Abstract
The conversion of putrescine to spermidine in the biosynthetic pathway of plant polyamines is catalyzed by two closely related spermidine synthases, SPDS1 and SPDS2, in Arabidopsis. In the yeast two-hybrid system, SPDS2 was found to interact with SPDS1 and a novel protein, SPMS (spermine synthase), which is homologous with SPDS2 and SPDS1. SPMS interacts with both SPDS1 and SPDS2 in yeast and in vitro. Unlike SPDS1 and SPDS2, SPMS failed to suppress the speΔ3 deficiency of spermidine synthase in yeast. However, SPMS was able to complement the speΔ4 spermine deficiency in yeast, indicating that SPMS is a novel spermine synthase. The SPDS and SPMS proteins showed no homodimerization but formed heterodimers in vitro. Pairwise coexpression of hemagglutinin- and c-Myc epitope–labeled proteins in Arabidopsis cells confirmed the existence of coimmunoprecipitating SPDS1-SPDS2 and SDPS2-SPMS heterodimers in vivo. The epitope-labeled SPDS and SPMS proteins copurified with protein complexes ranging in size from 650 to 750 kD. Our data demonstrate the existence of a metabolon involving at least the last two steps of polyamine biosynthesis in Arabidopsis.
INTRODUCTION
Polyamines are abundant DNA and RNA binding organic cations that are implicated in the regulation of morphogenesis, stress responses, senescence, and other basic processes in higher plants (Tiburcio et al., 1990; Kumar et al., 1997; Malmberg et al., 1998). The diamine putrescine is synthesized by either Orn or Arg decarboxylase (ODC and ADC, respectively) and converted to spermidine and spermine through the sequential addition of aminopropyl residues by spermidine and spermine synthases (SPDS and SPMS, respectively) (Tabor and Tabor, 1984; Pegg, 1986; Cohen, 1998). Both latter reactions require decarboxylated S-adenosyl Met as an aminopropyl donor that is produced by SAM decarboxylase (SAMDC). In alkaloid-producing plants, putrescine also is N-methylated by putrescine N-methyltransferases, which catalyze the first committed steps in the biosynthesis of nicotine and tropane alkaloids (Kutchan, 1995).
Mutations in genes that control polyamine biosynthesis have different effects on cell proliferation and viability in diverse organisms (Balasundaram et al., 1994; Hamasaki-Katagiri et al., 1997, 1998). Recent analysis of the acaulis5 (acl5) mutation, which prevents stem elongation, demonstrates the important function of one spermine synthase gene (ACL5) in Arabidopsis (Hanzawa et al., 2000). Spermidine plays an important role as an aminobutyryl donor in post-translational modification of a lysyl residue of eukaryotic translation factor eIF-5A (Coffino, 2001). Although the levels of spermidine seem to be regulated primarily through its recycling to putrescine by acetyltransferases in animals (Pietilä et al., 1997), spermidine production correlates with the tissue-specific regulation of SAMDC and SPDS in plants (Malmberg et al., 1998). A coordinated reduction of spermidine, SAMDC, and SPDS levels during stress suggests that the regulation of SPDS activity plays an important role in the proper adjustment of plant polyamine levels (Yamanoha and Cohen, 1985; Tiburcio et al., 1993). Duplicated genes that encode two closely related and differentially regulated SPDS isologs (SPDS1 and SPDS2) have been identified in pea, Datura, and Hyoscyamus (Hashimoto et al., 1998; Alabadí and Carbonell, 1999). Three Arabidopsis SPDS-like gene sequences have been deposited in GenBank, although only one gene, AtSPDS (SPDS2), has been characterized (Hashimoto et al., 1998). The SPDS orthologs from Escherichia coli, rat, bovine, and human sources are thought to be dimers (Lakanen et al., 1995). Recently, the first crystal structure of a SPDS from Thermotoga maritima was solved (Korolev et al., 2002). This enzyme is a tetramer in the crystal and in solution, suggesting that the tetramer represents the functional oligomeric form. By contrast, the structural resolution of native plant SPDS remains enigmatic.
Enzymes that execute sequential metabolic transformations are known to form complexes called metabolons (Srere, 1987). Complex formation between two successive enzymes effectively channels substrates from one active site to the other and raises their local concentration in the vicinity of the active site of the second enzyme, ensuring a high rate of metabolic transformation. It also prevents the loss or dilution of substrate by diffusion and protects chemically labile intermediates (Ovadi and Srere, 2000). An early report on the purification of SPDS from alfalfa suggested that this enzyme might produce different polyamines, including spermine and other uncommon compounds, in addition to spermidine (Bagga et al., 1997). The fact that the different products were produced at different rates indicated that these reactions cannot be catalyzed by a single SPDS enzyme; most likely, an enzyme complex is required. However, these data did not provide evidence for the occurrence of such a multienzyme complex. In this work, we have shown that Arabidopsis SPDS isologs interact with a novel SPMS in the yeast two-hybrid system and in vitro, and that these aminopropyl transferases occur in multiprotein complexes in vivo.
RESULTS
Interactions between Arabidopsis Aminopropyl Transferases in the Yeast Two-Hybrid System and in Vitro
SPDS was identified as a homodimeric enzyme (73 kD) in E. coli (Bowman et al., 1973) and was isolated from human organs as a heterodimeric protein consisting of two different subunits with identical mass (each 35 kD) but slightly different pI (Kajander et al., 1989). Soybean SPDS was reported to be a monomer of 74 kD, whereas maize SPDS was purified as a monomer of 43 kD (for review, see Yoon et al., 2000). Recently, molecular studies identified highly conserved duplicated genes in Datura, Hyoscyamus, and pea that code for closely related SDPS proteins of 36 to 39 kD (Hashimoto et al., 1998; Alabadí and Carbonell, 1999). To determine whether plant SPDS homologs are capable of interacting by forming homodimers and/or heterodimers, and to possibly identify additional protein-interacting partners, we performed a protein interaction screen using the Arabidopsis SPDS2 protein as bait in fusion with the Gal4 DNA binding domain in a yeast two-hybrid system (Durfee et al., 1993; Harper et al., 1993). Transformation of the yeast strain Y190 carrying the bait construct pAS2.1-SPDS2 with a pACT2 Arabidopsis cell suspension cDNA library yielded 3.6 × 107 transformants, from which 86 grew in the presence of 50 mM of the His3 inhibitor 3-aminotriazole and displayed the LacZ+ phenotype, indicating protein interactions.
Characterization of the pACT2 cDNA clones by DNA sequencing identified two cDNA classes that code for SPDS2-interacting proteins. Both identified sequences were already annotated in the databases. The first class included full-length SPDS1 cDNAs (gene At1g70310), whereas the other class involved full-length cDNAs (gene At5g53120) coding for a gene product that had homology with aminopropyl transferases, which upon functional characterization (see below) we named SPMS. Recent data regarding protein structure from bacterial spermidine synthase has allowed the elucidation of residues involved in multisubstrate adduct recognition (Korolev et al., 2002). Comparison by ClustalW analysis of deduced protein sequences of our two-hybrid bait (SPDS2) and isolated preys (SPDS1 and SPMS) together with the structurally resolved sequence of T. maritima SPDS (Korolev et al., 2002) showed a high degree of conservation for all residues involved in the recognition of the artificial inhibitor 5-adenosyl-1,8-diamino-3-thiooctane (Figure 1A). Moreover, the apparently conserved D101 allows the discrimination of putative methyl transferases, reducing the possibility that either SPDS2 or SPMS might code for this type of enzyme.
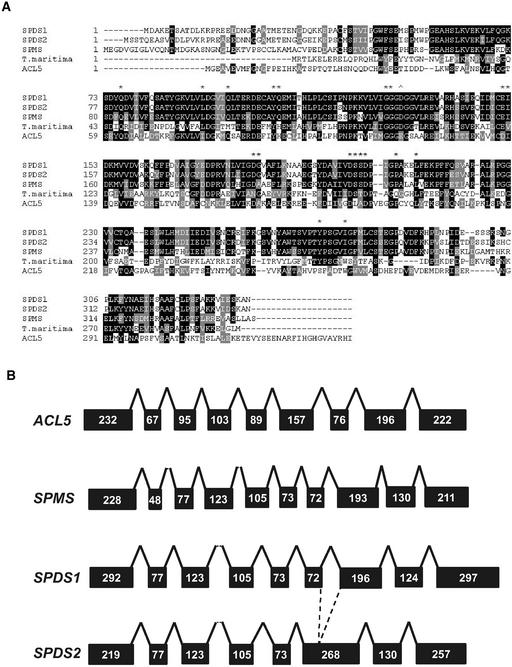
Figure 1.
Amino Acid Sequence Comparison and Genomic Organization of Arabidopsis Aminopropyl Transferase Proteins.
(A) Comparison of amino acid sequences of Arabidopsis and T. maritima SPDS proteins. The SPDS protein sequences were aligned and compared by Baylor College of Medicine Search Launcher (http://searchlauncher.bcm.tmc.edu). Black shading indicates identical amino acid residues, whereas similarities are highlighted in gray. Asterisks indicate the positions of residues that interact with the artificial inhibitor 5-adenosyl-1,8-diamino-3-thiooctane. The arrowhead indicates a residue of putrescine N-methyltransferase that deviates from the conserved residues of SPDS.
(B) Scheme of the intron/exon structure of the genomic regions of the SPDS1, SPDS2, SPMS, and ACL5 genes. Black boxes and solid lines represent exons and introns, respectively. Numbers refer to the nucleotides in each exon.
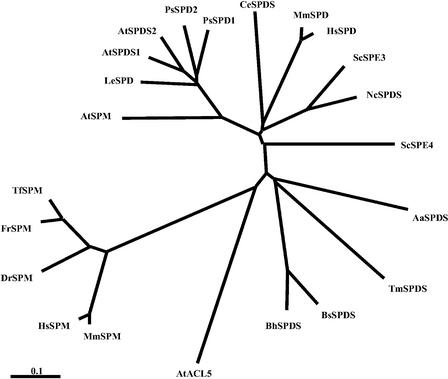
Estimation of pairwise comparisons yielded the highest similarity for SPDS1 and SPDS2 (82.7% amino acid identity), whereas SPMS showed only 56.7 and 56.6% identity with SPDS1 and SPDS2, respectively. The pI (5.73) and charge (−8.55 at pH 7.0) of SPMS differed considerably from those of SPDS1 (4.61 and −18.29) and SPDS2 (4.62 and −18.32). Comparison of the genomic sequences of spermidine and spermine synthase genes showed that although exon structure is conserved between SPDS1, SPDS2, and SPMS, the ACL5 genomic sequence is organized in a completely different pattern (Figure 1B). This observation, together with tree analysis (Figure 2), suggests that SPMS defines a new family close to the known plant SPDS that is divergent from the spermine synthase cluster, which includes Arabidopsis ACL5 sequences, indicating a possible paralog for the SPMS gene. BLAST searches identified SPDS1, SPDS2, SPMS, and ACL5 as the only members of the conserved aminopropyl transferase gene family in the sequenced Arabidopsis genome.
Figure 2.
Distance Relationship among Spermidine and Spermine Synthases of Diverse Origins.
Protein sequences were aligned using ClustalX, and the tree was generated using the neighbor-joining method. Origins of the proteins are as follows: AtACL5 (Arabidopsis), AtSPDS1 (Arabidopsis), AtSPDS2 (Arabidopsis), AtSPMS (Arabidopsis), ScSPE3 (Saccharomyces cerevisiae), ScSPE4 (S. cerevisiae), MmSPDS (Mus musculus), MmSPMS (M. musculus), HsSPDS (Homo sapiens), HsSPMS (H. sapiens), DrSPMS (Danio rerio), FrSPMS (Fugu rubripes), TfSPMS (Tetraodon fluviatilis), PsSPDS1 (Pisum sativum), PsSPDS2 (P. sativum), LeSPDS (Lycopersicon esculentum), TmSPDS (T. maritima), BsSPDS (Bacillus subtilis), BhSPDS (Bacillus holodurans), AaSPDS (Aquifex aeolicus), CeSPDS (Caenorhabditis elegans), and NcSPDS (Neurospora crassa).
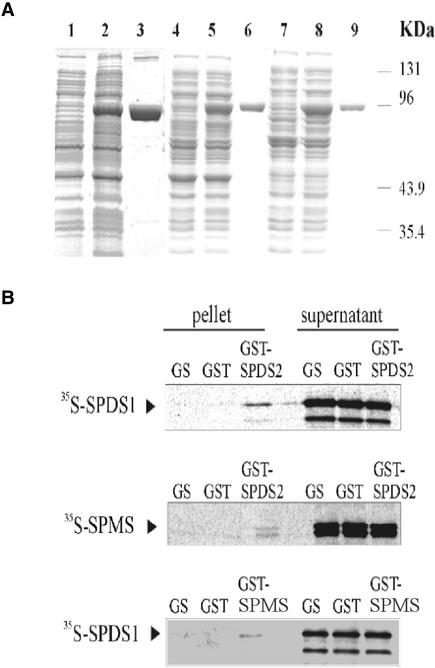
To confirm the results of primary two-hybrid screens, the coding regions of SPDS1, SPDS2, and SPMS cDNAs were inserted into the bait (pAS2.1) and prey (pACT2) vectors (Durfee et al., 1993; Harper et al., 1993), and the interaction of each bait was tested with each prey in the yeast two-hybrid system using LacZ+ filter-lift assays (Figure 3). The results showed that none of the proteins was capable of interacting with itself to form a homodimer, but all combinations of different baits and preys activated the LacZ reporter gene, indicating heterodimerization capabilities for all tested proteins. To support these results, in vitro protein interaction assays were performed. The coding domains of SPDS1, SPDS2, and SPMS cDNAs were fused to N-terminal glutathione S-transferase (GST) tags in pGEX expression vectors (Ausubel et al., 1989), and each fusion protein was purified from E. coli BL21 to apparent homogeneity by affinity chromatography on glutathione-Sepharose (GS matrix; Figure 4A). GST-SPDS2, GST-SPMS, and control GST proteins were immobilized on GS matrix, whereas SPDS1 and SPMS were labeled with 35S-Met by coupled transcription-translation in vitro. Equal amounts of labeled SPDS1 protein were incubated with GST-SPDS2 and GST beads as well as with control GS matrix. After removal of the supernatant and stringent washes, the matrix-bound proteins were eluted, size separated by SDS-PAGE together with the supernatant fractions, and visualized by autoradiography. Similar pulldown assays were performed with GST-SPDS2 and 35S-labeled SPMS and with GST-SPMS and 35S-labeled SPDS1 (Figure 4B). The results of these GST pulldown experiments demonstrated that GST-SPDS2 recruited SPDS1 and SPMS, whereas GST-SPMS bound SPDS1 in vitro. Translation of SPDS1 and SPMS proteins resulted in several N-terminally truncated products, the sizes of which corresponded to the positions of internal Met codons in the sequences. In addition to full-length translation products, GST-SPDS2 bound weakly the truncated forms of SPDS1 and SPMS in vitro, suggesting that the heterologous N-terminal domains of SPDS proteins may not be essential for their interactions.
Figure 3.
Yeast Two-Hybrid Interactions between SPDS and SPMS.
Yeast transformants containing either GAL4 binding domain (GBD) or GAL4 activation domain (GAD) fusion proteins were grown on nylon filters, placed on synthetic medium, and tested for β-galactosidase activity with the LacZ filter-lift assay. Photographs were taken after 12 h of β-galactosidase enzymatic reaction.
Figure 4.
Interactions of SPDS and SPMS Proteins in Vitro.
(A) The SPDS and SPMS proteins were fused to N-terminal GST tags using pGEX vectors and purified from E. coli using glutathione-Sepharose affinity chromatography. Quality tests of total protein extracts from E. coli before (lanes 1, 4, and 7) and after (lanes 2, 5, and 8) induction of GST-SPDS/SPMS expression with isopropylthiogalactoside, and that of purified GST-SPDS/SPMS proteins (lanes 3, 6, and 9), were performed by SDS-PAGE. Lanes 1 to 3, GST-SPDS1; lanes 4 to 6, GST-SPMS; and lanes 7 to 9, GST-SPDS2.
(B) SDPS1 and SPMS labeled with 35S-Met were subjected to pulldown assay with GST-SPDS2. Beads carrying immobilized GST-SPDS2 and control GST proteins, as well as the control glutathione-Sepharose matrix (GS), were incubated with equal amounts of labeled SPDS proteins. The eluted matrix-bound proteins and aliquots from the supernatants were separated by SDS-PAGE and detected by autoradiography. A similar pulldown assay was performed using 35S-SPDS1 and GST-SPMS.
Aminopropyl Transferase Activities Determined by Functional Yeast Complementation
Mutations in genes that affect spermidine and spermine biosynthesis have been studied in Saccharomyces cerevisiae. The speΔ3 mutation yields yeast cells with a growth dependence on spermidine supplementation (Hamasaki-Katagiri et al., 1997). This phenotypic consequence of the speΔ3 mutation facilitated the identification by heterologous complementation of pea SPDS cDNAs (Alabadí and Carbonell, 1999), demonstrating that plant SPDS enzymes are functional orthologs of yeast Spe3p. To perform a similar assay, the coding domains of Arabidopsis SPDS cDNAs were inserted downstream of a constitutive actin promoter in YEpACT and expressed in the speΔ3 yeast mutant as well as in the isogenic SPE3 control strain 2602. After selection of transformants using the LEU2 marker, the strains were grown in complete medium, cultures were diluted in spermidine-free medium every 24 h, and their growth was monitored by measuring OD600 (Figure 5A). Strains carrying the SPDS1 and SPDS2 constructs grew as well as the wild-type control, whereas transformants harboring YEpACT-SPMS showed growth arrest in the speΔ3 mutant with the control empty vector after 3 to 4 days in culture. Supplementation of these cultures with spermidine resulted in fully restored growth.
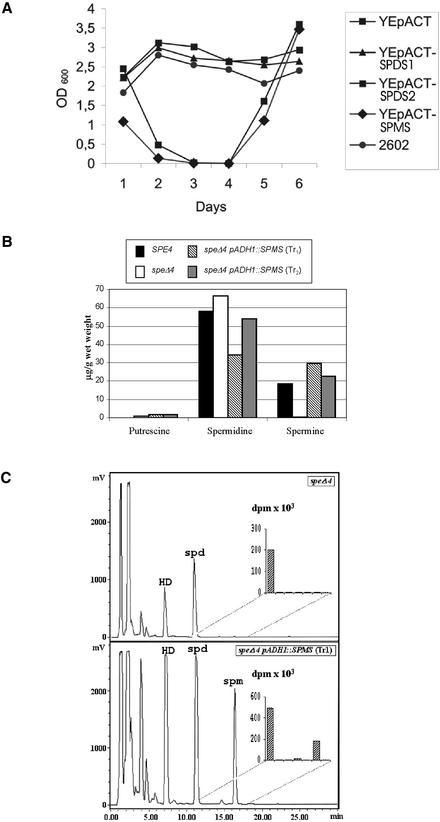
Figure 5.
Yeast Complementation Studies.
(A) Complementation test with the yeast speΔ3 mutant. The growth of the speΔ3 yeast strain carrying the Arabidopsis SPDS expression vectors YEpACT-SPDS1, YEpACT-SPDS2, and YEpACT-SPMS, and the empty control vector YEpACT, was monitored after diluting the cultures 1:500 every 24 h and measuring OD600 as described (Alabadí and Carbonell, 1999). SPE3 strain 2602 was used as a control. After 4 days in culture, 100 μM spermidine was added to the medium of YEpACT and YEpACT-SPMS transformants. The OD600 values correspond to the mean of three independent assays.
(B) Complementation test with the yeast speΔ4 mutant. Putrescine, spermidine, and spermine contents were determined, as described in Methods, in the extracts of the yeast wild type (strain 2602), the spe4 mutant (Y504), and the speΔ4 mutant transformed with the Arabidopsis SPMS cDNA under the control of the yeast ADH1 promoter.
(C) Conversion of 14C-spermidine into 14C-spermine in the yeast speΔ4 mutant transformed with the Arabidopsis SPMS cDNA under the control of the yeast ADH1 promoter. Yeast cells were incubated with 14C-spermidine, and radioactivity was quantified in fractions after HPLC separation of dansylated polyamines. The inset shows the radioactivity of the HPLC fractions collected every 1 min during the interval 11 to 18 min. HD, 1,6-diamine hexane; spd, spermidine; spm, spermine.
These data demonstrated that SPMS was incapable of suppressing the speΔ3 mutation, whereas both SPDS1 and SPDS2 complemented the deficiency of SPDS activity in yeast. The possibility that SPMS might encode a protein with spermine synthase activity was investigated using the yeast speΔ4 mutant defective in spermine synthesis (Hamasaki-Katagiri et al., 1998). Because spermine is not essential for yeast growth, polyamine content was determined in extracts of the wild type, speΔ4 mutant, and spe4 mutant transformed with the Arabidopsis SPMS gene under the control of the yeast ADH1 promoter. As shown in Figure 5B, SPMS complemented the lack of spermine synthesis in speΔ4 mutants. Incubation of the transformants with 14C-spermidine resulted in the production of 14C-spermine, indicating that the spermine detected in the extracts was the result of the enzymatic conversion of spermidine (Figure 5C). All of these results confirmed that both SPDS1 and SPDS2 are capable of providing spermidine synthase activity, whereas SPMS can be considered a functional spermine synthase.
Assembly of Enzyme Heterodimers and Formation of Multiprotein Complexes in Vivo
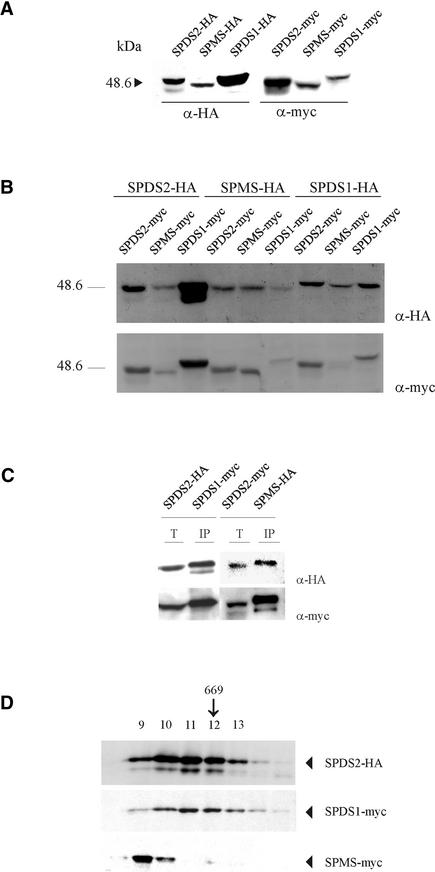
To validate the results obtained by yeast two-hybrid screens and in vitro protein binding assays, we exploited a recently developed technology for the expression of double epitope-tagged proteins in plant cells (Ferrando et al., 2000, 2001). The coding regions of SPDS1, SPDS2, and SPMS cDNAs were inserted into plant intron-tagged epitope-labeling vectors to generate protein fusions with C-terminal hemagglutinin (HA) and c-Myc epitope tags. Each individual HA and c-Myc fusion protein was expressed in Arabidopsis cells by Agrobacterium tumefaciens–mediated transformation (Ferrando et al., 2001). After 6 days of Agrobacterium infection, the expression and purification of epitope-tagged proteins was monitored by protein gel blot analysis and immunopurification. To test the specificity of immunoassays, the SPDS1, SPDS2, and SPMS-HA epitope-tagged proteins were expressed individually in Arabidopsis cells, and cell extracts were prepared and bound to an anti-c-Myc IgG matrix. The matrix-bound proteins were eluted specifically with c-Myc peptide and, together with aliquots of total cell extracts, were immunoblotted with anti-HA and anti-c-Myc antibodies. All HA-tagged proteins were detectable readily with anti-HA IgG, but no artificial binding to the anti-c-Myc IgG matrix and no cross-reaction of anti-c-Myc IgG with HA-tagged proteins were observed in this control experiment (data not shown). Subsequently, combinations of SPDS1-myc and SPDS2-HA, SPDS2-myc and SPMS-HA, and SPMS-myc and SPDS1-HA proteins were coexpressed in Arabidopsis cells.
A comparison of the protein expression levels for single or double transformations is shown in Figures 6A and 6B. Unlike the similar expression patterns obtained with single transformations, variable amounts were obtained in some co-transformation experiments, as was evident for the SPDS1-SPMS combination with a reduced amount of expressed tagged proteins. Protein extracts prepared from these transformed cell cultures were subjected to immunoaffinity purification on anti-c-Myc IgG matrix. Immunoblotting of the eluted matrix-bound proteins and samples of total protein extracts with anti-HA and anti-c-Myc antibodies showed that SPDS1-myc copurified with an equimolar amount of SPDS2-HA (Figure 6C). Similarly, SPDS2-myc immunoprecipitated an equimolar amount of SPMS-HA. The appearance of additional bands probably reflects the presence of degradation products, as reported previously (Ferrando et al., 2001). The association of SPMS-myc with SPDS1-HA was not detected, possibly as a result of the low expression levels for this pair combination.
Figure 6.
Immunological Detection of Epitope-Labeled SPDS and SPMS.
The formation of heterodimers and protein complexes in Arabidopsis cell extracts is shown.
(A) Individual expression of epitope-tagged SPDS and SPMS in plant cells. Total protein extracts from Arabidopsis cells transformed with the indicated constructs were immunoblotted with anti-HA and anti-c-Myc antibodies.
(B) Effect of double transformations on protein expression levels. Arabidopsis cells were transformed simultaneously as indicated, and total protein extracts were used for immunoblot analysis with anti-HA and anti-c-Myc antibodies. Remarkably, all pairwise combinations of SPDS1 and SPMS yielded very low protein expression levels.
(C) Detection of SPDS heterodimers formed in vivo. Epitope-labeled SPDS1-myc and SPDS2-HA, as well as SPDS2-myc and SPMS-HA, proteins were coexpressed in Arabidopsis cells. The cell extracts were subjected to immunoaffinity purification on anti-c-Myc IgG beads. Aliquots from the total protein extracts (T) and eluted matrix-bound proteins (IP) were immunoblotted with anti-HA and anti-c-Myc antibodies.
(D) Superose 6 size fractionation of protein extracts prepared from Arabidopsis cells expressing SPDS2-HA, SPDS1-myc, and SPMS-myc proteins. Protein complexes carrying the epitope-labeled proteins were assayed by immunoblotting with anti-HA and anti-c-Myc antibodies. The arrow indicates the position of the molecular mass marker thyroglobulin (669 kD).
To determine the sizes of native SPDS complexes, the cell extracts were subjected to size fractionation on a Superose 6 column, which was calibrated previously with molecular mass standards (Figure 6D). Immunoblotting of column fractions with anti-HA and anti-c-Myc antibodies revealed that SPDS2-HA and SPDS1-myc eluted between 650 to 750 kD, whereas SPMS-myc was detected in overlapping fractions between 700 and 750 kD. Because the expected size of protein dimers was ∼80 kD, these results indicate that the Arabidopsis spermidine and spermine synthases described here occur in multiprotein complexes in vivo.
DISCUSSION
Our current knowledge of the regulation of polyamine biosynthesis in higher plants is based largely on the characterization of gene expression patterns and the physiological measurement of polyamine distribution in diverse plant organs. Because of the pharmaceutical importance of polyamine-derived alkaloids, biotechnological approaches aim to modify the accumulation and distribution of polyamines using overexpression and antisense inhibition of ADC, ODC, SPDS, SPMS, and SAMDC genes in various species (for review, see Kumar et al., 1997; Malmberg et al., 1998). These approaches revealed that the altered expression of polyamine biosynthesis genes often results in growth retardation, sterility, and other developmental defects. An important role played by polyamines in the regulation of plant development was illustrated by the recent characterization of a mutation in the Arabidopsis SPMS gene ACL5 (Hanzawa et al., 2000). Several genes that control plant polyamine biosynthesis appear to be controlled stringently by stress signaling (Watson and Malmberg, 1996; for review, see Shoji et al., 2000). Polyamines, chiefly spermidine, also were found to modulate the activity of KAT1-like inward-rectifying K+ channels and thereby to regulate the closure of gas-exchange cells (i.e., stomata) during stress (Liu et al., 2000).
Several lines of evidence suggest that in plants the regulation of SPDS function during stress could play an important role. It has been suggested that plant SPDS may occur in the form of a multienzyme complex (Bagga et al., 1997). Thus, characterization of SPDS in crude alfalfa enzyme extracts showed that although alfalfa SPDS was highly specific for putrescine as the initial substrate, extended enzymatic reaction produced, besides spermidine, spermine and several uncommon polyamines (Bagga et al., 1997). These authors postulated the existence of an enzymatic complex to explain the formation of spermine and other minor polyamines.
Substrate channeling is the process of direct transfer of an intermediate between the active sites of two enzymes that catalyze sequential reactions in a biosynthetic pathway (Srere, 1987; Spiney and Ovady, 1999). The active sites can be located either on separate domains in a multifunctional enzyme or on separate subunits of a multienzyme complex named metabolon. The potential advantages of substrate channeling include the following: (1) it prevents or impedes the loss of intermediates by diffusion; (2) it decreases the transit time required for an intermediate to reach the active site of the next enzyme; (3) it reduces the transit time for the system to reach the new steady state; (4) it protects chemically labile intermediates; (5) it circumvents unfavorable equilibria; and (6) it segregates the intermediates of competing chemical and enzymatic reactions and provides new means of metabolic regulation by the modulation of enzyme associations (Spiney and Ovady, 1999; Ovadi and Srere, 2000). Substrate channeling has been well documented, with many enzymes from prokaryotic and eukaryotic organisms involved in different metabolic pathways (reviewed by Ovadi and Srere, 2000). There are several examples of both multifunctional proteins and multienzyme complexes within enzymes involved in amino acid metabolism (Srere, 1987). Therefore, it is likely that substrate channeling also occurs in sequential enzymatic reactions of the polyamine biosynthetic pathway.
We performed a two-hybrid screen with Arabidopsis SPDS2 and found that it can interact with SPDS1 in yeast. Unexpectedly, the two-hybrid screen also identified a novel SPDS-like protein, SPMS, which shows different pI and charge compared with SPDS1 and SPDS2. SPMS interacts with both SPDS1 and SPDS2 in yeast and in vitro. SPMS, unlike SPDS1 and SPDS2, does not complement the yeast speΔ3 mutation, but it restores the spermine-deficient content caused by the yeast speΔ4 mutation. This finding demonstrates that SPDS1 and SPDS2 are enzymatically active SPDS isologs, whereas SPMS is a new spermine synthase. ClustalX analysis and intron/exon structures indicate that SPMS is related more closely to SPDS1 and SPDS2 than to the ACL5 isolog of SPMS. In fact, ACL5 was not identified in the original two-hybrid screen using SPDS2 as a bait under screening saturation conditions. Moreover, no interaction was found in a direct two-hybrid test with ACL5 against both SPDS1 and SPDS2 (data not shown). Thus, it seems that spermine synthases in Arabidopsis are encoded by genes of different origins, with one of them, SPMS, being the result of a relatively recent duplication of an ancestral spermidine synthase gene. The yeast two-hybrid and in vitro protein interaction tests indicated that the Arabidopsis SPDS and SPMS proteins cannot form homodimers but heterodimerize with each other in all combinations.
To determine whether the dimerization of SPDS and SPMS subunits occurs in vivo, we coexpressed combinations of HA- and c-Myc epitope–labeled subunits in Arabidopsis cells. The detection of both HA and c-Myc epitopes after immunoaffinity purification of c-Myc epitope–labeled subunits showed that SPDS1-SPDS2 and SPDS2-SPMS heterodimers are formed in vivo but failed to demonstrate the existence of an SPDS1-SPMS complex, probably as a result of technical limitations of the system, such as low protein expression levels. In fact, an evident reduction in the protein levels for the pair combination SPDS1-SPMS was observed independently of the epitope-tagged subunit (Figure 6B). Although the reason of this observation remains unclear, it may account for the limitation in the coimmunopurification of both proteins. Intriguingly, size fractionation of epitope-labeled SPDS and SPMS proteins revealed that they all copurify within large multiprotein complexes of 650 to 750 kD. Together, these results indicate the existence of a metabolon involving at least the last two steps of polyamine biosynthesis in Arabidopsis.
There are some examples of polyamine multifunctional enzymes. In Plasmodium falciparum, ODC and SAMDC activities are located on a single polypeptide, with the SAMDC domain in the N-terminal part connected to the C-terminal ODC domain through a hinge region (Müller et al., 2000). It was discussed that the bifunctional organization might provide a biological advantage compared with monofunctional proteins because it could either facilitate the synthesis of products or exhibit regulatory functions on the activity of a partner enzyme domain (Müller et al., 2000). In a previous report, the existence of a multifunctional enzyme involved in the synthesis of putrescine was proposed in Lathyrus sativus and Cucumis sativus (Srivenugopal and Adiga, 1981). However, experimental data indicating the existence of a metabolon carrying multiple enzymes from the polyamine biosynthesis pathway were missing. The data presented here provide evidence for such supramolecular organization.
Spatial organization is a fundamental aspect of most cellular processes. This includes metabolic systems in which catalytic efficiency and control of end-product specificity can be enhanced by the assembly of enzymes into macromolecular complexes (Burbulis and Winkel-Shirley, 1999). Therefore, it is possible that spermidine formed by the SPDS-catalyzed reaction may be channeled effectively to SPMS, modulating the formation of the end product of the pathway. This finding suggests the existence of a tight connection between SPDS and SPMS that may favor the formation of spermine.
Resolution of the first crystal structure of an aminopropyl transferase has revealed that T. maritima SPDS is a tetramer (Korolev et al., 2002). It was suggested that tetramerization of SPDS monomers possibly underlies their thermostability, indicating that these enzymes may need some type of quaternary organization to increase protein stability (Korolev et al., 2002). The analysis of immunopurified protein complexes by matrix-assisted laser-desorption ionization is expected to reveal additional elements of the SPDS/SPMS metabolon.
METHODS
Analysis of Protein Interactions in the Yeast Two-Hybrid System
A full-length SPDS2 cDNA was amplified by PCR from a previously described Arabidopsis thaliana cDNA library (Németh et al., 1998) using the primer pair SPDS2F1 (5′-GGGGAATTCATGTCTTCAACACAAGAA-3′) and SPDS2R1 (5′-TTCCTGCAGGTTGGCTTTCGAATC-AAT-3′). The cDNA was sequenced and inserted as an EcoRI-PstI fragment into the yeast two-hybrid vector pAS2.1 (Harper et al., 1993) to generate an SPDS2 bait in fusion with the Gal4 DNA binding domain (GBD). After transformation of yeast Y190 with the pAS2.1-SPDS2 construct, the expression of the GBD-SDPS2 protein was monitored by immunoblot analysis with anti-Gal4p-DB antibody (Clontech, Palo Alto, CA). Subsequently, the yeast strain was transformed with 0.3 mg of DNA from a pACT2 Arabidopsis cell suspension cDNA library (Németh et al., 1998). A total of 3.6 × 107 transformants were selected on synthetic dextrose medium containing 50 mM 3-aminotriazole and lacking Leu, Trp, and His (Durfee et al., 1993), inoculated on nylon filters, and grown on synthetic dextrose plates with 25 mM 3-aminotriazole to verify their LacZ+ phenotype using β-galactosidase filter-lift assays (Durfee et al., 1993). pACT2 plasmids were isolated from the LacZ+ clones and transformed into yeast Y190 strains that carried no bait or harbored pAS2.1-SPDS2 or a negative control plasmid, such as pAS2.1-lamin (Matchmaker System; Clontech) or pAS2.1-Skp1 (Farràs et al., 2001).
To perform other confirmatory protein interaction tests, the SPMS coding region was cloned from pACT2-SPMS in pAS2.1 as a BamHI-XhoI fragment. Yeast strain Y190 carrying the pAS2.1-SPMS bait was transformed with pACT2-SPDS1, pACT2-SPDS2, or pACT2-SPMS to test for homodimerization and heterodimerization of SPMS. Analogously, the SPDS1 coding sequence was cloned in pAS2.1 as a PstI-EcoRI fragment after PCR amplification with the primer pair SPDS1F1 (5′-GCAGAATTCATGGACGCTAAAGAAACC-3′) and SPDS1R1 (5′-GGGCTGCAGACGATCCTCCAGATTAGT-3′). The yeast strain expressing the GBD-SPDS1 bait was transformed with pACT2-SPDS1, pACT2-SPDS2, and pACT2-SPMS. Finally, the SPDS2 coding region was amplified by PCR using the primer pair SPDS2F2 (5′-GCGGAATTCCCATGTCTTCAACCAAGAA-3′) and SPDS2R2 (5′-GGGCTC-GAGGGGCAAGCAGAAAGCAGC-3′), cloned as an EcoRI-XhoI fragment in pACT2, and tested for interaction with the bait constructs pAS2.1-SPDS1, pAS2.1-SPDS2, and pAS2.1-SPMS.
The ACL5 coding sequence was amplified by PCR with the primer pair ACLF (5′-GGGGAATTCATGGGTGAAGCCGTAGAGG-3′) and ACLR (5′-CCCCAGCTGAATATGCCGGTACGCCACAC-3′) and cloned into pGEM vector (Promega). Subsequently, the EcoRI-SalI fragment containing the ACL5 coding sequence was moved into pAS2.1, and the resulting NcoI-SalI fragment was subcloned further into pACT2 digested with NcoI and XhoI to yield the pACT2-ACL5 construct. A direct two-hybrid test was performed with pACT2-ACL5 against pAS2.1-SPDS1 and pAS2.1-SPDS2.
Analysis of Protein Interactions in Vitro
The coding domain of SPDS2 was cloned from pAS2.1-SDPS2 in pGEX-5X-2 (Pharmacia) as a XhoI-NotI fragment to express a glutathione S-transferase (GST)–SPDS2 fusion protein, which was purified from Escherichia coli BL21 to apparent homogeneity using glutathione-Sepharose (Ausubel et al., 1989). The SPDS2 and SPMS coding regions were moved as EcoRI-XhoI fragments from the pACT2-SPDS clones into pBluescriptII SK+ and used as templates to synthesize 35S-Met–labeled proteins by coupled in vitro transcription and translation (Promega). GST-SPDS2 and control GST proteins were immobilized on glutathione-Sepharose. Equal aliquots of 35S-labeled SPDS1 and SPMS proteins were incubated with GST-SPDS2, GST, and control Sepharose beads in binding buffer (20 mM Tris-HCl, pH 7.0, 137 mM NaCl, and 0.1% Igepal CA-630; Sigma, St. Louis, MO) for 1 h at 4°C. After washing the beads three times with binding buffer, the matrix-bound proteins were eluted with SDS sample buffer (Ausubel et al., 1989) and, together with the supernatant fractions, were size fractionated on SDS-PAGE gels to detect the labeled SPDS1 and SPMS proteins by autoradiography. The SPMS coding region was moved as an EcoRI-XhoI fragment from pACT2-SPMS into pGEX-4T-2 (Pharmacia) to similarly purify a GST-SPMS fusion protein. In vitro binding assays with GST-SPMS and 35S-labeled SPDS1 proteins were performed as described above.
Complementation Assays with the Yeast speΔ3 and speΔ4 Null Mutants
To express the Arabidopsis SPDS proteins in the yeast spermidine synthase null mutant speΔ3 (Hamasaki-Katagiri et al., 1997), the SPDS1 and SPMS coding sequences were moved from pACT2-SPDS plasmids as EcoRI-XhoI fragments into pBluescriptII SK. The resulting plasmids were linearized by XhoI and then treated with a DNA polymerase Klenow fragment and PstI to obtain cDNA fragments that were cloned into PstI and filled-in HindIII sites downstream of a yeast actin promoter in YEpACT (Németh et al., 1998). The SPDS2 coding sequence was cloned similarly in pBluescriptII SK+ to isolate a cDNA fragment with SalI and filled-in HindIII ends that was cloned into XhoI and filled-in NotI sites of YEpACT. The SPDS expression vectors and control empty vector YEpACT were introduced into yeast strain YN158 (MATα his6 leu2 ura3-52 speΔ3::URA3; kindly provided by N. Hamasaki-Katagiri [Hamasaki-Katagiri et al., 1997] and H. Tabor [National Institutes of Health, Bethesda, MD]) using the lithium acetate transformation method (Gietz and Sugino, 1988). Transformants were selected in minimal medium (2% Suc and 0.7% yeast nitrogen base without amino acids [Difco]) supplemented with His (30 mg/L) and spermidine (100 μM; free of other polyamines) at 30°C. In the suppressor assays, the transformants carrying YEpACT-SPDS and YEpACT vectors were grown in minimal medium without spermidine and diluted 1:500 in fresh medium every 24 h. To revert the phenotype, the medium was supplemented with 100 μM spermidine (Alabadí and Carbonell, 1999). An isogenic wild-type yeast strain, 2602 (MATα his6 leu2 ura3-52 SPE3), was used as a control (Hamasaki-Katagiri et al., 1997).
For the complementation of speΔ4 mutants, the EcoRI-XhoI fragment containing the SPMS cDNA was cloned into the filled-in HindIII restriction site in pAN10 (Navas et al., 1993) and transformed into Y504 (MATα his6 leu2 ura3-52 speΔ4::LEU2) (Hamasaki-Katagiri et al., 1998). Aliquots of 200 mg (wet weight) from yeast cultures were centrifuged, and the pellet was resuspended in 500 μL of 0.2 M perchloric acid containing 1,6-diaminohexane as an internal standard (0.5 μmol/g). Detection and quantification of polyamines was performed by HPLC (Carbonell and Navarro, 1989). Conversion of spermidine into spermine by SPMS was assayed by incubating 25 mL of a yeast culture (7 mg/mL yeast wet weight) with 5 μCi of 14C-spermidine (Amersham) for 6 h. Culture density after this time was 16 mg/mL. Aliquots of 400 mg of yeast cells were collected by centrifugation, and polyamines were extracted, dansylated, and fractionated by HPLC. Radioactivity in fractions was determined with a liquid scintillation counter.
Double Epitope Labeling and Expression of SPDS and SPMS Proteins in Arabidopsis Cells
The expression cassette of the intron-tagged c-Myc epitope-labeling vector pGIGI (Ferrando et al., 2001) was inserted as a blunt-ended NotI fragment into filled-in EcoRI-BamHI sites of the Agrobacterium tumefaciens binary vector pPCV002 (Koncz et al., 1994) to yield pPCV002-GIGI. The expression cassette of the intron-tagged hemagglutinin (HA) epitope-labeling vector pMENCHU (Ferrando et al., 2000) was cloned as a blunt-ended Tth111-SfiI fragment into filled-in EcoRI-SacI sites of the binary vector pPCV812 (Koncz et al., 1994) to yield pPCV812-MENCHU. To generate SPDS2 fusions with C-terminal c-Myc and HA epitopes, the SPDS2 coding sequence was amplified by PCR using the primer pair SPDS2F3 (5′-GTTCCCGGGATG-TCTTCAACACAAGAA-3′) and SPDS2R3 (5′-GGGTCTAGAGTT-GGCTTTCGAATCAAT-3′) and cloned as an SmaI-XbaI fragment in the expression cassettes of pPCV002-GIGI and pPCV812-MENCHU. Analogously, the SPMS coding sequence was amplified by PCR using the primer pair SPMSF1 (5′-GGGAAGCTTATGGAGGGA-GACGTCGGA-3′) and SPMSR1 (5′-GTTAGATCTCTTCTCTCC-GCAGGATGT-3′) and cloned as an HindIII-BglII fragment in pLOLA (Ferrando et al., 2001). The SPMS-myc expression cassette from pLOLA was inserted as an XhoI-SacI fragment into SalI-SacI sites of pPCV812. The PCR-amplified SPMS HindIII-BglII fragment was cloned in pPCV812-MENCHU to generate an SPMS-HA fusion. The SPDS1 coding sequence was amplified by PCR using the primers SPDS1F2 (5′-GTTCCCGGGATGGACGCTAAAGAAACC-3′) and SPDS1R2 (5′-GCCTCTAGAACGATCCTCCAGATTAGT-3′) and inserted as a SmaI-XbaI fragment into the expression cassettes of pPCV002-GIGI and pPCV821-MENCHU to label SPDS1 with C-terminal c-Myc and HA epitopes. The binary vectors were introduced into Agrobacterium strain GV3101 (pMP90RK) by electroporation (Main et al., 1995) and used in cotransformation experiments to express pairwise combinations of SPDS-myc and SPDS-HA proteins in cultured Arabidopsis cells, as described previously (Ferrando et al., 2001).
Immunoaffinity Purification and Size Fractionation of Double Epitope-Labeled Protein Complexes from Transformed Arabidopsis Cells
Arabidopsis cells expressing the individual SPDS-myc constructs alone or in combination with SPDS-HA constructs were harvested at 6 days after Agrobacterium-mediated transformation. The frozen cells were homogenized to powder with a mortar and pestle in the presence of liquid nitrogen (Ferrando et al., 2000, 2001; Farràs et al., 2001) and resuspended on ice in extraction buffer (1 mL/g) containing 50 mM Tris-HCl, pH 7.6, 10% glycerol, 0.5% Igepal CA-630, 1 mM EDTA, 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 2 μg/mL pepstatin, 0.5 μg/mL aprotinin, 0.5 μg/mL leupeptin, and 0.5 μg/mL antipain. After preparation of a cleared lysate by centrifugation (6000g for 15 min at 4°C), the protein concentration in the supernatant was determined, and the extract was supplemented with 150 mM NaCl. The protein extract was precleared with 50 μL of Sepharose–G matrix (Sigma; equilibrated in IPW buffer: 50 mM Tris-HCl, pH 7.6, 150 mM NaCl, and 0.1% Igepal CA-630; Sigma) by continuous shaking at 4°C for 2 h. The protein extract was subjected to immunoaffinity purification and immunodetection as described (Ferrando et al., 2001). An aliquot from the cell extracts (2 mg of protein) was size fractionated on a Superose 6 column (PC 3.2/30; Amersham Pharmacia Biotech) equilibrated with 50 mM Tris-Cl, pH 7.5, and 150 mM NaCl. The column was calibrated with the following molecular mass standards (Amersham Pharmacia Biotech): blue dextran (2000 kD), thyroglobulin (669 kD), apoferritin (443 kD), and BSA (67 kD). The flow rate was set to 0.25 mL/min, and 1-mL fractions were collected every 4 min. Molecular mass markers eluted at 8 mL (blue dextran), 12.5 mL (thyroglobulin), 14.5 mL (apoferritin), and 16.5 mL (BSA). The eluted fractions were separated by SDS-PAGE and subjected to immunoblot analysis to detect the epitope-labeled proteins, as described above.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The accession numbers for the genes/proteins mentioned in this article are as follows: AtSPDS (AB006693 and AAC98040), AtSPDS1 (AJ251296 and CAB61614), At5g53120 (AY040013), AtSPMS (AAK64170 and BAB08415), AtSPDS2 (AJ251297 and CAB61615), AtACL5 (AF184094), ScSPE3 (Saccharomyces cerevisiae; U27519), ScSPE4 (AF067970), MmSPDS (Mus musculus; NM_009272), MmSPMS (NM_009214), HsSPDS (Homo sapiens; M64231), HsSPMS (NM_004595), DrSPMS (Danio rerio; AJ009633), FrSPMS (Fugu rubripes; AJ009865), TfSPMS (Tetraodon fluviatilis; AJ009863), PsSPDS1 (Pisum sativum; AF043108), PsSPDS2 (AF043109), LeSPDS (Lycopersicon esculentum; AJ006414), TmSPDS (Thermotoga maritima; TM0654), BsSPDS (Bacillus subtilis; 16,077,068), BhSPDS (Bacillus holodurans; 15,612,563), AaSPDS (Aquifex aeolicus; AE000672), CeSPDS (Caenorhabditis elegans; AJ306734), and NcSPDS (Neurospora crassa; AB001598).
Acknowledgments
The authors thank H. Tabor and N. Hamasaki-Katagiri for providing the yeast speΔ3 and speΔ4 mutants and A. Pegg for supplying data before publication, as well as members of the Arabidopsis Genetics Group at the Max-Planck-Institut für Züchtungsforschung and N. Jovanovic (Universitat de Barcelona) for their help in this study. This work was supported by grants from the Ministerio de Ciencia y Tecnología (BIO-99-453 to A.F.T. and Bio99-1201-CO2-1 to J.C.) and the Deutsch Forschungsgemeinschaft (KO 1438/3-2 to C.K.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.004077.
References
- Alabadí, D., and Carbonell, J. (1999). Differential expression of two spermidine synthase genes during early fruit development and in vegetative tissues of pea. Plant Mol. Biol. 39, 933–943. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1989). Current Protocols in Molecular Biology. (New York: Greene/Wiley).
- Bagga, S., Rochford, J., Klaene, Z., Kuehn, G.D., and Phillips, G.C. (1997). Putrescine aminopropyl transferase is responsible for biosynthesis of spermidine, spermine, and multiple uncommon polyamines in osmotic stress-tolerant alfalfa. Plant Physiol. 114, 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasundaram, D., Dinman, J.D., Tabor, C.W., and Tabor, H. (1994). SPE1 and SPE2: Two essential genes in the biosynthesis of polyamines that modulate +1 ribosomal frameshifting in Saccharomyces cerevisiae. J. Bacteriol. 176, 7126–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, W.H., Tabor, C.W., and Tabor, H. (1973). Spermidine biosynthesis: Purification and properties of propylamine transferase from Escherichia coli. J. Biol. Chem. 248, 2480–2486. [PubMed] [Google Scholar]
- Burbulis, I.E., and Winkel-Shirley, B. (1999). Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc. Natl. Acad. Sci. USA 96, 12929–12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell, J., and Navarro, J.L. (1989). Correlation of spermine levels with ovary senescence and with fruit set and development in Pisum sativum L. Planta 178, 482–487. [DOI] [PubMed] [Google Scholar]
- Coffino, P. (2001). Regulation of cellular polyamines by antizyme. Nat. Rev. Mol. Cell Biol. 2, 188–194. [DOI] [PubMed] [Google Scholar]
- Cohen, S. (1998). A Guide to the Polyamines. (Oxford, UK: Oxford University Press).
- Durfee, T., Becherer, K., Chen, P.L., Yeh, S.H., Yang, Y., Kilburn, A.E., Lee, W.H., and Elledge, S.J. (1993). The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Farràs, R., Ferrando, A., Jásik, J., Kleinow, T., Ökrész, L., Tiburcio, A.F., Salchert, K., del Pozo, C., Schell, J., and Koncz, C. (2001). SKP1-SnRK protein kinase interactions mediate proteosomal binding of a plant SCF ubiquitin ligase. EMBO J. 20, 2742–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando, A., Farràs, R., Jásik, J., Schell, J., and Koncz, C. (2000). Intron-tagged epitope: A tool for facile detection and purification of proteins expressed in Agrobacterium-transformed cells. Plant J. 22, 553–560. [DOI] [PubMed] [Google Scholar]
- Ferrando, A., Koncz-Kálmán, Z., Farràs, R., Tiburcio, A.F., Schell, J., and Koncz, C. (2001). Detection of in vivo protein interactions between Snf1-related kinase subunits with intron-tagged epitope-labelling in plant cells. Nucleic Acids Res. 29, 3685–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Hamasaki-Katagiri, N., Katagiri, Y., Tabor, C.W., and Tabor, H. (1998). Spermine is not essential for growth of Saccharomyces cerevisiae: Identification of the SPE4 gene (spermine synthase) and characterization of a spe4 deletion mutant. Gene 210, 195–201. [DOI] [PubMed] [Google Scholar]
- Hamasaki-Katagiri, N., Tabor, C.W., and Tabor, H. (1997). Spermidine biosynthesis in Saccharomyces cerevisiae: Polyamine requirement of a null mutant of the SPE3 gene (spermidine synthase). Gene 187, 35–43. [DOI] [PubMed] [Google Scholar]
- Hanzawa, Y., Takahashi, T., Michael, A.J., Burtin, D., Long, D., Pineiro, M., Coupland, G., and Komeda, Y. (2000). ACAULIS5, an Arabidopsis gene required for stem elongation, encodes a spermine synthase. EMBO J. 19, 4248–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.W., Adami, G.R., Wei, N., Keyomarsi, K., and Elledge, S.J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Hashimoto, T., Tamaki, K., Suzuki, K., and Yamada, Y. (1998). Molecular cloning of plant spermidine synthases. Plant Cell Physiol. 39, 73–79. [DOI] [PubMed] [Google Scholar]
- Kajander, E.O., Kauppinen, L.I., Pajula, R.L., Karkola, K., and Eloranta, T.O. (1989). Purification and partial characterization of human polyamine synthases. Biochem. J. 259, 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., Martini, N., Szabados, L., Hrouda, M., Bachmair, A., and Schell, J. (1994). Specialized vectors for gene tagging and expression studies. In Plant Molecular Biology Manual, Vol. B2, S.B. Gelvin, R.A. Schilperoort, and D.P.S. Verma, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–22.
- Korolev, S., Ikeguchi, Y., Skarina, T., Beasley, S., Arrowsmith, C., Edwards, A., Joachimiak, A., Pegg, A.E., and Savchenko, A. (2002). The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor. Nat. Struct. Biol. 9, 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., Altabella, T., Taylor, M., and Tiburcio, A.F. (1997). Recent advances in polyamine research. Trends Plant Sci. 2, 124–130. [Google Scholar]
- Kutchan, T.M. (1995). Alkaloid biosynthesis: The basis for metabolic engineering of medicinal plants. Plant Cell 7, 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakanen, J.R., Pegg, A.E., and Coward, J.K. (1995). Synthesis and biochemical evaluation of adenosylspermidine, a nucleoside-polyamine adduct inhibitor of spermidine synthase. J. Med. Chem. 38, 2714–2727. [DOI] [PubMed] [Google Scholar]
- Liu, K., Fu, H., Bei, Q., and Luan, S. (2000). Inward potassium channel in guard cells as a target for polyamine regulation of stomatal movements. Plant Physiol. 124, 1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main, G.D., Reynolds, S., and Gartland, J.S. (1995). Electroporation protocols for Agrobacterium. In Methods in Molecular Biology, Vol. 44, Agrobacterium Protocols, K.M.A. Gartland and M.R. Davey, eds (Totowa, NJ: Humana Press), pp. 405–412. [DOI] [PubMed]
- Malmberg, R.L., Watson, M.B., Galloway, G.L., and Yu, W. (1998). Molecular genetic analysis of plant polyamines. Crit. Rev. Plant Sci 17, 199–224. [Google Scholar]
- Müller, S., Da'dara, A., Lüersen, K., Wrenger, C., Das Gupta, R., Madhubala, R., and Walter, R.D. (2000). In the human malaria parasite Plasmodium falciparum, polyamines are synthesized by a bifunctional ornithine decarboxylase, S-adenosylmethionine decarboxylase. J. Biol. Chem. 275, 8097–8102. [DOI] [PubMed] [Google Scholar]
- Navas, M.A., Cerdán, S., and Gancedo, J.M. (1993). Futile cycles in Saccharomyces cerevisiae strains expressing the gluconeogenic enzymes during growth on glucose. Proc. Natl. Acad. Sci. USA 90, 1290–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh, K., et al. (1998). Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12, 3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovadi, J., and Srere, P.A. (2000). Macromolecular compartmentation and channeling. Int. Rev. Cytol. 192, 255–280. [DOI] [PubMed] [Google Scholar]
- Pegg, A.E. (1986). Recent advances in the biochemistry of polyamines in eukaryotes. Biochem. J. 234, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietilä, M., Alhonen, L., Halmekytö, M., Kanter, P., Jänne, J., and Porter, C.W. (1997). Activation of polyamine catabolism profoundly alters tissue polyamine pools and affects hair growth and female fertility in transgenic mice overexpressing spermidine/spermine N-acetyltransferase. J. Biol. Chem. 272, 18746–18751. [DOI] [PubMed] [Google Scholar]
- Shoji, T., Yamada, Y., and Hashimoto, T. (2000). Jasmonate induction of putrescine N-methyltransferase genes in the root of Nicotiana sylvestris. Plant Cell Physiol. 41, 831–839. [DOI] [PubMed] [Google Scholar]
- Spiney, H.O., and Ovady, J. (1999). Substrate channeling. Methods 19, 306–321. [DOI] [PubMed] [Google Scholar]
- Srere, P.A. (1987). Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 56, 89–124. [DOI] [PubMed] [Google Scholar]
- Srivenugopal, K.S., and Adiga, P.R. (1981). Enzymic conversion of agmatine to putrescine in Lathyrus sativus seedlings: Purification and properties of a multifunctional enzyme (putrescine synthase). J. Biol. Chem. 256, 9532–9541. [PubMed] [Google Scholar]
- Tabor, C.W., and Tabor, H. (1984). Polyamines. Annu. Rev. Biochem. 53, 749–790. [DOI] [PubMed] [Google Scholar]
- Tiburcio, A.F., Kaur-Sawhney, R., and Galston, A.W. (1990). Polyamine metabolism. In The Biochemistry of Plants, J.B. Miflin and P.J. Lea, eds (New York: Academic Press), pp. 283–325.
- Tiburcio, A.F., Kaur-Sawhney, R., and Galston, A.W. (1993). Spermidine biosynthesis as affected by osmotic stress in oat leaves. Plant Growth Regul. 13, 103–109. [Google Scholar]
- Watson, M.B., and Malmberg, R.L. (1996). Regulation of Arabidopsis thaliana (L.) Heynh. arginine decarboxylase by potassium deficiency stress. Plant Physiol. 111, 1077–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanoha, B., and Cohen, S.S. (1985). S-Adenosylmethionine decarboxylase and spermidine synthase from Chinese cabbage. Plant Physiol. 78, 784–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, S.O., Lee, Y.S., Lee, S.H., and Cho, Y.D. (2000). Polyamine synthesis in plants: Isolation and characterization of spermidine synthase from soybean (Glycine max) axes. Biochim. Biophys. Acta 1475, 17–26. [DOI] [PubMed] [Google Scholar]