Abstract
Flavonoids are a group of polyphenolic plant secondary metabolites important for plant biology and human nutrition. In particular flavonols are potent antioxidants, and their dietary intake is correlated with a reduced risk of cardiovascular diseases. Tomato fruit contain only in their peel small amounts of flavonoids, mainly naringenin chalcone and the flavonol rutin, a quercetin glycoside. To increase flavonoid levels in tomato, we expressed the maize transcription factor genes LC and C1 in the fruit of genetically modified tomato plants. Expression of both genes was required and sufficient to upregulate the flavonoid pathway in tomato fruit flesh, a tissue that normally does not produce any flavonoids. These fruit accumulated high levels of the flavonol kaempferol and, to a lesser extent, the flavanone naringenin in their flesh. All flavonoids detected were present as glycosides. Anthocyanins, previously reported to accumulate upon LC expression in several plant species, were present in LC/C1 tomato leaves but could not be detected in ripe LC/C1 fruit. RNA expression analysis of ripening fruit revealed that, with the exception of chalcone isomerase, all of the structural genes required for the production of kaempferol-type flavonols and pelargonidin-type anthocyanins were induced strongly by the LC/C1 transcription factors. Expression of the genes encoding flavanone-3′-hydroxylase and flavanone-3′5′-hydroxylase, which are required for the modification of B-ring hydroxylation patterns, was not affected by LC/C1. Comparison of flavonoid profiles and gene expression data between tomato leaves and fruit indicates that the absence of anthocyanins in LC/C1 fruit is attributable primarily to an insufficient expression of the gene encoding flavanone-3′5′-hydroxylase, in combination with a strong preference of the tomato dihydroflavonol reductase enzyme to use the flavanone-3′5′-hydroxylase reaction product dihydromyricetin as a substrate.
INTRODUCTION
Flavonoids form a large group of polyphenolic compounds that occur naturally in plants. Based on their core structure, the aglycone, they can be grouped into different classes, such as chalcones, flavanones, dihydroflavonols, flavonols, and anthocyanins (Figure 1). To date, >4000 different flavonoids have been identified. This large diversity is attributable to single or combinatorial modifications of the aglycone, such as glycosylation, methylation, and acylation. As a group, flavonoids are involved in many aspects of plant growth and development, such as pathogen resistance, pigment production, UV light protection, pollen growth, and seed coat development (Harborne, 1986).
Figure 1.
Scheme of the Flavonoid Biosynthesis Pathway.
Only the flavonoid classes relevant to this article are shown (in boxes). ANS, anthocyanidin synthase; C, cyanidin; CHI, chalcone isomerase; CHS, chalcone synthase; D, delphinidin; DFR, dihydroflavonol reductase; DK, dihydrokaempferol; DM, dihydromyricetin; DQ, dihydroquercetin; F3H, flavanone-3-hydroxylase; F3′H, flavanone-3′-hydroxylase; F3′5′H, flavanone-3′5′-hydroxylase; FLS, flavonol synthase; K, kaempferol; M, myricetin; P, pelargonidin; PAL, Phe-ammonia lyase; Q, quercetin; 4CL, 4-coumarate:coenzyme A ligase; C4H, cinnamic acid 4-hydroxylase.
There is increasing evidence to suggest that flavonoids, in particular those belonging to the class of flavonols (such as kaempferol and quercetin), are potentially health-protecting components in the human diet as a result of their high antioxidant capacity (Rice Evans et al., 1995, 1997; Sugihara et al., 1999; Dugas et al., 2000; Duthie and Crozier, 2000; Ng et al., 2000) and their ability, in vitro, to induce human protective enzyme systems (Cook and Samman, 1996; Manach et al., 1996; Janssen et al., 1998; Choi et al., 1999; Frankel, 1999; Hollman and Katan, 1999; Shih et al., 2000). Based on these findings, it was postulated that flavonoids may offer protection against major diseases such as coronary heart diseases and cancer (Hertog and Hollman, 1996; Steinmetz and Potter, 1996; Trevisanato and Kim, 2000). In addition, several epidemiological studies have suggested a direct relationship between cardioprotection and consumption of flavonols from dietary sources such as onion, apple, and tea (Hertog et al., 1993; Keli et al., 1996). These studies suggest that a systematic increase in the daily intake of certain flavonoids could lead to a 30 to 40% reduction in death by coronary heart diseases.
Based on studies of this type, there is growing interest in the development of food crops enriched with health-protective flavonoids. An excellent candidate for such an approach is tomato, one of the most important food crops worldwide. In tomato, the main flavonol is rutin (quercetin-3-O-rutinoside) (Woeldecke and Herrmann, 1974; Herrmann, 1979). Quantitative analysis of hydrolyzed extracts revealed that the levels of the quercetin aglycone range from 4 to 11 mg/kg fresh weight in large tomato varieties (Crozier et al., 1997). These levels are low compared with those in onion (300 mg quercetin/kg fresh weight) and broccoli (30 mg kaempferol/kg fresh weight) (Hertog et al., 1992; Crozier et al., 1997; Price et al., 1998).
Previously, we had characterized the flavonoid content of fruit of the processing tomato variety FM6203 (Muir et al., 2001). Red fruit peel contained naringenin chalcone, the flavonol glycosides rutin and kaempferol-3-O-rutinoside, and small amounts of a quercetin trisaccharide. In the fruit flesh (pericarp), flavonoids were absent, except for traces of rutin. These biochemical data correlated well with the expression of the flavonoid biosynthesis genes in these fruit. In the peel, significant levels could be detected of the transcripts encoding chalcone synthase (CHS), flavanone-3-hydroxylase (F3H), and flavonol synthase (FLS), enzymes required for the production of flavonols. By contrast, chalcone isomerase (CHI) mRNA levels were barely detectable, explaining the accumulation of the CHI substrate naringenin chalcone (Figure 1). In the flesh, there was no significant accumulation of any of these transcripts compared with that in the peel. Subsequent overexpression of the petunia CHI-A gene in the tomato fruit resulted in a dramatic increase of flavonols (mainly rutin) in the peel at the expense of naringenin chalcone. In the flesh, however, no detectable increase of any flavonoids was observed (Muir et al., 2001). These results indicated that the flavonoid biosynthesis pathway in tomato fruit is active only in the peel and that the flavonoid levels found are determined, at least in part, by the expression of flavonoid biosynthesis genes. Because the peel constitutes only 5% of the total fruit weight, we aimed to upregulate the flavonoid biosynthesis pathway in the fruit flesh by overexpressing regulatory genes.
In the past decade, a number of regulatory genes involved in the production of flavonoids have been isolated. In general, these genes belong to one of two families of transcription factors, the MYB-type C1 family and the basic helix-loop-helix, MYC-type R family (Dooner et al., 1991). Mutant analysis and gene expression studies in several plant species (maize, petunia, and Antirrhinum) revealed that these two gene families are required for the production of anthocyanins in the plant. In maize, the encoded R and C1 transcription factors control the expression of several structural genes in the pathway leading to anthocyanins, starting with the first step, CHS, up to anthocyanidin-3-glucosyltransferase (Dooner et al., 1991). Two of the best-studied examples of these transcription factor genes from maize, the MYB-type C1 and the MYC-type LC genes, have been expressed ectopically in various transgenic plant species, such as tobacco, Arabidopsis (Lloyd et al., 1992), petunia (Bradley et al., 1998), and tomato (Goldsbrough et al., 1996). Although there was no detectable effect when C1 was introduced, expression of the LC gene led to an enhanced anthocyanin pigmentation of those tissues that normally are capable of producing anthocyanins. Similar results were obtained with the ectopic expression of the Antirrhinum Delila gene, another member of the MYC transcription factor family (Mooney et al., 1995). However, the effects of LC and C1 on flavonoids other than anthocyanins have not been studied to date.
We have expressed the maize LC and C1 genes specifically in the fruit of transgenic tomatoes. In this article, we show that the expression of both genes is required and sufficient to upregulate the flavonoid pathway in tomato flesh, a tissue that normally does not produce any flavonoids. This resulted in a strong accumulation of flavonols and a more modest increase in flavanones, but remarkably, not in the accumulation of anthocyanins. We provide a mechanism to explain the accumulation of these types of flavonoids compared with the anthocyanins reported in other plants.
RESULTS
Approach, Gene Constructs, and Plant Transformation
Because we had shown previously (Muir et al., 2001) that the flavonoid biosynthesis pathway is not active in tomato fruit flesh, based on both metabolite and gene expression studies, we aimed to upregulate the entire flavonoid pathway in this tissue by expressing the maize transcription factor genes LC and C1 in transgenic tomato plants.
The LC gene was expressed under the control of the fruit-specific tomato E8 promoter, whereas the C1 gene was expressed under the control of either the E8 promoter or the constitutive double 35S promoter of Cauliflower mosaic virus (CaMV). LC and C1 are thought to act in a coordinated manner (Mol et al., 1998); therefore, this approach was expected to lead to an increase of flavonoids specifically in the fruit. As shown in Figure 2, seven binary constructs were made containing fusions of either the C1 or LC gene alone (pBBC10, pBBC20, and pBBC30) or both LC and C1 together (pBBC200, pBBC250, pBBC300, and pBBC350). Two versions of the LC gene were used: an LC cDNA with its 5′ leader (LC+) and an LC cDNA lacking its 5′ untranslated leader (LC−). The 5′ leader contains a small open reading frame that represses LC translation; hence, higher levels of LC protein are obtained when the 5′ leader is removed (Damiani and Wessler, 1993). A detailed description of the construction of all plasmids is given in Methods.
Figure 2.
Constructs Used in this Study.
Fusion constructs consisted of the following gene fusions: pBBC10, 35S-C1; pBBC20, E8-LC−; pBBC30, E8-LC+; pBBC200, 35S-C1/E8-LC−; pBBC300, 35S-C1/E8-LC+; pBBC250, E8-C1/E8-LC−; and pBBC350, E8-C1/E8-LC+. C1, maize C1 cDNA; LC, maize LC cDNA (LC−, version minus the 5′ leader; LC+, version plus the 5′ leader); Pd35s, double 35S promoter of CaMV; Pe8, tomato E8 promoter; Tnos, Agrobacterium nopaline synthase terminator, Trbc: pea ribulose bisphosphate carboxylase small subunit terminator.
The constructs referred to above were used to transform FM6203 tomato tissue by Agrobacterium tumefaciens–mediated transformation. Transgenic plants obtained after transformation with construct pBBC10, pBBC20, pBBC30, pBBC200, pBBC250, pBBC300, or pBBC350 were numbered from 100, 200, 300, 2000, 2500, 3000, and 3500 onward, respectively.
Analysis of Flavonoid Levels in the Fruit of Transgenic Plants
Red fruit from both LC/C1 (2000, 2500, 3000, and 3500 numbers) and wild-type tomato plants was harvested, and the flesh was analyzed by HPLC for the presence of flavonoids. In hydrolyzed LC/C1 extracts (Figure 3), a large increase was observed in the level of kaempferol compared with that seen in wild-type extracts. In addition to kaempferol, we observed a significant increase in the level of naringenin. Because flavonoids generally are present in a conjugated form (i.e., as O-glycosides), we determined whether different kaempferol and naringenin glycosides were produced by analyzing nonhydrolyzed flesh extracts of LC/C1 and wild-type fruit. Detailed spectral analysis of these HPLC results (Figure 4) revealed that at least five different kaempferol-O-glycosides (e.g., kaempferol-3-O-rutinoside) and at least five different naringenin-O-glycosides (e.g., naringin) accumulated in the flesh of red LC/C1 fruit. These compounds were barely detectable in the flesh of unripe green fruit, but they increased rapidly upon ripening of the fruit, reflecting the ripening-dependent increase in the activity of the E8 promoter used in the gene constructs (data not shown).
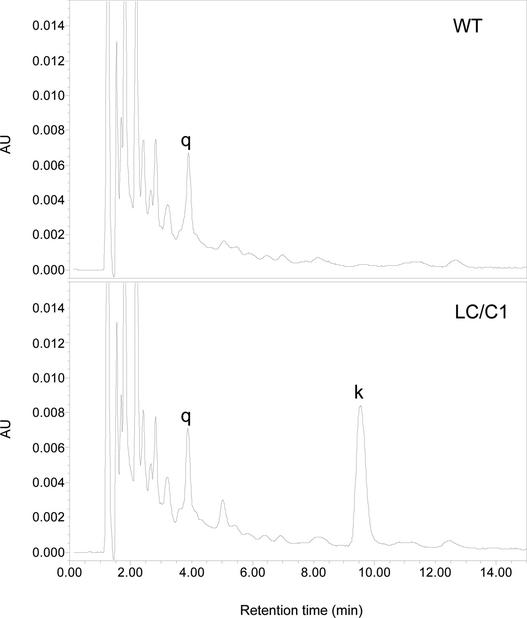
Figure 3.
HPLC Results, Recorded at 370 nm, of Hydrolyzed Extracts from the Flesh of Red Fruit of Wild-Type and Transgenic (LC/C1) Tomato Plants. q, quercetin; k, kaempferol; AU, absorption units.
Figure 4.
HPLC Results, Recorded at 280 nm, of Nonhydrolyzed Extracts from the Flesh of Red Fruit of Wild-Type and Transgenic (LC/C1) Tomato Plants.
AU, absorption units; K, kaempferol; N, naringenin; WT, wild type.
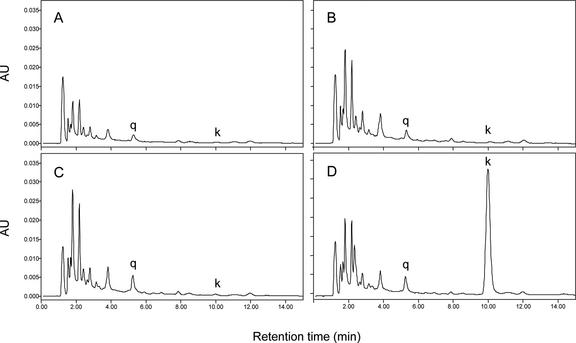
To determine whether both LC and C1 were required to upregulate the flavonoid pathway, hydrolyzed extracts from whole red fruit of plants transformed with either C1 or LC alone and wild-type plants were analyzed by HPLC. As shown in Figure 5, the chromatograms of plants transformed with only one of these two transcription factor genes showed no differences compared with untransformed wild-type plants. By contrast, whole fruit extracts of plants transformed with both LC and C1 revealed a clear accumulation of kaempferol. Detailed spectral analysis of the chromatogram peaks in nonhydrolyzed extracts confirmed that in LC/C1 plants, both kaempferol and naringenin glycosides were increased, and that in LC or C1 single-gene transgenic plants, no significant difference in the levels of any tested flavonoids was observed compared with the wild type (data not shown).
Figure 5.
HPLC Results, Recorded at 370 nm, of Hydrolyzed Extracts from Whole Red Fruit of Tomato Plants.
Samples were from wild-type (A), LC (B), C1 (C), and LC/C1 (D) tomato plants. AU, absorption units; k, kaempferol; q, quercetin.
Hydrolyzed extracts of whole red fruit were used to quantify the levels of quercetin, kaempferol, and naringenin in LC/C1 plants. Figure 6A shows only the results obtained for 35S-C1/E8-LC+ plants (3000 numbers), because this series was analyzed most extensively. In fruit of transformed plants, kaempferol levels were increased up to 60-fold relative to the mean level in wild-type fruit, whereas quercetin was increased slightly (up to 3-fold) in fruit of only a few plants. When expressed on a whole-fruit basis, the level of naringenin was increased up to twofold in only a few plants (Figure 6A), although all fruit accumulated naringenin in the flesh (Figure 4). This apparent discrepancy is explained by the fact that the naringenin level measured in the whole LC/C1 fruit represents the increased amount of naringenin present in flesh plus the amount of naringenin formed during hydrolysis from naringenin chalcone that is always present in the peel, the latter being highly variable between fruit samples. From the data in Figure 6A, it can be concluded that the accumulation of kaempferol accounted predominantly for the increase in total flavonols (quercetin plus kaempferol) in the fruit (up to 20-fold).
Figure 6.
Quantification of Flavonoids in Hydrolyzed Extracts of Whole Red Fruit Obtained from Plants Transformed with LC/C1 Constructs.
(A) Kaempferol, quercetin, and naringenin levels are shown for plants transformed with 35S-C1/E8-LC+ (3000 series). WT, untransformed control wild-type fruit (n = 10).
(B) Kaempferol levels are shown in plants transformed with constructs 35S-C1/E8-LC+ (3000 series), 35S-C1/E8-LC− (2000 series), and E8-C1/E8-LC− (2500 series).
A significant accumulation of kaempferol also was observed for the other three types of LC/C1 plants (data not shown). To compare the effect of the LC leader and the type of promoter controlling C1 gene expression, we determined the levels of kaempferol in hydrolyzed red fruit extracts of 35S-C1/E8-LC− (2000 series), 35S-C1/E8-LC+ (3000 series), and E8-C1/E8-LC− (2500 series). Unfortunately, for E8-C1/E8-LC+ plants (3500 series), only three transformants were obtained; therefore, these plants were not used in this comparison. As shown in Figure 6B, all three types of constructs tested resulted in plants with kaempferol levels ranging from 2 mg/kg fresh weight (equal to control levels) to 40 to 80 mg/kg fresh weight. Apparently, neither the LC leader (LC− versus LC+) nor the type of promoter driving C1 expression (35S versus E8) was a critical determinant for the extent of flavonoid induction in tomato fruit. Nevertheless, the highest kaempferol levels were found in plants transformed with E8-C1/E8-LC− (2500 series). However, we cannot yet conclude that this is attributable to a significant promoter effect, because there was a rather large (twofold to threefold) variation in kaempferol levels within fruit of a single transgenic plant during the year and between clones of the same line (data not shown).
Anthocyanin formation, which has been reported to be induced by LC in various plant species, such as tomato (Goldsbrough et al., 1996), tobacco (Lloyd et al., 1992), and petunia (Bradley et al., 1998), occasionally was visible by eye but not by HPLC (i.e., anthocyanin level <1 mg/kg fresh weight) in the peel of some young green 35S-C1/E8-LC fruit (2000/3000 series). This reddish-purple pigmentation disappeared when ripening progressed: anthocyanins never were detectable in red fruit, either by eye or by HPLC.
In summary, these results show that both regulatory genes (LC and C1) are required and sufficient to induce the biosynthesis of flavonoids in tomato fruit. This leads to a marked accumulation of kaempferol and naringenin glycosides in the flesh, a tissue that normally does not produce significant levels of any flavonoids.
Stability of the High-Flavonol Phenotype
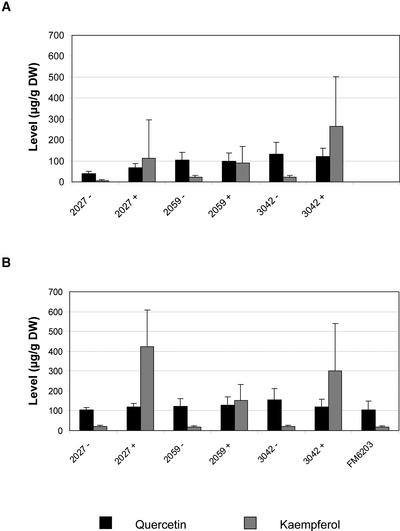
To determine whether the high-flavonol phenotype was retained in later generations, flavonol levels were determined in red fruit of segregating T1 and homozygous T2 populations of three single-copy 35S-C1/E8-LC lines (Figure 7). For each line, plants were grouped as either transgenic (+) or azygous (−) based on NPT-II PCR analysis. The azygous lines are progeny of transgenic parent lines that have lost the transgene through segregation. Azygous plants are considered ideal controls because they have been through the entire process of generating transgenic plants, exactly like their transgenic “sibling” plants. In all three lines, the transgene was inherited in a 3:1 ratio, as expected for single-copy transgene insertions. Transgenic T1 plants had a strongly increased kaempferol content compared with their azygous relatives, whereas the quercetin content did not differ significantly between transgenic and azygous plants (Figure 7A). Similar results were obtained in the T2 generation (Figure 7B). It also was shown that there was no difference in the results obtained for azygous lines and FM6203 plants that had not been through the tissue culture process at all. These results indicate that the high-flavonol phenotype is inherited stably in later generations.
Figure 7.
Stability of the High-Kaempferol Phenotype.
Red fruit were harvested from segregating T1 (A) and homozygous T2 (B) populations of three independent transgenic lines. Three fruit of each plant were pooled, hydrolyzed extracts were prepared, and quercetin and kaempferol levels were determined. The results shown represent mean values obtained for 5 to 10 transgenic plants for each transgenic (+) and azygous (−) line. For comparison, analysis of hydrolyzed extracts from fruit of FM6203 parent plants that did not go through the tissue culture procedure is shown. DW, dry weight.
Flavonoids in Leaves of Transgenic Plants
The flavonoid content in mature leaves of homozygous T2 populations of the above-mentioned LC/C1 lines was determined by HPLC analysis of hydrolyzed extracts. As shown in Figure 8, the level of the main flavonol in leaves, quercetin, was not altered significantly in transgenic leaves (+) compared with azygous leaves (−). However, there was a clear increase in the levels of both kaempferol and naringenin in transgenic leaves compared with their azygous relatives. In leaves of primary transformants containing either LC or C1 alone, there was no significant increase in the level of kaempferol. This finding indicates that, as in fruit, the expression of both LC and C1 is required and sufficient to upregulate the flavonoid biosynthesis pathway in leaves. In all plants analyzed for flavonoids in leaves, the C1 gene was expressed under the control of the constitutive double 35S promoter of CaMV, whereas the LC gene was expressed under the control of the fruit-specific tomato E8 promoter. The requirement for LC in addition to C1 expression to upregulate the flavonoid pathway in leaves suggests that the activity of the E8 promoter is not confined strictly to the fruit but also is significant in leaves. Leaves of LC/C1 plants expressing both LC and C1 under the control of the E8 promoter were not analyzed in this experiment.
Figure 8.
Flavonol Levels in Leaves of Homozygous LC/C1 T2 Lines and T0 Lines Transformed with LC or C1 Alone.
Leaves were harvested from homozygous T2 populations of three independent transgenic lines. Three leaves of each plant were pooled, hydrolyzed extracts were prepared, and quercetin (A) and kaempferol and naringenin (B) levels were determined. The results shown represent mean values obtained for 5 to 10 transgenic plants for each transgenic (+) and azygous (−) line. Similarly, kaempferol levels were determined in T0 lines transformed with the single-gene constructs pBBC10, pBBC20, and pBBC30 (C). DW, dry weight.
Analysis of Anthocyanins in Tomato Leaves
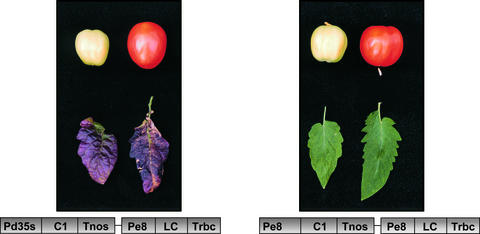
Although all transgenic plants were phenotypically similar to the wild type, we observed a clearly increased reddish-purple anthocyanin pigmentation in nodes and old leaves of some LC/C1 plants containing 35S-C1/E8-LC compared with LC/C1 plants containing E8-C1/E8-LC, plants expressing LC or C1 alone, or wild-type plants (Figure 9).
Figure 9.
Phenotypical Analysis of Leaves and Green and Red Fruit of LC/C1 Plants.
In plants containing 35S-C1/E8-LC, anthocyanins accumulate in older leaves. In plants expressing both C1 and LC under the control of the fruit-specific E8 promoter, this effect is not seen. Fruit of either type of LC/C1 plants do not accumulate anthocyanins.
To compare the anthocyanins produced in LC/C1 leaves with those accumulating in wild-type tomato leaves in response to abiotic stress, the anthocyanin content and composition in extracts of 3-week-old, light-stressed LC/C1 and wild-type seedlings was determined using HPLC/photodiode array/mass spectrometry. In both extracts, the same three major anthocyanin peaks could be identified, corresponding to petunidin 3-(caffeoyl)rutinoside-5-glucoside, petunidin 3-(p-coumaroyl)rutinoside-5-glucoside, and malvidin 3-(p-coumaroyl)rutinoside-5-glucoside (Table 1). Quantification of these peaks, using malvidin 3-glucoside as a standard, revealed that light-stressed LC/C1 seedlings contained threefold to fourfold higher levels of these anthocyanins than light-stressed wild-type seedlings. In agreement with published data (Bongue-Bartelsman and Philips, 1995), our HPLC/photodiode array/mass spectrometry measurements revealed that all anthocyanins detected in these colored leaves were of the petunidin and malvidin type and thus were derived solely from the precursor dihydromyricetin. In contrast to our results with mature leaves, in which we observed a clear increase of kaempferol and, occasionally, anthocyanins in LC/C1 plants relative to wild-type plants, these light-stressed LC/C1 and wild-type seedlings contained similarly high levels of both quercetin and kaempferol. This finding suggests that the LC/C1 effect observed in mature leaves and fruit mimics an integral part of the natural induction of the flavonoid pathway in response to abiotic stress.
Table 1.
Amounts of Anthocyanins and Flavonols Detected in Light-Stressed Seedlings
| Compound | Control (mg/kg fresh wt) |
LC/C1 (mg/kg fresh wt) |
|---|---|---|
| Petunidin 3-(caffeoyl) rutinoside-5-glucoside |
7.5 | 30.8 |
| Petunidin 3-(p-coumaroyl) rutinoside-5-glucoside |
76.0 | 266.0 |
| Malvidin 3-(p-coumaroyl) rutinoside-5-glucoside |
46.0 | 117.1 |
| Quercetin | 279.2 | 277.2 |
| Kaempferol | 67.1 | 76.6 |
Analysis of LC and C1 Gene Expression
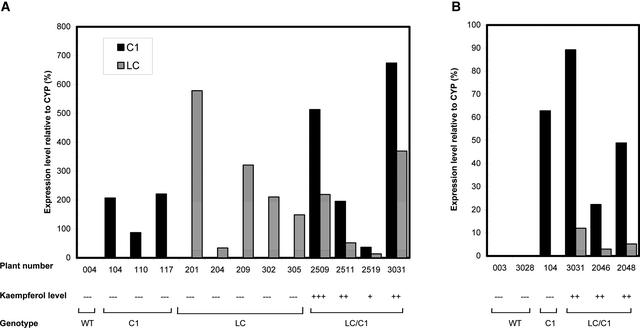
The expression of the introduced LC and C1 genes in transgenic plants was analyzed by real-time quantitative reverse transcriptase–mediated (RT) PCR. For these experiments, we made use of the fluorogenic TaqMan assay on the ABI PRISM7700 sequence detection system. Using this technology, we analyzed transgene expression in fruit of single-gene plants (LC or C1 alone) and double-gene plants (LC and C1 together). As an internal control, we used the tomato cyclophylin gene (CYP), which is expressed constitutively in turning and red tomato fruit, as observed by RNA gel blot analysis (data not shown). Total RNA was isolated from red fruit of the tomato plants mentioned above, and first-strand cDNA was synthesized by reverse transcription. Aliquots of 100 ng of cDNA were used in three TaqMan PCRs with probes for LC, C1, and CYP, respectively. LC and C1 mRNA expression levels were quantified relative to the amount of CYP mRNA. The absolute amount of CYP mRNA varied by <2.5-fold between the different transgenic plants (mean ± sd of Ct values [see Methods] was 20.44 ± 0.34; n = 13), indicating that this gene can be used as an authentic internal control. As shown in Figure 10, the control plant 004 failed to give a signal with either the C1 probe or the LC probe. By contrast, all transgenic plants tested showed clear expression of the introduced transgenes at the transcriptional level. The C1 single-gene plants (104, 109, and 117) showed only C1 gene expression, and the LC single-gene plants (201, 204, 209, 302, and 305) showed only LC gene expression, whereas all tested LC/C1 double-gene plants (2509, 2511, 2519, and 3031) showed expression of both LC and C1 in their fruit. Although the expression levels of the individual LC and C1 genes in some single-gene transgenic plants were higher than the levels found in some double-gene plants, there was no detectable increase in kaempferol levels in hydrolyzed extracts of any single-gene plant compared with the wild type (Figure 8). These results confirm our previous conclusion that the expression of both LC and C1 together is required to upregulate the flavonoid pathway in tomato fruit.
Figure 10.
TaqMan Analysis of LC and C1 Gene Expression in Red Fruit and Leaves of Wild-Type and Transgenic Tomato Plants.
Results are shown separately for red fruit (A) and leaves (B). Total RNA was reverse transcribed, and aliquots were amplified using primer pairs specific for LC, C1, and CYP (cyclophylin). RNA levels for each gene were expressed relative to the amount of CYP RNA, as described in Methods. The mean value for CYP expression (determined as Ct values) was 20.44 ± 0.34 for all fruit samples (mean ± sd; n = 13) and 18.09 ± 0.22 for all leaf samples (n = 6).
In the same way, LC and C1 mRNA levels were determined in mature leaves of transgenic and wild-type plants (Figure 10). In the control plant 003, there was no detectable expression of either LC or C1. In the leaves of the C1 single-gene plant 104, there was high C1 expression and, as expected, no LC expression. In leaves of LC/C1 plants containing 35S-C1/E8-LC, high C1 expression and low, but significant, LC expression were observed. These results are consistent with our previous observation that in leaves, as in fruit, the expression of both LC and C1 is required to upregulate the flavonoid pathway. Although the LC gene is expressed under the control of the E8 promoter, its expression in leaves remains sufficient to induce the biosynthesis of flavonoids, provided that C1 is expressed simultaneously by the 35S promoter.
Effect of LC/C1 on the Expression of Flavonoid Pathway Genes
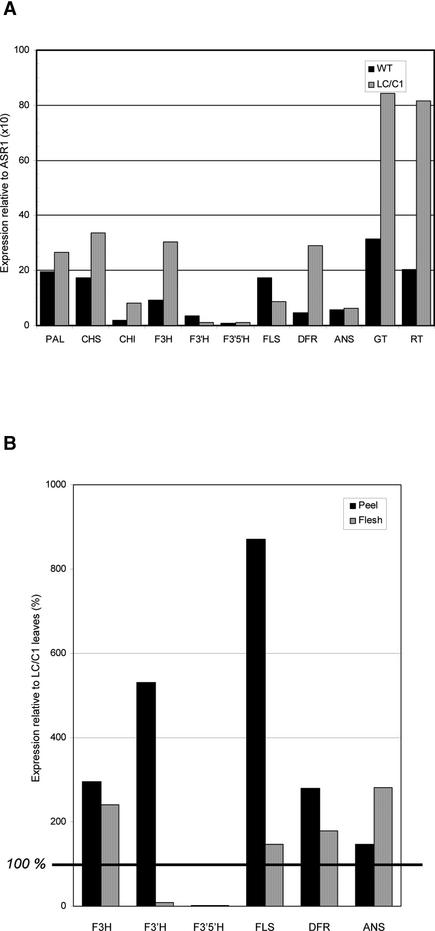
The effect of the introduced LC and C1 transcription factors on the expression of flavonoid biosynthesis genes was examined by real-time quantitative SYBR-Green RT-PCR, as described in Methods. Fruit samples of representative LC/C1 and wild-type plants, harvested in the green, turning, and red stages, were separated into peel and flesh, and total RNA was isolated from each tissue. Expression levels for Phe-ammonia lyase (PAL), the flavonoid pathway genes chalcone synthase (CHS), chalcone isomerase (CHI), flavanone-3-hydroxylase (F3H), flavanone-3′-hydroxylase (F3′H), flavanone-3′5′-hydroxylase (F3′5′H), flavonol synthase (FLS), dihydroflavonol reductase (DFR), and anthocyanidin synthase (ANS), and the glycosyltransferases flavonol-3-glucosyltransferase (GT) and flavonol-3-glucoside-rhamnosyltransferase (RT) were determined and expressed relative to the ASR1 gene encoding an abscisic stress ripening protein (Iusem et al., 1993). This gene was chosen as an internal control because DNA microarray analysis (data not shown) and SYBR-Green RT-PCR analysis showed that it has a high and stable mRNA expression level in all three ripening stages and fruit tissues tested (mean ± sd of Ct values was 15.69 ± 0.33; n = 6).
As shown in Figure 11, there was a clear expression of PAL, F3H, F3′H, FLS, GT, and RT in the peel of wild-type green fruit. Transcript levels of these genes increased during ripening, reaching a maximum level in the turning stage, and then decreased again in the red stage. Expression levels for CHS and CHI were low in green fruit peel and thus may be responsible for the relatively low flavonoid levels found in green fruit (Muir et al., 2001). Upon ripening, expression of CHS increased dramatically (>100-fold), whereas CHI transcript levels remained low. The genes required for the production of tomato leaf anthocyanins (F3′5′H, DFR, and ANS) were not expressed in tomato fruit peel. Together, the expression profiles of the above-mentioned genes correlated well with the accumulation of naringenin chalcone and quercetin in tomato fruit peel during ripening (Muir et al., 2001). In flesh of wild-type fruit, there was very low expression of all genes tested, in accordance with the observation that flavonoids are not detected in flesh. In LC/C1 flesh (Figure 11), however, we observed a >100-fold induction of the genes encoding CHS, F3H, and DFR and a 5- to 15-fold induction of the genes encoding FLS, ANS, GT, and RT relative to wild-type flesh. Neither PAL nor CHI, F3′H, or F3′5′H were induced by LC/C1 in flesh. In LC/C1 peel (Figure 11A), the same transcripts were induced by LC/C1, but the induction ratios for CHS, F3H, FLS, GT, and RT were much lower than those in flesh as a result of the clear expression of these genes in wild-type peel. In conclusion, these results indicate that the expression of the maize LC and C1 transcription factor genes in tomato leads to the induction of all of the flavonoid biosynthesis genes required for the production of kaempferol-type flavonols and pelargonidin-type anthocyanins.
Figure 11.
SYBR-Green RT-PCR Analysis of Flavonoid Pathway Gene Expression in Fruit Peel and Flesh during Ripening of Wild-Type and LC/C1 Tomato Fruit.
Total RNA was reverse transcribed, and aliquots were amplified using primer pairs specific for PAL, CHS, CHI, F3H, F3′H, F3′5′H, FLS, DFR, ANS, GT, RT, and the internal control ASR1. RNA levels for each gene were expressed relative to the amount of ASR1 RNA, as described in Methods, and multiplied by 1000. The mean value for ASR1 expression (determined as Ct values) of all fruit samples was 15.69 ± 0.33 (mean ± sd; n = 6). WT, wild type.
Because mature leaves of some LC/C1 plants contain both kaempferol- and quercetin-type flavonols and delphinidin-type anthocyanins (Figure 1), this tissue is expected to produce adequate transcript levels of all of the flavonoid structural genes involved. Expression levels in green wild-type and purple LC/C1 mature leaves of all genes tested are shown in Figure 12A. Compared with that in fruit, the LC/C1 effect on flavonoid gene expression is much more modest in leaves (ranging from twofold to sixfold). This is most likely the result of the fact that the flavonoid pathway is already active in wild-type leaves. In addition, we observed much less variation in the transcript levels between genes in leaves (Figure 12A) and fruit (Figure 11), suggesting that all of the genes are expressed at a sufficient level to produce the flavonols and anthocyanins found. To compare gene expression levels in leaves and fruit, we determined the RNA levels of the genes encoding enzymes acting at and around the branch point between the formation of flavonols and anthocyanins (Figure 1) in peel and flesh of turning LC/C1 fruit relative to the levels found in LC/C1 leaves (Figure 12B). This was possible because the two internal controls used showed constant expression levels in leaves and green fruit (CYP) and in the three ripening stages tested (ASR1). In LC/C1 flesh, expression levels of F3H, FLS, DFR, and ANS were in the same range as in leaves, whereas expression levels of F3′H and F3′5′H were 10- to 100-fold lower than those in LC/C1 leaves. This finding suggests that the expression of the latter two genes may be limiting steps in the partitioning of the pathway toward dihydroquercetin and dihydromyricetin in LC/C1 flesh.
Figure 12.
Comparative Analysis of Flavonoid Pathway Gene Expression in Leaves and Fruit of WT and LC/C1 Tomatoes.
(A) SYBR-Green RT-PCR analysis of flavonoid pathway gene expression in green wild-type and purple LC/C1 tomato leaves, as described in Figure 11. RNA levels for each gene were expressed relative to the amount of ASR1 RNA and multiplied by 10. Because the ASR1 gene has a 100-fold lower expression in leaves than in fruit, multiplication of the ratios by 10 instead of 1000 (as for fruit) results in values that can be compared directly with those given for fruit. The raw Ct values for ASR1 expression in leaves were 22.67 (wild type) and 22.47 (LC/C1). WT, wild type.
(B) Comparison of RNA expression levels of F3H, F3′H, F3′5′H, FLS, DFR, and ANS in turning LC/C1 peel and flesh relative to the levels in LC/C1 leaves. Expression levels in leaves were extrapolated to the levels found in fruit, based on the constitutive expression of CYP in leaves and green fruit (mean ± sd of Ct values is 19.47 ± 0.27; n = 4) and of ASR1 in the three ripening stages tested (mean ± sd of Ct values is 15.69 ± 0.33; n = 6). Expression of each gene is given as a percentage of the expression in leaves. Experimental details are described in Methods.
In LC/C1 peel, we also observed a low expression level of F3′5′H but a very strong expression of F3′H and FLS compared with that in leaves. This finding suggests that in LC/C1 peel, the production of dihydromyricetin is limited because of the low F3′5′H expression and that there is a strong pull of the pathway toward the production of dihydroquercetin, quercetin, and kaempferol because of the high F3′H and FLS expression.
In combination with the apparent substrate preference of the DFR enzyme for dihydromyricetin and of the FLS enzyme for dihydrokaempferol and dihydroquercetin, these expression data suggest that F3′5′H expression is a key factor in the accumulation of anthocyanins in LC/C1 leaves and their absence in fruit.
DISCUSSION
In this article, we show that the simultaneous expression of the maize transcription factor genes LC, a member of the MYC-type R gene family, and C1, a member of the MYB-type C1 gene family, in fruit of transgenic tomato plants leads to a strong induction of flavonoid biosynthesis in the flesh of the fruit, a tissue that normally lacks significant levels of any flavonoids. On a whole-fruit basis, this resulted in an up to 60-fold increase in the level of the flavonol kaempferol, which was conjugated with different sugars. In addition to these kaempferol glycosides, a significant increase was observed in the level of several naringenin glycosides in the flesh. Both LC and C1 were required and sufficient to induce the biosynthesis of these flavonoids, because plants containing either LC or C1 alone did not differ from control plants with respect to flavonoid levels.
Two versions of the LC cDNA were used in our gene constructs: one with and one without the 5′ untranslated region (UTR). The LC 5′ UTR contains a small upstream open reading frame that is involved in the translational control of LC expression. It was shown by transient expression studies in maize kernels that the removal of this 5′ UTR results in 25- to 30-fold enhanced LC protein expression (Damiani and Wessler, 1993). Although we did not investigate whether the same mechanism exists in tomato, accumulation of the end product kaempferol in tomato fruit was not influenced significantly by the presence or absence of the LC 5′ UTR. In addition, there was no clear correlation between LC or C1 gene expression levels and kaempferol levels, except for the requirement of a basal expression of both genes.
We aimed to induce the flavonoid biosynthesis pathway specifically in the fruit by expressing either the LC gene or both LC and C1 under the control of the fruit-specific E8 promoter to prevent the possible confounding effects of increased flavonoid levels during tissue culture or subsequent plant growth. Nevertheless, in leaves of E8-LC/35S-C1 plants, a small but clear increase in flavonoids was detectable, whereas in leaves of plants expressing E8-LC or 35S-C1 alone, no increase of flavonoids was detected compared with control leaves. This suggests that in leaves, as in fruit, the expression of both LC and C1 is required to induce the biosynthesis of certain flavonoids. TaqMan gene expression analysis showed that a low but significant expression of the LC gene was detectable in E8-LC/35S-C1 leaves, indicating that the E8 promoter is somewhat leaky or not restricted fully to fruit.
Genes that encode homologs of the maize R and C1 transcription factor families have been expressed ectopically in maize cell lines (Grotewold et al., 1998) and in several dicotylenous plant species, such as petunia (Bradley et al., 1998), tomato (Goldsbrough et al., 1996), tobacco, and Arabidopsis (Lloyd et al., 1992; Mooney et al., 1995). In all studies reported to date, ectopic expression of an R gene, either alone or in combination with C1, resulted in an increased production of anthocyanins. None of these studies mention an effect of these transgenes on flavonoids other than anthocyanins. In agreement with these observations, we also observed anthocyanins in our LC/C1 plants, but only in vegetative tissues and occasionally in some young green fruit. By contrast, we never detected anthocyanins at later stages of LC/C1 fruit ripening, despite the fact that LC/C1 strongly induced the accumulation of other flavonoids.
To provide an explanation for the accumulation of flavonols and the lack of anthocyanins in our LC/C1 fruit, we investigated the partitioning of the flavonoid pathway toward flavonols and anthocyanins in more detail by combining metabolic information with gene expression data. As shown in Figure 1, dihydrokaempferol (DK) is produced as a product of the F3H enzyme. Using DK as a substrate, the action of the two cytochrome P450 hydroxylases F3′H and F3′5′H leads to the production of two other dihydroflavonols, dihydroquercetin (DQ) and dihydromyricetin (DM). These three types of dihydroflavonols (DK, DQ, and DM) serve as common precursors for the formation of the three types of flavonols (kaempferol, quercetin, and myricetin) through the action of FLS as well as the three types of anthocyanins (pelargonidin, cyanidin, and delphinidin) through the action of DFR. In LC/C1 leaves, all three types of dihydroflavonols (DK, DQ, and DM) must be present, because significant amounts of the derived reaction products kaempferol, quercetin (both flavonols), and delphinidin (anthocyanin) are present. This is in agreement with our observations that in leaves, all genes required for the biosynthesis of DK, DQ, and DM, including F3′H and F3′5′H, are expressed. However, and in accordance with previous findings (Bongue-Bartelsman and Philips, 1995), only delphinidin-type anthocyanins, which are derived from DM, were observed. These results indicate (1) that the tomato DFR enzyme, which is encoded by a single gene in tomato (Goldsbrough et al., 1994; Yoder et al., 1994), is restricted in its substrate specificity to DM, because only delphinidin is formed, and (2) that the tomato FLS enzyme preferentially uses DQ and DK as substrates, because the flavonol myricetin was not detected in any tomato tissue examined. A substrate preference for specific dihydroflavonol precursors of DFR and FLS has been shown in other solanaceous species as well (Oud et al., 1995).
In LC/C1 fruit peel, at least two types of dihydroflavonols (DK and DQ) must be formed, because their respective reaction products (kaempferol and quercetin) were present in this tissue. This is in agreement with our gene expression data showing that all genes, except CHI, required for the production of DK and DQ, plus the flavonols and anthocyanins derived from them, were expressed. The lack of anthocyanins in fruit peel can be explained by the observation that DFR cannot use DK and DQ as substrates and that the only substrate that can be used by DFR (DM) is not produced, as a result of a very low F3′5′H expression in fruit peel compared with leaves.
A similar situation was found in LC/C1 fruit flesh. Despite the induction of DFR, ANS, and FLS, only kaempferol was detected in this tissue. This finding suggests that of the dihydroflavonols, only DK is produced in fruit flesh. The absence of DQ and DM, and thus their respective reaction products quercetin (from FLS activity) and delphinidin (from DFR activity), most likely is attributable to the very low, and LC/C1-insensitive, expression of both F3′H and F3′5′H in fruit flesh compared with leaves.
Together, these results lead to the conclusion that expression of the LC and C1 transcription factors in tomato leads to the induction of all of the genes required for the production of flavonols and anthocyanins, except for CHI and the two B-ring hydroxylases F3′H and F3′5′H. The endogenous expression pattern of the F3′5′H gene (a 10- to 100-fold higher level in leaves than in fruit), in combination with the substrate specificity of the tomato DFR enzyme (i.e., DM), are the primary factors determining the absence of anthocyanins in fruit and their presence in leaves of these LC/C1 tomato lines. The observation of anthocyanins in some young green LC/C1 fruit suggests that, at least in these fruit, all flavonoid pathway genes, including F3′5′H, were expressed sufficiently to enable anthocyanin formation at early stages of fruit development. Our results also indicate that the types of flavonols found in fruit peel and flesh and in leaves are determined mainly by the expression of the F3′H gene, in combination with the substrate specificity of the FLS enzyme. In addition to the low F3′5′H gene expression, a strong pull of the pathway toward DQ, quercetin, and kaempferol by F3′H and FLS may be of significance for the absence of anthocyanins in fruit peel as well.
In contrast to our results with E8-LC and E8-LC/35S-C1 plants, Goldsbrough et al. (1996) reported the accumulation of anthocyanins in leaves and red fruit of cherry tomato plants expressing the LC gene under the control of the constitutive 35S promoter. For E8-LC leaves, this discrepancy between results can be explained by a LC gene dosage effect resulting from the relatively high activity of the 35S promoter compared with the E8 promoter in this tissue (Figure 10). Apparently, the activity of the E8 promoter is too weak in leaves to produce sufficient LC to stimulate the flavonoid pathway. Only when C1 was expressed coordinately at high levels (E8-LC/35S-C1) was E8-driven LC expression sufficient to induce the flavonoid pathway in leaves. In ripe fruit, the activity of the E8 promoter was on the same order as that of the 35S promoter (Figure 10A, E8-C1 in plant 2509 versus 35S-C1 in plant 3031). Nevertheless, we did not observe anthocyanin production in ripe LC or LC/C1 fruit because of the low F3′5′H activity, as argued above. This finding suggests that anthocyanins present in ripe 35S-LC cherry tomato fruit must be produced primarily at very early stages of ripening. Strikingly, and in agreement with this idea, we detected low levels of anthocyanins in some young green LC/C1 fruit, suggesting that in these early ripening stages, F3′5′H expression is sufficient to make anthocyanin production possible. Differences in the activities of the E8 and 35S promoters in these early stages of fruit development (up to the green stage) may explain the apparently contradictory observations with respect to anthocyanin accumulation in ripe cherry tomato versus ripe FM6203 tomato, because the activity of the E8 promoter was relatively low at the green stage compared with later stages of development (e.g., as reflected by the expression of the target genes; Figure 11).
Other explanations also are possible. For example, we cannot exclude the possibility that the F3′5′H gene is expressed in ripe cherry tomato fruit as a result of genotypical or physiological differences. Alternatively, the DFR enzyme in cherry tomatoes may have different substrate specificity than the one in our tomato line, such that it is able to use DK or DQ as substrate. It is noteworthy, in this respect, that alteration of a single amino acid in the substrate binding domain of the petunia DFR enzyme was sufficient to alter its specificity for the three dihydroflavonol substrates (Johnson et al., 2001).
In conclusion, these results clearly show that ectopic expression of combinations of transcription factor genes is a powerful way to influence metabolic pathways. However, the effects on metabolite content are not easy to predict and can vary considerably between plant species and varieties as a result of gene dosage effects, the endogenous and transgene-induced expression profiles of structural genes, and the substrate specificities of the enzymes involved, which may differ from plant to plant.
METHODS
Plant Growth Conditions
Tomato (Lycopersicon esculentum) line FM6203 and transgenic plants were grown in a containment greenhouse with a 16-h photoperiod and 21/17°C day/night temperatures.
Cloning Procedures and Plasmid Constructions
All DNA manipulations, including PCR, restriction digestion, agarose gel electrophoresis, ligation, and transformation into Escherichia coli DH5α, were performed as described previously (Sambrook et al., 1989).
In general, all promoters, structural genes, and terminators used were amplified by PCR with primers containing the desired restriction enzyme site attached to their 5′ ends. Restriction sites were chosen in such a way that all promoters were cloned as KpnI-BamHI fragments, all structural genes were cloned as BamHI-SalI fragments, and all terminators were cloned as SalI-ClaI fragments, unless stated otherwise.
Plasmid pFLAP10 (P35S-C1-Tnos) was constructed as follows. First, the Agrobacterium tumefaciens nopaline synthase (NOS) terminator was amplified from plasmid pBI121 (Jefferson et al., 1987) and cloned as a 0.2-kb SalI-ClaI fragment in the pUCAP (van Engelen et al., 1995) derivative pUCM2, in which suitable restriction sites for cloning were introduced. This resulted in plasmid pFLAP1. Second, the C1 gene was isolated as a 1.6-kb EcoRI-PacI fragment from plasmid pAL71 (Lloyd et al., 1992), suitable adapters were ligated, and the C1 gene was cloned as a 1.6-kb BamHI-SalI fragment into pFLAP1, resulting in plasmid pFLAP2. Third, the double 35S promoter of Cauliflower mosaic virus was amplified from plasmid pMOG18 (Sijmons et al., 1990) and cloned as a 0.85-kb KpnI-BamHI fragment into plasmid pFLAP3, resulting in plasmid pFLAP10.
To construct plasmids pFLAP20 (Pe8-LC−-Trbc) and pFLAP30 (Pe8-LC+-Trbc), the LC gene and the pea ribulose bisphosphate carboxylase terminator (Trbc) were isolated from plasmid pAL144 (LC−) and pAL65 (LC+), respectively (Lloyd et al., 1992) and cloned as a BamHI-ClaI fragment in plasmid pUCM2. This resulted in plasmid pFLAP4(LC−) and pFLAP5 (LC+). Next, a 2-kb tomato E8 promoter fragment was amplified by PCR from plasmid pT7E8 (Bovy et al., 1999) and cloned as a KpnI-BamHI fragment upstream of the LC genes of pFLAP4 and pFLAP5, resulting in plasmids pFLAP20 and pFLAP30, respectively. Plasmid pFLAP15 (Pe8-C1-Tnos) was constructed by replacing the 35S promoter in pFLAP10 with the E8 promoter from pFLAP20.
To make the two-gene constructs pFLAP200 (P35S-C1-Tnos/Pe8-LC−-Trbc) and pFLAP300 (P35S-C1-Tnos/Pe8-LC+-Trbc), the inserts of plasmids pFLAP10 and either pFLAP20 or pFLAP30 were cloned behind each other in plasmid pUCM3, a derivative of plasmid pUCAP (van Engelen et al., 1995) in which the multiple cloning site was modified in such a way that it contained suitable cloning sites. First, the insert of pFLAP10 was cloned as a KpnI-ClaI fragment in pUCM3. This resulted in plasmid pFLAP100. Then, the inserts of plasmids pFLAP20 and pFLAP30 were cloned as a NotI-AscI fragment behind the C1 gene in plasmid pFLAP100, resulting in plasmids pFLAP200 and pFLAP300, respectively.
To make the two-gene constructs pFLAP250 (Pe8-C1-Tnos/Pe8-LC−-Trbc) and pFLAP300 (Pe8-C1-Tnos/Pe8-LC+-Trbc), the inserts of plasmids pFLAP15 and either pFLAP20 or pFLAP30 were cloned behind each other in plasmid pUCM3. First, the insert of pFLAP15 was cloned as a KpnI-ClaI fragment in pUCM3. This resulted in plasmid pFLAP150. Then, the inserts of plasmids pFLAP20 and pFLAP30 were cloned as a NotI-AscI fragment behind the C1 gene in plasmid pFLAP150, resulting in plasmids pFLAP250 and pFLAP350, respectively.
For plant transformation, we made use of the binary vector pBBC3, a derivative of plasmid pGPTV-KAN (Becker et al., 1992). To construct pBBC3, pGPTV-KAN was digested with EcoRI-HindIII, and the gusA-Tnos gene was replaced by a small multiple cloning site consisting of PacI-EcoRI-HindIII-AscI restriction sites. The inserts of plasmids pFLAP10, pFLAP20, pFLAP30, pFLAP200, pFLAP250, pFLAP300, and pFLAP350 were transferred as PacI-AscI fragments to plasmid pBBC3. This resulted in plasmids pBBC10, pBBC20, pBBC30, pBBC200, pBBC250, pBBC300, and pBBC350, respectively.
Plant Transformation
Binary plasmids were transformed into Agrobacterium strain LBA4404 (Hoekema et al., 1985) by the freeze-thaw method (Gynheung et al., 1988). The presence of the plasmids was tested by restriction enzyme analysis after back transformation to E. coli.
Tomato variety FM6203 was transformed by Agrobacterium-mediated transformation of cotyledons (Fillatti et al., 1987). Plants transformed with pBBC10, pBBC20, pBBC30, pBBC200, pBBC250, pBBC300 and pBBC350 were numbered from 100, 200, 300, 2000, 2500, 3000 and 3500 onward, respectively.
Harvest of Plant Material
For both flavonoid (HPLC) and RNA (TaqMan) analyses, fruit, fully expanded mature leaves, and germinating seedlings were harvested and frozen immediately in liquid nitrogen. The material was stored frozen at −80°C. Extracts were made of pools of at least three fruit of each plant to minimize sample variation. Likewise, the first, second, and eighth fully expanded leaves of each plant were pooled before extraction.
Flavonoid Extraction and HPLC Analyses
Flavonoids were determined as aglycones or as their glycosides by preparing hydrolyzed and nonhydrolyzed extracts, respectively. Hydrolyzed extracts were prepared and analyzed by HPLC with photodiode detection essentially according to Hertog et al. (1992). tert-Butylated hydroxyquinon was left out of the extraction protocol because it coeluted with naringenin during isocratic HPLC (25% acetonitrile in 0.1% trifluoroacetic acid). Dose–response curves of quercetin, naringenin, and kaempferol (0 to 20 μg/mL) were established to quantify these compounds in the hydrolyzed extracts. Nonhydrolyzed extracts were prepared in 75% aqueous methanol with 10 min of sonication. Subsequent HPLC of the flavonoid species extracted was with a gradient of 5 to 50% acetonitrile in 0.1% trifluoroacetic acid.
Absorbance spectra and retention times of eluting peaks were compared with those of commercially available flavonoid standards (Apin Chemicals, Abingdon, UK). It was established that during the hydrolysis step, 100% of aglycones were released from their respective glycosides, whereas >95% of naringenin chalcone was converted chemically into naringenin. Recoveries of quercetin, kaempferol, and naringenin standards added to peel or flesh extracts just before hydrolysis were >90%. The lowest detection limit for flavonoids in the tomato extracts was ∼0.1 μg/mL, corresponding to 1 mg/kg fresh weight. Variation between replicate extractions and injections was <5%.
Anthocyanins in seedlings were extracted in acid methanol (95% methanol plus 5% 1 N HCl; 0.5 g fresh weight in 10 mL). After sonication and centrifugation at 3500g, the residue was extracted twice. The supernatants were combined, methanol was evaporated after adding 5 mL of water, and the remaining water extract was washed with hexane to remove lipid-soluble compounds. Analysis of anthocyanins in the extracts was performed by reverse-phase HPLC (Hewlett-Packard 1100 HPLC, equipped with a Phenomenex Aqua C18 4.6 × 150-mm column, 5 μm, at 35°C; Phenomenex, Torrence, CA) with photodiode array and mass detection. The mass spectrometer was a Finnigan LCQ (ThermoQuest, San Jose, CA) with an electrospray ionization interface. Both the auxiliary and the sheath gas were a mixture of nitrogen and helium. The capillary voltage was 3 V, and the capillary temperature was 195°C. Spectra were recorded in positive ion mode between mass-charge ratios of 120 and 1500. The mass spectrometer was programmed to perform a series of three scans: a full mass spectrum, an MS2 of the most abundant ion in the first scan using a collision energy of 30, and an MS3 of the most abundant ion in the second scan using a collision energy of 30. An aliquot of the aqueous anthocyanin extract was subjected to hydrolysis by mixing (1:1) with 6 N HCl and heating at 100°C in a closed vial purged with nitrogen for 45 min. Separation of this hydrolyzed extract was on a Waters Spherisorb ODS2 column (3 μm; 4.6 × 150 mm; 35°C) using a gradient of acetonitrile in 4.5% formic acid. Quantification of anthocyanins was made from the areas of their peaks recorded at 520 nm compared with an external standard of malvidin 3-glucoside.
TaqMan Analysis
The expression of the introduced LC and C1 genes in the obtained transgenic plants was analyzed by real-time quantitative reverse transcriptase–mediated (RT) PCR using the fluorogenic TaqMan assay on the ABI PRISM7700 sequence detection system (Perkin-Elmer Applied Biosystems). The principle of this procedure is as follows. Total RNA, extracted from the source of interest, is converted into cDNA by reverse transcription. The expression of the target gene is monitored by PCR with a fluorogenic TaqMan probe that hybridizes specifically between the forward and reverse primers. The single-stranded TaqMan probe is labeled with two fluorescent dyes: a reporter dye (6-carboxyfluorescein) at the 5′ end and a quencher dye (6-carboxytetramethylrhodamine) at the 3′ end. When the probe is intact, the signal from the reporter dye is absorbed by the quencher dye. During PCR, however, the probe is cleaved as a result of the 5′ nuclease activity of Taq DNA polymerase, leading to uncoupling of the reporter dye and the quencher dye. This results in an increase in reporter fluorescence. With each cycle, additional reporter dye molecules are cleaved from their respective probes, and the increase in fluorescence intensity is monitored continuously during PCR. The PCR cycle at which the fluorescence reaches a certain threshold value, Ct, which is defined as the PCR cycle at which a statistically significant increase of reporter fluorescence is first detected, is a measure for the starting copy number of the target RNA. Relative quantitation of the target RNA expression level was performed using the comparative Ct method (User Bulletin 2, ABI PRISM7700 Sequence Detection System, December 1997; Perkin-Elmer Applied Biosystems) in which the differences in the Ct for the target RNA and an endogenous control RNA, called ΔCt, were calculated to normalize for the differences in the total amount of cDNA present in each reaction and the efficiency of the RT step. Finally, the target RNA expression level was expressed as a percentage of the control RNA expression level using the equation 2 EXP ΔCt × 100%.
Total RNA was isolated from red fruit and leaves of tomato plants as described (Bovy et al., 1995). First-strand cDNA was synthesized from 350 ng of total RNA by reverse transcription. Aliquots of 100 ng of cDNA were used in three TaqMan PCR procedures (according to the manufacturer's protocol) with a C1, a LC, and a CYP probe, respectively. The tomato cyclophylin gene (CYP) is expressed constitutively in turning and red tomato fruit, as observed by RNA gel blot analysis (data not shown); therefore, it can be used as an internal control. The sequences of the TaqMan primers and probes used are listed in Table 2. The LC and C1 transcript levels were expressed relative to the amount of CYP mRNA, as described above.
Table 2.
Overview of TaqMan/SYBR-Green Primers and Probes Used
| Primer/probe | Gene | Sequence (5′ to 3′) |
|---|---|---|
| C1F | C1 | GCCCTGGCGTCGTTTCT |
| C1R | C1 | TGGACATCTATACGTGTACTTGTTGTCTAC |
| C1Ta | C1 | CTCCGCTGTCAGACGGCCGG |
| LCF | LC | CGGGAGCAGCACAGGAAAT |
| LCR | LC | GTCGCTTTCGCTCCGACAT |
| LCTa | LC | TGGCACTGGCACCAAGAACCACG |
| CYPF | CYP | GAGTGGCTCAACGGAAAGCA |
| CYPR | CYP | CCAACAGCCTCTGCCTTCTTA |
| CYPTa | CYP | ACATCCATGCCTTCAACAACTTGTCCAA |
| PAL_TOM_F | PAL | ATTGGGAAATGGCTGCTGATT |
| PAL_TOM_R | PAL | TCAACATTTGCAATGGATGCA |
| CHS_TOM_F | CHS | TGGTCACCGTGGAGGAGTATC |
| CHS_TOM_R | CHS | GATCGTAGCTGGACCCTCTGC |
| CHI_TOM_F | CHI | GTTTTTCACAAACCAACAGTTCTGAT |
| CHI_TOM_R | CHI | GAAGCAGTGCTCGATTCCATAAT |
| F3H_TOM_F | F3H | CACACCGATCCAGGAACCAT |
| F3H_TOM_R | F3H | GCCCACCAACTTGGTCTTGTA |
| F3′H_TOM_F | F3′H | GCACCACGAATGCACTTGC |
| F3′H_TOM_R | F3′H | CGTTAGTACCGTCGGCGAAT |
| F3′5'H_TOM_F | F3′5′H | GGCAATTGGACGAGATCCTG |
| F3′5'H_TOM_R | F3′5′H | AAGGAACCTCTCGGGAGTGAA |
| FLS_TOM_F | FLS | GAGCATGAAGTTGGGCCAAT |
| FLS_TOM_R | FLS | TGGTGGGTTGGCCTCATTAA |
| DFR_TOM_F | DFR | TCCGAAGACGACAACGGTTT |
| DFR_TOM_R | DFR | TGACAAGCCAAGAGCCGATAA |
| ANS_TOM_F | ANS | GAACTAGCACTTGGCGTCGAA |
| ANS_TOM_R | ANS | TTGCAAGCCAGGCACCATA |
| GT_TOM_F | GT | CGAACGACGAAACACTGTTGA |
| GT_TOM_R | GT | TGCAGCATAGATGGCATTGG |
| RT_TOM_F | RT | CTGGCAATGCAAACAGAGTGA |
| RT_TOM_R | RT | TCGACTTGCGGAAGAGTGAGA |
| ASR1_TOM_F | ASR1 | CCTGTTCCACCACAAGGACAA |
| ASR1_TOM_R | ASR1 | GTGCCAAGTTTACCGATTTGC |
Fluorescently labeled TaqMan probes.
SYBR-Green Analysis
As an alternative to the use of TaqMan probes, the fluorescent intercalating dye SYBR-Green can be used to monitor the RT-PCR on line with the ABI PRISM7700 sequence detection system. This dye gives a specific fluorescent signal when bound to double-stranded DNA, so fluorescence increases with the formation of PCR product. Although the sensitivity of SYBR-Green is at least as good as that for TaqMan probes, it is necessary that the PCR be specific, because the dye will bind equally well to aspecific PCR amplification products. Therefore, for each primer combination, the specificity of the amplification must be confirmed on an agarose gel. As for TaqMan probes, quantitation of target mRNA levels can be performed relative to an endogenous control RNA using the comparative Ct method.
Using sequence information from randomly chosen or specifically isolated tomato cDNAs (data not shown) or from ESTs and cDNAs present in the public domain databases, gene-specific primers were developed for tomato homologs of the genes encoding Phe-ammonia lyase, the flavonoid pathway genes chalcone synthase, chalcone isomerase, flavanone-3-hydroxylase, flavanone-3′-hydroxylase, flavanone-3′5′-hydroxylase, flavonol synthase, dihydroflavonol reductase, and anthocyanidin synthase, and the flavonol glycosyltransferases flavonol-3-glucosyltransferase and flavonol-3-glucoside-rhamnosyltransferase. All of these tomato homologs showed >90% amino acid identity to the established petunia genes. The sequences of all of the primers used are shown in Table 2.
Total RNA was isolated from turning fruit of tomato plants as described previously (Bovy et al., 1995). First-strand cDNA was synthesized from 1.5 μg of total RNA by reverse transcription. Aliquots of 100 ng of cDNA were used in SYBR-Green PCR (according to the manufacturer's protocol) with the primers mentioned above. As a control, we used primers specific for abscisic stress ripening gene1 (ASR1). Using the comparative Ct method, all genes were expressed relative to ASR1.
To compare the expression levels in leaves with those found in fruit, we made use of the constitutive expression of our internal controls in leaves and green fruit (CYP) and in the three ripening stages tested (ASR1). First, expression levels of each gene in turning fruit were expressed relative to their expression in green fruit using ASR1 as a control, according to the equation ΔΔCt(T-G) = ΔCt (turning fruit) − ΔCt (green fruit). Second, expression levels of each gene in leaves were expressed relative to their expression in green fruit using CYP as a control, according to the equation ΔΔCt(L-G) = ΔCt (leaves) − ΔCt (green fruit). Third, all genes can be expressed relative to leaves using the equation ΔΔCt(T-L) = ΔΔCt(T-G) − ΔΔCt(L-G). Finally, RNA expression in leaves was expressed as a percentage of the level of the corresponding gene in leaves, according to the equation 2 EXP ΔΔCt(T-L) × 100%.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Acknowledgments
We thank Ronald Davis for the kind provision of constructs pAL69, pAL144, and pAL71; Bob Cowper for organizing and performing the T1 and T2 trials; Carl Jarman for PCR screening of T1 and T2 seeds for segregation analysis; and Dirk Bosch for critically reading the manuscript.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.004218.
References
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20, 1195–1197. [DOI] [PubMed] [Google Scholar]
- Bongue-Bartelsman, M., and Philips, D.A. (1995). Nitrogen stress regulates gene expression of enzymes in the flavonoid biosynthetic pathway of tomato. Plant Physiol. Biochem. 33, 539–546. [Google Scholar]
- Bovy, A., van den Berg, C., de Vrieze, G., Thompson, W.F., Weisbeek, P., and Smeekens, S. (1995). Light-regulated expression of the Arabidopsis thaliana ferredoxin gene requires sequences upstream and downstream of the transcription initiation site. Plant Mol. Biol. 27, 27–39. [DOI] [PubMed] [Google Scholar]
- Bovy, A.G., Hijden, H.T., Hughes, S.G., Muir, S.R., Tunen, A.J., Verhoezen, M.E., and Vos, C.H. (1999). Methods and composition for modulating flavonoid content. W09937794 A 19990729.
- Bradley, J.M., Davies, K.M., Deroles, S.C., Bloor, S.J., and Lewis, D.H. (1998). The maize Lc regulatory gene up-regulates the flavonoid biosynthetic pathway of petunia. Plant J. 13, 381–392. [Google Scholar]
- Choi, S.U., Ryu, S.Y., Yoon, S.K., Jung, N.P., Park, S.H., Kim, K.H., Choi, E.J., and Lee, C.O. (1999). Effects of flavonoids on the growth and cell cycle of cancer cells. Anticancer Res. 19, 5229–5233. [PubMed] [Google Scholar]
- Cook, N.C., and Samman, S. (1996). Flavonoids: chemistry, metabolism, cardioprotective effects, and dietary sources. J. Nutr. Biochem. 7, 66–76. [Google Scholar]
- Crozier, A., Lean, M.E.J., McDonald, M.S., and Black, C. (1997). Quantitative analysis of the flavonoid content of commercial tomatoes, onions, lettuces, and celery. J. Agric. Food Chem. 45, 590–595. [Google Scholar]
- Damiani, R.D., Jr., and Wessler, S.R. (1993). An upstream open reading frame represses expression of Lc, a member of the R/B family of maize transcriptional activators. Proc. Natl. Acad. Sci. USA 90, 8244–8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K., Robbins, T.P., and Jorgensen, R.A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25, 173–199. [DOI] [PubMed] [Google Scholar]
- Dugas, A.J., Castaneda Acosta, J., Bonin, G.C., Price, K.L., Fischer, N.H., and Winston, G.W. (2000). Evaluation of the total peroxyl radical-scavenging capacity of flavonoids: Structure-activity relationships. J. Nat. Prod. 63, 327–331. [DOI] [PubMed] [Google Scholar]
- Duthie, G., and Crozier, A. (2000). Plant-derived phenolic antioxidants. Curr. Opin. Lipidol. 11, 43–47. [DOI] [PubMed] [Google Scholar]
- Fillatti, J.J., Kiser, J., Rose, R., and Comai, L. (1987). Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Bio. Technol. 5, 726–730. [Google Scholar]
- Frankel, E.N. (1999). Food antioxidants and phytochemicals: Present and future perspectives. Fett Lipid 101, 450–455. [Google Scholar]
- Goldsbrough, A., Belzile, F., and Yoder, J.I. (1994). Complementation of the tomato anthocyanin without (aw) mutant using the dihydroflavonol 4-reductase gene. Plant Physiol. 105, 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsbrough, A.P., Tong, Y., Yoder, J.I., and Tong, Y.S. (1996). Lc as a non-destructive visual reporter and transposition excision marker gene for tomato. Plant J. 9, 927–933. [Google Scholar]
- Grotewold, E., Chamberlin, M., Snook, M., Siame, B., Butler, L., Swenson, J., Maddock, S., St. Clair, G., and Bowen, B. (1998). Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10, 721–740. [PMC free article] [PubMed] [Google Scholar]
- Gynheung, A.N., Ebert, P.R., Mitra, A., and Ha, S.B. (1988). Binary vectors. In Plant Molecular Biology Manual, S.B. Gelvin, R.A. Schilperoort, and D.P.S. Verma, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. A3/1–A3/19.
- Harborne, J.B. (1986). The Flavonoids. Advances in Research Since 1986, 1st ed. (London: Chapmand Hall).
- Herrmann, K. (1979). Uebersicht ueber die Inhaltsstoffe der Tomaten. Z. Lebensm. Unters. Forsch. 169, 179–200. [Google Scholar]
- Hertog, M.G., Feskens, E.J., Hollman, P.C., Katan, M.B., and Kromhout, D. (1993). Dietary antioxidant flavonoids and risk of coronary heart disease: The Zutphen Elderly Study. Lancet 342, 1007–1011. [DOI] [PubMed] [Google Scholar]
- Hertog, M.G.L., and Hollman, P.C.H. (1996). Potential health effects of the dietary flavonol quercetin. Eur. J. Clin. Nutr. 50, 63–71. [PubMed] [Google Scholar]
- Hertog, M.G.L., Hollman, P.C.H., and Katan, M.B. (1992). Content of potentially anticarcinogenic flavonoids of 28 vegetables and 9 fruits commonly consumed in The Netherlands. J. Agric. Food Chem. 40, 2379–2383. [Google Scholar]
- Hoekema, A., van Haaren, M., Fellinger, A., Hooykaas, P., and Schilperoort, R. (1985). Non-oncogenic vectors: Plant vectors for use in the Agrobacterium binary system. Plant Mol. Biol. 5, 85–89. [DOI] [PubMed] [Google Scholar]
- Hollman, P.C.H., and Katan, M.B. (1999). Health effects and bioavailability of dietary flavonols. Free Radical Res. 31 (suppl. S), S75.–S80. [DOI] [PubMed] [Google Scholar]
- Iusem, N.D., Bartholomew, D.M., Hitz, W.D., and Scolnik, P.A. (1993). Tomato (Lycopersicon esculentum) transcript induced by water deficit and ripening. Plant Physiol. 102, 1353–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, K., Mensink, R.P., Cox, F.J., Harryvan, J.L., Hovenier, R., Hollman, P.C., and Katan, M.B. (1998). Effects of the flavonoids quercetin and apigenin on hemostasis in healthy volunteers: Results from an in vitro and a dietary supplement study. Am. J. Clin. Nutr. 67, 255–262. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusion: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E.T., Ryu, S., Yi, H., Shin, B., Cheong, H., Choi, G., Ryu, S.H., Yi, H.K., Shin, B.C., Cheong, H.S., and Choi, G.T. (2001). Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J. 25, 325–333. [DOI] [PubMed] [Google Scholar]
- Keli, S.O., Hertog, M.G., Feskens, E.J., and Kromhout, D. (1996). Dietary flavonoids, antioxidant vitamins, and incidence of stroke: The Zutphen Study. Arch. Intern. Med. 156, 637–642. [PubMed] [Google Scholar]
- Lloyd, A.M., Walbot, V., and Davis, R.W. (1992). Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258, 1773–1775. [DOI] [PubMed] [Google Scholar]
- Manach, C., Regerat, F., Texier, O., Agullo, G., Demigne, C., and Remesy, C. (1996). Bioavailability, metabolism and physiological impact of 4-oxo-flavonoids. Nutr. Res. 16, 517–544. [Google Scholar]
- Mol, J., Grotewold, E., and Koes, R. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3, 212–217. [Google Scholar]
- Mooney, M., Desnos, T., Harrison, K., Jones, J., Carpenter, R., and Coen, E. (1995). Altered regulation of tomato and tobacco pigmentation genes caused by the delila gene of Antirrhinum. Plant J. 7, 333–339. [Google Scholar]
- Muir, S.R., Collins, G.J., Robinson, S., Hughes, S.G., Bovy, A.G., de Vos, C.H., van Tunen, A.J., and Verhoeyen, M.E. (2001). Overexpression of petunia chalcone isomerase in tomato results in fruit containing dramatically increased levels of flavonols. Nat. Biotechnol. 19, 470–474. [DOI] [PubMed] [Google Scholar]
- Ng, T.B., Liu, F., and Wang, Z.T. (2000). Antioxidative activity of natural products from plants. Life Sci. 66, 709–723. [DOI] [PubMed] [Google Scholar]
- Oud, J.S.N., Schneiders, H., Kool, A.J., and van Grinsven, M. (1995). Breeding of transgenic orange Petunia hybrida varieties. Euphytica 84, 175–181. [Google Scholar]
- Price, K.R., Casuscelli, F., Colquhoun, I.J., and Rhodes, M.J.C. (1998). Composition and content of flavonol glycosides in broccoli florets (Brassica oleracea) and their fate during cooking. J. Sci. Food Agric. 77, 468–472. [Google Scholar]
- Rice Evans, C.A., Miller, N.J., Bolwell, P.G., Bramley, P.M., and Pridham, J.B. (1995). The relative antioxidant activities of plant-derived polyphenolic flavonoids. Free Radical Res. 22, 375–383. [DOI] [PubMed] [Google Scholar]
- Rice Evans, C.A., Miller, N.J., Paganga, G., and Miller, N. (1997). Antioxidant properties of phenolic compounds: The polyphenolic content of fruit and vegetables and their antioxidant activities. What does a serving constitute? Trends Plant Sci. 2, 152–159. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Shih, H., Pickwell, G.V., and Quattrochi, L.C. (2000). Differential effects of flavonoid compounds on tumor promoter-induced activation of the human CYP1A2 enhancer. Arch. Biochem. Biophys. 373, 287–294. [DOI] [PubMed] [Google Scholar]
- Sijmons, P.C., Dekker, B.M.M., Schrammeijer, B., Verwoerd, T.C., van der Elzen, P.J.M., and Hoekema, A. (1990). Production of correctly processed human serum albumin in transgenic plants. Biol. Technol. 8, 217–221. [DOI] [PubMed] [Google Scholar]
- Steinmetz, K.A., and Potter, J.D. (1996). Vegetables, fruit, and cancer prevention: A review. J. Am. Diet. Assoc. 96, 1027–1039. [DOI] [PubMed] [Google Scholar]
- Sugihara, N., Arakawa, T., Ohnishi, M., and Furuno, K. (1999). Anti- and pro-oxidative effects of flavonoids on metal-induced lipid hydroperoxide-dependent lipid peroxidation in cultured hepatocytes loaded with alpha-linolenic acid. Free Radical Biol. Med. 27, 1313–1323. [DOI] [PubMed] [Google Scholar]
- Trevisanato, S.I., and Kim, Y.I. (2000). Tea and health. Nutr. Rev. 58, 1–10. [DOI] [PubMed] [Google Scholar]
- van Engelen, F.A., Molthoff, J.W., Conner, A.J., Nap, J.P., Pereira, A., and Stiekema, W.J., (1995). pBINPLUS: An im-proved plant transformation vector based on pBIN19. Transgenic Res. 4, 288–290. [DOI] [PubMed] [Google Scholar]
- Woeldecke, M., and Herrmann, K. (1974). Flavonole und Flavone der Gemuesearten. III. Flavonole und Flavone der Tomaten und des Gemuesepaprikas. Z. Lebensm. Unters. Forsch. 155, 216–219. [DOI] [PubMed] [Google Scholar]
- Yoder, J.I., Belzile, F., Tong, Y., and Goldsbrough, A. (1994). Visual markers for tomato derived from the anthocyanin biosynthetic pathway. Euphytica 79, 163–167. [Google Scholar]