).
Real-Time Quantitative Reverse Transcriptase–Mediated PCR
For the cDNA clones to be analyzed, gene-specific oligonucleotides were prepared. These primers had a Tm of >55°C and were designed to produce a PCR product of 180 to 230 bp. Amplification mixtures (20 μL per reaction) consisted of 1 × Taq buffer (Gibco BRL), 3.5 mM MgCl2, 0.2 mM deoxynucleotide triphosphate, 16 pmol of each primer, 2.5 × SYBR Green I (Molecular Probes, Eugene, OR), 0.8 units of Platinum Taq DNA polymerase (Gibco BRL), and cDNA corresponding to 25 ng of total RNA. Reactions were run on a Rotor-Gene 2000 Real-Time Cycler (Corbett Research, Sydney, Australia). Cycling conditions were as follows: 5 min at 94°C, 40 cycles of 15 s at 94°C, 15 s at 53°C, and 20 s at 72°C, 300 s at 40°C, and 60 s at 55°C. This was followed by a melting-curve program (55 to 99°C, with a 5-s hold at each temperature). Fluorescence data were acquired at the 72°C step and during the melting-curve program. An 18S rRNA cDNA clone was used as a template to produce a standard curve. 18S rRNA and ribulose bisphosphate (At1g67090) cDNA clones were used in control reactions to correct for uneven amounts of sample and control cDNA templates.
Motif Searches
The 5′ regions of the clustered genes were retrieved by performing BLASTN queries of the respective cDNA clones against the complete Arabidopsis genome sequence. A detailed description of these methods can be found in the supplemental data online. The sequences of the 5′ regions (up to 2000 bp) were used to obtain shared motifs by finding common short sequences (6 to 8 bp) that are overrepresented in the 5′ regions within a gene cluster compared with all genes outside of the cluster. These motif search algorithms, which are based on stochastic optimization procedures, were performed using the Motif Sampler algorithms, which can be accessed through the PlantCARE database World Wide Web site (http://sphinx.rug.ac.be:8080/PlantCARE/cgi/index.html; see Lawrence et al., 1993).
We used MatInspector to detect consensus matches for known transcription factor binding sites in the motifs we found (http://transfac.gbf.de/programs/matinspector/matinspector.html; Quandt et al., 1995). MatInspector uses information about known transcription factor binding sites as present in the TRANSFAC database (http://www.gene-regulation.de; Hehl and Wingender, 2001).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Supplementary Material
Acknowledgments
We thank Dr. Russell Jones for critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.004747.
Footnotes
Online version contains Web-only data.
References
- Armstrong, W. (1979). Aeration in higher plants. Adv. Bot. Res. 7, 225–232. [Google Scholar]
- Bailey-Serres, J., and Freeling, M. (1990). Hypoxic stress-induced changes in ribosomes of maize seedling roots. Plant Physiol. 94, 1237–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani, O., Cuartero, J., Fernandez, J.A., Valpuesta, V., and Botella, M.A. (2001). Identification of two loci in tomato reveals distinct mechanisms for salt tolerance. Plant Cell 13, 873–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, W.W., Huang, L., Shen, M., Webster, C., Burlingame, A.L., and Roberts, J.K. (2000). Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low-oxygen environment, and identification of proteins by mass spectrometry. Plant Physiol. 122, 295–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, S.C., Schenk, P., Kazan, K., and Manners, J.M. (2002). Using biplots to interpret gene expression patterns in plants. Bioinformatics 18, 202–204. [DOI] [PubMed] [Google Scholar]
- Chen, W., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruxelles, G.L., Peacock, W.J., Dennis, E.S., and Dolferus, R. (1996). Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiol. 111, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, E.S., Dolferus, R., Ellis, M., Rahman, M., Wu, Y., Hoeren, F.U., Grover, A., Ismond, K.P., Good, A.G., and Peacock, W.J. (2000). Molecular strategies for improving waterlogging tolerance in plants. J. Exp. Bot. 51, 89–97. [PubMed] [Google Scholar]
- Dolferus, R., Jacobs, M., Peacock, W.J., and Dennis, E.S. (1994). Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol. 105, 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus, R., Marbaix, G., and Jacobs, M. (1985). Alcohol dehydrogenase in Arabidopsis: Analysis of the induction phenomenon in plantlets and tissue cultures. Mol. Gen. Genet. 199, 256–264. [Google Scholar]
- Douce, R., and Neuburger, M. (1999). Biochemical dissection of photorespiration. Curr. Opin. Plant Biol. 2, 214–222. [DOI] [PubMed] [Google Scholar]
- Drew, M.C., He, I., and Morgan, P.W. (2000). Programmed cell death and aerenchyma formation in roots. Trends Plant Sci. 5, 123–127. [DOI] [PubMed] [Google Scholar]
- Ellner, S.P. (2001). Review of R, version 1.1.1. Bull. Ecol. Soc. Am. 82, 127–128. [Google Scholar]
- Fennoy, S.L., and Bailey-Serres, J. (1995). Post-transcriptional regulation of gene expression in oxygen-deprived roots of maize. Plant J. 7, 287–295. [DOI] [PubMed] [Google Scholar]
- Ferl, R.J., and Laughner, B.H. (1989). In vivo detection of regulatory factor binding sites of Arabidopsis thaliana ADH. Plant Mol. Biol. 12, 357–366. [DOI] [PubMed] [Google Scholar]
- Gabriel, K.R. (1971). The biplot graphic display of matrices with application to principal component analysis. Biometrika 58, 453–467. [Google Scholar]
- Grace, S.C., and Logan, B.A. (2000). Energy dissipation and radical scavenging by the plant phenylpropanoid pathway. Phil. Trans. R. Soc. Lond. 355, 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehl, R., and Wingender, E. (2001). Database-assisted promoter analysis. Trends Plant Sci. 6, 251–255. [DOI] [PubMed] [Google Scholar]
- Hoeren, F., Dolferus, R., Wu, Y., Peacock, W.J., and Dennis, E.S. (1998). Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase (ADH1) gene by low oxygen. Genetics 149, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, E.A., Walker, J.C., Dennis, E.S., and Peacock, W.J. (1987). Regulated expression of an alcohol dehydrogenase-1 chimeric gene introduced into maize protoplasts. Planta 170, 535–540. [DOI] [PubMed] [Google Scholar]
- Hua, J., and Meyerowitz, E.M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94, 261–271. [DOI] [PubMed] [Google Scholar]
- Ibrahim, R.K., Bruneau, A., and Bantignies, B. (1998). Plant O-methyltransferases: Molecular analysis, common signature and classification. Plant Mol. Biol. 36, 1–10. [DOI] [PubMed] [Google Scholar]
- Ihaka, R., and Gentleman, R. (1996). R: A language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314. [Google Scholar]
- Kennedy, R.A., Rumpho, M.E., and Fox, T.C. (1992). Anaerobic metabolism in plants. Plant Physiol. 84, 1204–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C.E., Altschul, S.F., Boguski, M.S., Liu, J.S., Neuwald, A.F., and Wootton, J.C. (1993). Detecting subtle sequence signals: A Gibbs sampling strategy for multiple alignment. Science 262, 208–214. [DOI] [PubMed] [Google Scholar]
- Manjunath, S., Williams, A.J., and Bailey-Serres, J. (1999). Oxygen deprivation stimulates Ca2+-mediated phosphorylation of mRNA cap-binding protein eIF4E in maize roots. Plant J. 19, 21–30. [DOI] [PubMed] [Google Scholar]
- Muench, D.G., and Good, A.G. (1994). Hypoxically inducible barley alanine aminotransferase: cDNA cloning and expression analysis. Plant Mol. Biol. 24, 417–427. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Olive, M.R., Peacock, W.J., and Dennis, E.S. (1991). The anaerobic responsive element contains two GC-rich sequences essential for binding a nuclear protein and hypoxic activation of the maize Adh1 promoter. Nucleic Acids Res. 19, 7053–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Mendez, A., Aguilar, R., Briones, E., and De Sanchez, J.E. (1993). Characterisation of ribosomal protein phosphorylation in maize axes during germination. Plant Sci. 94, 71–79. [Google Scholar]
- Potuschak, T., Stary, S., Schlogelhofer, P., Becker, F., Nejinskaia, V., and Bachmair, A. (1998). PRT1 of Arabidopsis thaliana encodes a component of the plant N-end rule pathway. Proc. Natl. Acad. Sci. USA 95, 7904–7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt, K., Frech, K., Karas, H., Wingender, E., and Werner, T. (1995). MatInd and MatInspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, P., Weber, H., Damond, M., and Farmer, E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond, T., and Somerville, S. (2000). Chasing the dream: Plant EST microarrays. Curr. Opin. Plant Biol. 3, 108–116. [DOI] [PubMed] [Google Scholar]
- Ruan, Y., Gilmore, J., and Conner, T. (1998). Towards Arabidopsis genome analysis: Monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J. 15, 821–833. [DOI] [PubMed] [Google Scholar]
- Russell, D.A., and Sachs, M.M. (1991). The maize glyceraldehyde-3-phosphate dehydrogenase gene family: Organ-specific expression and genetic analysis. Mol. Gen. Genet. 229, 219–228. [DOI] [PubMed] [Google Scholar]
- Saab, I.N., and Sachs, M.M. (1996). A flooding-induced xyloglucan endo-transglycosylase homolog in maize is responsive to ethylene and associated with aerenchyma. Plant Physiol. 112, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, M.M., Freeling, M., and Okimoto, R. (1980). The anaerobic proteins of maize. Cell 20, 761–767. [DOI] [PubMed] [Google Scholar]
- Sachs, M.M., Subbaiah, C.C., and Saab, I.N. (1996). Anaerobic gene expression and flooding tolerance in maize. J. Exp. Bot. 47, 1–15. [Google Scholar]
- Schena, M., Shalon, D., Heller, R., Chai, A., Brown, P.O., and Davis, R.W. (1996). Parallel human genome analysis: Microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y., and Shinozaki, K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp, B.J., Bown, A.W., and McLean, M.D. (1999). Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 4, 446–451. [DOI] [PubMed] [Google Scholar]
- Shiao, T.L., Ellis, M.H., Dolferus, R., Dennis, E.S., and Doran, P.M. (2002). Overexpression of alcohol dehydrogenase or pyruvate decarboxylase improves growth of hairy roots at reduced oxygen concentrations. Biotechnol. Bioeng. 77, 455–461. [DOI] [PubMed] [Google Scholar]
- Singer, T., Yordan, C., and Martienssen, R.A. (2001). Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev. 15, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah, C.C., Bush, D.S., and Sachs, M.M. (1994. a). Elevation of cytosolic calcium precedes anoxic gene expression in maize suspension cultured cells. Plant Cell 6, 1747–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah, C.C., Zhang, J., and Sachs, M.M. (1994. b). Involvement of intracellular calcium in anaerobic gene expression and survival of maize seedlings. Plant Physiol. 105, 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis, B., Watts, R.A., Andersson, C.R., Llewellyn, D.J., Hargrove, M.S., Olson, J.S., Dennis, E.S., and Peacock, W.J. (1997). Two hemoglobin genes in Arabidopsis thaliana: The evolutionary origins of leghemoglobins. Proc. Natl. Acad. Sci. USA 94, 12230–12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers, M., and Jorgensen, P. (2000). Proteolysis and the cell cycle: With this RING I do thee destroy. Curr. Opin. Genet. Dev. 10, 54–64. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe, G.C., Feil, R., and Turpin, D.H. (1990). Anaerobic metabolism in the N-limited green alga Selenastrum minutum. I. Regulation of carbon metabolism and succinate as a fermentation product. Plant Physiol. 94, 1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartapetian, B.B., and Jackson, M.B. (1997). Plant adaptations to anaerobic stress. Ann. Bot. 79 (suppl. A), 3.–20. [Google Scholar]
- Walker, J.C., Howard, E.A., Dennis, E.S., and Peacock, W.J. (1987). DNA sequences required for anaerobic expression of the maize alcohol dehydrogenase 1 gene. Proc. Natl. Acad. Sci. USA 84, 6624–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, C., Gaut, R.L., Browning, K.S., Ravel, J.M., and Roberts, J.K.M. (1991). Hypoxia enhances phosphorylation of eukaryotic elongation factor 4A in maize root tips. J. Biol. Chem. 266, 23341–23346. [PubMed] [Google Scholar]
- Wingler, A., Lea, P.J., Quick, W.P., and Leegood, R.C. (2000). Photorespiration: Metabolic pathways and their role in stress protection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1517–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.H., Dudoit, S., Luu, P., and Speed, T.P. (2001). Normalization for cDNA microarray data. Available at www.stat.berkeley.edu/users/terry/zarray/Html/normspie.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.