Abstract
The COP9 signalosome (CSN) is an evolutionarily conserved multiprotein complex that mediates the repression of photomorphogenesis in the dark in Arabidopsis through the degradation of transcription factors such as HY5 and HYH. CSN-mediated HY5 and HYH degradation also requires the activity of the putative E3 ubiquitin ligase (E3) component COP1 and the E2-conjugating enzyme variant COP10. Recently, it was shown that CSN also is required for auxin responses mediated by the SCF-type E3 SCFTIR1. To determine whether Arabidopsis CSN is required for E3-mediated processes in a more general manner, we generated plants with reduced E3 function by suppressing AtRBX1, an essential core subunit of SCF-type E3s. We observed that AtRBX1 transgenic plants share multiple phenotypes with CSN reduced-function plants, such as morphological defects and reduced responses to auxin, jasmonic acid, and cold stress, suggesting that CSN is required for multiple AtRBX1-mediated processes. Furthermore, we observed that mutants with defects in AXR1, a protein that had been described only as a regulator of SCFTIR1 function, also is required for other E3-mediated processes and for the COP1/COP10/CSN-mediated repression of photomorphogenesis in the dark. We conclude that CSN and AXR1 are of general importance for different pathways that are controlled by E3-mediated protein degradation.
INTRODUCTION
The COP9 signalosome (CSN) is a multiprotein complex that is conserved in most eukaryotes. Arabidopsis mutants with defects in individual CSN subunits fail to accumulate CSN and display photomorphogenic phenotypes when grown in the dark (Wei et al., 1994; Chamovitz et al., 1996). Genetic and molecular studies have revealed that CSN together with COP1 and COP10 controls photomorphogenesis through the light-dependent degradation of the transcription factors HY5 and HYH (Kwok et al., 1996; Ang et al., 1998; Holm et al., 2002). HY5 and HYH degradation is impaired in cop1 and cop10 mutants as well as in mutants of CSN (Kwok et al., 1996; Ang et al., 1998; Osterlund et al., 2000; Holm et al., 2002; Suzuki et al., 2002). Substrate-specific protein degradation in eukaryotes is preceded by substrate ubiquitylation and requires the activity of an E1 ubiquitin–activating enzyme (E1), an E2 ubiquitin–conjugating enzyme (E2), and an E3 ubiquitin ligase (E3) (Hershko and Chiechanover, 1998). COP1 contains a RING finger domain, which is found in many E3 ubiquitin ligases, and it can interact with HY5 and HYH via its WD40 domain (Ang et al., 1998; Deshaies, 1999; Osterlund et al., 2000). Recently, it was shown that the COP1-interacting partner CIP8, a RING-H2 motif protein, possesses E3 activity toward HY5 in vitro (Hardtke et al., 2002). COP10 encodes an E2 variant protein that interacts physically with several CSN subunits and the RING domain of COP1 (Suzuki et al., 2002). Based on these characteristics, it is thought that COP1, CIP8, and COP10 are part of an E2/E3 system that mediates HY5 and HYH ubiquitylation and degradation.
CSN also was found to interact with the cullin and RBX1 subunits of SCF-type E3s (Lyapina et al., 2001; Schwechheimer et al., 2001). In SCF-type E3s, cullin and RBX1 typically form a core complex that can associate with distinct combinations of a SKP1 homolog and an F-box protein, which functions as a degradation substrate receptor (Deshaies, 1999). In Arabidopsis, CSN interacts with SCFTIR1, an SCF-type E3 that mediates auxin response (Schwechheimer et al., 2001). Plants with reduced CSN function and SCFTIR1 loss-of-function mutants show similar phenotypes. Based on these observations, it was concluded that CSN acts as a positive regulator of SCFTIR1-mediated processes (Schwechheimer et al., 2001). AXR1 is another positive regulator of auxin response that modulates SCFTIR1 activity (Leyser et al., 1993). AXR1 is a subunit of the heterodimeric Nedd8/RUB1-activating enzyme that mediates the first step in the conjugation of the ubiquitin-like modification Nedd8/RUB1 to the cullin subunit of SCF-type E3s (neddylation) (del Pozo et al., 1998; del Pozo and Estelle, 1999; Hochstrasser, 2000). Interestingly, an antagonistic biochemical mechanism, Nedd8/RUB1 deconjugation (deneddylation), is one of the biochemical activities that has been attributed to CSN (Lyapina et al., 2001; Schwechheimer et al., 2001).
Because CSN interacts with the SCF core complex subunits cullin and RBX1, it has been hypothesized that CSN also may be essential for processes that are mediated by SCF complexes other than SCFTIR1 (Lyapina et al., 2001; Schwechheimer et al., 2001). This hypothesis finds support in the observation that Arabidopsis plants with reduced CSN function have many morphological phenotypes that cannot be attributed solely to the loss of SCFTIR1 (Peng et al., 2001a, 2001b; Schwechheimer et al., 2001). Because SCFTIR1 is the only SCF-type E3 from Arabidopsis that has been characterized at the physiological and molecular level to date, it has been difficult to attribute defects of plants with reduced CSN function to specific E3-mediated processes (Gray et al., 1999). To identify processes that require SCF-type E3 activity, we have generated transgenic Arabidopsis plants with reduced E3 function by suppressing the SCF core complex subunit AtRBX1. We found that transgenic plants with reduced AtRBX1 levels are defective in a number of processes that are attributable to a reduction of E3 activity. These processes are impaired in a similar manner in plants with reduced CSN function, suggesting that CSN is essential for these responses. Interestingly, we also found that AXR1 plays a general role in these E3-mediated processes and that AXR1 function is essential for the COP1/COP10/CSN-mediated repression of photomorphogenesis in the dark. Together, these results indicate that CSN and AXR1 are essential for the proper function of multiple E3-mediated developmental pathways.
RESULTS
AtRBX1 Is a Subunit of SCF-Type E3s and Copurifies with CSN
RBX1 is a conserved and essential subunit of SCF-type E3s and of the von Hippel–Lindau tumor-suppressor E3 complex (Kamura et al., 1999; Skowyra et al., 1999). RBX1 is essential for SCF-type E3 function, and as a result of the importance of SCF-type E3s in cell cycle control, yeast RBX1 deletion strains are not viable. The Arabidopsis genome contains two RBX1 orthologs that we designated AtRBX1a (At5g20570) and AtRBX1b (At3g42830). AtRBX1a and AtRBX1b share 84% identity at the amino acid level and are more closely related to each other than to any RBX1 ortholog from other eukaryotes (Figures 1A and 1B). Based on the high sequence conservation between AtRBX1a and AtRBX1b, both proteins may have redundant functions. Although we have been unable to amplify AtRBX1b transcripts from Arabidopsis RNA preparations by reverse transcriptase–mediated (RT) PCR, AtRBX1a-specific transcripts can be amplified readily, suggesting that AtRBX1a is the predominantly expressed RBX1 ortholog in Arabidopsis.
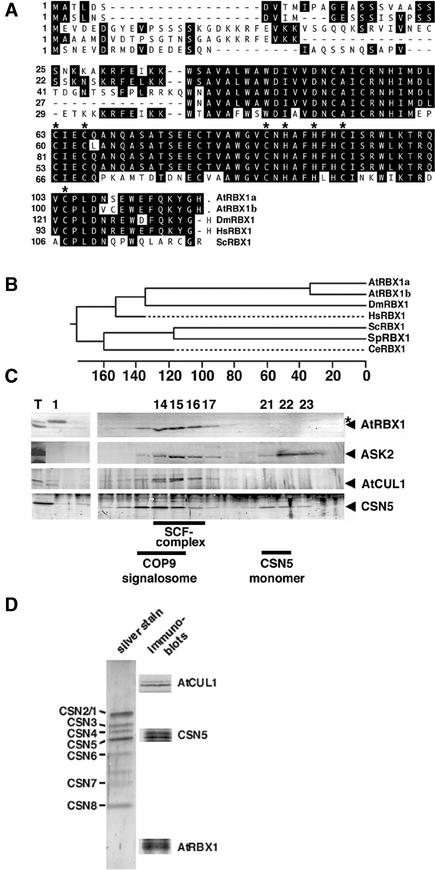
Figure 1.
The Arabidopsis RBX1 Ortholog AtRBX1 Is Part of an SCF-Type E3 Ubiquitin Ligase Complex and Copurifies with CSN.
(A) Sequence alignment of the two RBX1 orthologs AtRBX1a and AtRBX1b from Arabidopsis and their counterparts from human, Drosophila melanogaster, and Saccharomyces cerevisiae. Identical amino acids are boxed in black. Asterisks mark the conserved residues of the RING-H2 finger domain.
(B) Phylogenetic tree with the RBX1 orthologs from human, D. melanogaster, S. cerevisiae, Schizosaccharomyces pombe, and Caenorhabditis elegans. The sequence alignment and the phylogenetic tree were produced using CLUSTAL (DNAStar, Madison, WI).
(C) Immunoblots of Superose 6 gel filtration fractions probed with antibodies against CSN5, AtRBX1, ASK2, and AtCUL1. AtRBX1, AtCUL1, and ASK2 cofractionate as a complex that is slightly smaller than CSN. AtRBX1 and AtCUL1 but not the SCF complex subunit ASK2 or CSN also are present in the flow-through fraction (fraction 1). Fraction numbers are indicated. Lane T contained the total unfractionated extract.
(D) AtRBX1 and AtCUL1 copurify with immunoaffinity-purified CSN. Immunoblots against AtCUL1, CSN5, and AtRBX1 are shown in the right lane. The individual CSN subunits as visualized by silver staining are indicated at left.
To provide evidence for an association of AtRBX1 with SCF complexes, we examined the cofractionation of AtRBX1 with the individual SCF subunits using gel filtration chromatography (Figure 1C). Consistent with the notion that AtRBX1 is a subunit of SCF complexes, we found that AtRBX1, the cullin homolog AtCUL1, and the SKP1 homolog ASK2 cofractionate as a protein complex of ∼300 kD (Figure 1C). Previously, we had described a direct interaction between SCFTIR1 subunits and CSN using coimmunoprecipitation and immunoaffinity purification, and we had shown that AtRBX1 interacts with CSN subunits in the yeast two-hybrid system (Schwechheimer et al., 2001). Confirming and extending our previous results, we detected AtRBX1 in immunoaffinity-purified preparations of CSN (Figure 1D). In gel filtration chromatography, CSN eluted as a protein complex distinct from the SCF complex, suggesting that CSN and the SCF complex are not stably associated (Figure 1C). The loss of CSN in CSN mutants did not affect the integrity of the SCF complex (data not shown). Together, these data provide evidence that AtRBX1 functions in the context of SCF-type E3s in Arabidopsis.
Transgenic Lines with Reduced AtRBX1 Function Display Various Growth Defects
To obtain plants with defects in multiple E3-mediated pathways, we generated plants with reduced AtRBX1 function. Because of the essential role of SCF complexes in cell cycle control, we predicted that the loss of AtRBX1 function would be embryonic lethal and decided to use the antisense and cosuppression strategies to obtain plants with partially suppressed AtRBX1 function. We generated transgenic Arabidopsis plants that express either sense or antisense AtRBX1a cDNA from the constitutive 35S promoter of Cauliflower mosaic virus. Both approaches gave rise to transgenic plants with indistinguishable morphological phenotypes, which we classified according to their severity. Transgenic lines with a weak phenotype, such as antisense (AS) line RBX AS9, had reduced apical dominance characterized by reduced height of the primary inflorescence and an increased number of secondary inflorescences (Figure 2A). Sense (S) lines RBX S3 and antisense line RBX AS20 had a severe phenotype and were phenotypically indistinguishable from each other in that they had a dwarfed stature (maximum height of 5 cm), accumulated high levels of anthocyanin, failed to produce secondary inflorescences, and did not generate branches in the axils of their cauline leaves (Figure 2B). Even stronger phenotypes were observed in the transgenic T1 generation. The plants showing this phenotype were severely dwarfed (maximum height of 4 mm), failed to elongate their primary inflorescence, failed to produce secondary inflorescences, and accumulated high levels of anthocyanin (Figure 2D). Despite the fact that many of the plants with this phenotype produced at least one flower, these flowers were infertile, precluding further study. Other lines, such as RBX AS7, were indistinguishable from the wild type in the aboveground parts of the plants (data not shown).
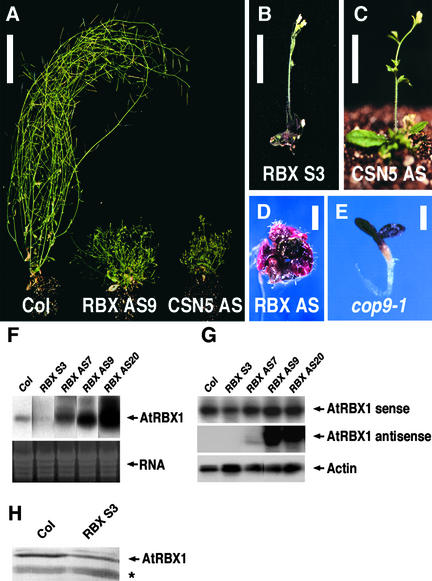
Figure 2.
Transgenic Plants with Reduced AtRBX1 Function Have Phenotypes Similar to Those of Plants with Reduced CSN Function.
(A) Four-week-old Arabidopsis plants (from left to right): Columbia wild-type plants (Col), RBX AS9 (AtRBX1 antisense line), and the CSN transgenic plant CSN5 AS (CSN5 antisense line; weak phenotype). The transgenic plants appear bushy as a result of the reduced height of the primary inflorescence and an increased number of secondary inflorescences. Bar = 5 cm.
(B) Four-week-old RBX S3 plant (AtRBX1 sense line; strong phenotype).
(C) Four-week-old CSN transgenic plant CSN5 AS (CSN5 antisense line).
Plants in (B) and (C) have reached maximum elongation growth. Bars = 2.5 cm.
(D) Three-week-old AtRBX1 antisense line RBX AS with a severe phenotype. Some transgenic plants with this phenotype produced flowers that were infertile. Bar = 2 mm.
(E) Ten-day old cop9-1 mutant seedling (CSN loss-of-function mutant). Bar = 1 mm.
(F) RNA gel blot showing reduced expression of the endogenous AtRBX1 gene in the RBX S3 cosuppression line and increased expression of the AtRBX1 gene in the RBX AS antisense lines. Equal amounts of total RNA (10 μg) were loaded.
(G) Semiquantitative RT-PCR analysis reveals that the high levels of AtRBX1 expression on the RNA gel blot are attributable to high levels of expression of the AtRBX1 antisense transgene. RT-PCR of an actin gene was used as an amplification control.
(H) Immunoblot with a polyclonal antibody against AtRBX1 shows that AtRBX1 transgenic plants with a severe phenotype have reduced levels of AtRBX1 protein. Cross-reaction of a second protein (asterisk) shows equal protein loading.
Expression analysis by RNA gel blotting and semiquantitative RT-PCR revealed that plants with a strong phenotype expressing AtRBX1a in the sense orientation had a 50% reduction in AtRBX1 mRNA levels compared with wild-type plants (Figures 2F and 2G). In plants expressing the AtRBX1 antisense cDNA, we detected a positive correlation between the expression levels of the antisense AtRBX1a transcript and the severity of their phenotypes (Figures 2F and 2G). Immunoblot analysis revealed that AtRBX1 protein levels in transgenic plants with a strong phenotype, such as RBX S3, were reduced to ∼60% of wild-type levels, whereas no obvious reduction of AtRBX1 protein levels was detectable in plants with a weak phenotype (Figure 2H). We conclude that plants expressing AtRBX1 antisense and sense cDNAs have partially reduced AtRBX1 function. Our inability to recover transgenic lines with more severely reduced AtRBX1 levels may be attributable to the crucial role of RBX-containing SCF complexes in cell cycle regulation, as demonstrated by the lethal phenotype of yeast RBX deletion mutants (Kamura et al., 1999; Skowyra et al., 1999).
Plants with Reduced AtRBX1 and CSN Function Have Similar Morphological Defects
Because we hypothesized that CSN is required for different E3-mediated processes whose functions may be reduced in AtRBX1 transgenic lines, we expected that at least some aspects of their phenotype would be observed in plants with reduced CSN function. Loss of apical dominance is one of the most obvious phenotypes that has been observed in transgenic plants with partially reduced CSN function (Figure 2A) (Peng et al., 2001a, 2001b; Schwechheimer et al., 2001). Many plants from the same transgenic series had more severely reduced height and failed to produce secondary inflorescences (Figure 2C). Finally, CSN loss-of-function mutants such as cop9-1 display a photomorphogenic seedling phenotype when grown in the dark, accumulate high levels of anthocyanins, and are seedling lethal (Figure 2E) (Wei et al., 1994; Kwok et al., 1996). As described above, AtRBX1 reduced-function plants show several morphological defects that are similar to the phenotypes seen in CSN reduced-function plants, and we took this as a first indication that AtRBX1 and CSN are required for a similar set of developmental pathways (Figures 2A to 2E).
AtRBX1 Is Required for Proper Auxin Response
The E3 SCFTIR1, CSN, and AXR1 act together to promote auxin response in Arabidopsis. To determine whether AtRBX1 is a positive regulator of SCFTIR1 function and auxin response, we examined whether plants with reduced AtRBX1 function show developmental defects that are comparable to the defects observed in the SCFTIR1 and AXR1 loss-of-function mutants (tir1-1 and axr1-3) and in transgenic plants with reduced CSN function (CSN5 AS) (Leyser et al., 1993; Ruegger et al., 1998; Schwechheimer et al., 2001). These mutants and transgenic lines are characterized by reduced sensitivity to auxin, reduced lateral root formation, and reduced root hair growth compared with the wild type (Ruegger et al., 1998; Gray et al., 1999; Schwechheimer et al., 2001). When we examined these aspects of plant growth in AtRBX1 transgenic plants, we found that they produced fewer lateral roots than the wild type, had reduced root hair growth, and had reduced inhibition by auxin (Figures 3A to 3D). The classification of the AtRBX1 lines based on the severity of their aboveground phenotypes was reflected in the severity of their root phenotypes. However, it is interesting that line RBX AS7, which did not show an apparent aboveground phenotype, produced fewer lateral roots than the wild type. This finding indicates that lateral root formation may be a very sensitive indicator for the degree of suppression of AtRBX1 function. By contrast, the axr1-3 mutant did not show any obvious aboveground phenotypes in our growth conditions but had very pronounced root and auxin-response phenotypes. This finding may indicate that AXR1 activity is restricted largely to the Arabidopsis root.
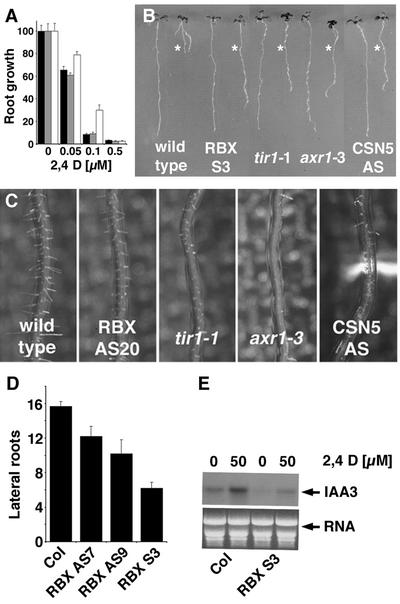
Figure 3.
Plants with Reduced AtRBX1 Function Show Reduced Response to Auxin.
(A) Relative root growth of wild-type seedlings (Columbia; black bar), the AtRBX1 antisense line RBX AS7 (gray bar), and the cosuppression line RBX S3 (white bar). The seedlings were grown for 4 days on unsupplemented medium and then transferred to medium containing 2,4-D. Relative root growth was measured using NIH Image software. Root growth in the absence of auxin was set at 100%.
(B) Similar phenotypes are found in tir1-1 and axr1-3 mutants as well as in transgenic seedlings with reduced CSN function (CSN5 AS). Seedlings were grown in the absence of auxin (left) or on 0.1 μM 2,4-D. The asterisks indicate the position of the root tip at the time of transfer.
(C) and (D) The rate of root hair elongation (C) and the number of lateral roots per root (D) were reduced in AtRBX1 transgenic lines. Reduced root hair elongation in axr1 mutants, tir1 mutants, and plants with reduced CSN function was reported previously (Schwechheimer et al., 2001). Col, Columbia wild type.
(E) Induction of the AUX/IAA gene IAA3 is reduced in AtRBX1 lines compared with the wild type. A total of 25 μg of total RNA was loaded per lane.
In wild-type plants, the expression of AUX/IAA genes is induced rapidly by auxin (Abel et al., 1995). It is thought that this induction is controlled at least in part by the degradation of AUX/IAA proteins that act as repressors of their own gene transcription via a negative feedback loop (Abel et al., 1994; Ulmasov et al., 1997; Gray et al., 2001). Genetic and molecular evidence suggests that at least some of the AUX/IAA repressor proteins are targeted for degradation by SCFTIR1 and that CSN and AXR1 also are required for this process (Gray et al., 2001; Schwechheimer et al., 2001). A characteristic feature of mutants with reduced auxin response is the reduced induction of AUX/IAA genes. Providing molecular evidence of the notion that AtRBX1 is required for proper auxin response, we found that the induction of the AUX/IAA gene IAA3 is compromised in AtRBX1 transgenic plants (Figure 3E). Together, these findings suggests that AtRBX1 is a positive regulator of an auxin response that supposedly functions in the context of SCFTIR1.
Proper Response to Jasmonic Acid and Cold Stress Requires AtRBX1, CSN, and AXR1 Function
The Arabidopsis F-box protein COI1 is required for the proper response to the phytohormone jasmonic acid (JA) (Xie et al., 1998). COI1 is related closely to TIR1, and it has been hypothesized that COI1 is the F-box subunit of a putative SCFCOI1 complex (Xie et al., 1998; del Pozo and Estelle, 2000; Xu et al., 2002). Based on the apparent role of AtRBX1 in SCFTIR1-mediated processes, we also investigated the possibility that AtRBX1 is involved in JA responses. COI1 loss-of-function mutants (coi1-1) display reduced sensitivity to root growth inhibition by exogenously applied JA and are insensitive to the induction of VSP gene expression by JA (Xie et al., 1998). When we subjected AtRBX1 transgenic seedlings to a root growth inhibition assay using biologically active methyl jasmonate (MeJA), we found that AtRBX1 seedlings were less sensitive to root growth inhibition by JA than wild-type seedlings (Figure 4A). Although root elongation in wild-type seedlings on 10 μM MeJA was reduced by >80%, root growth in the AtRBX1 lines and the coi1-1 mutants was reduced by ∼50% (Figure 4A). Consistent with the notion that the AtRBX1 plants are less sensitive to JA, JA induction of VSP transcript also was reduced in the AtRBX1 plants compared with wild-type plants (Figure 4B). Because AXR1 and CSN have been implicated in the SCFTIR1-mediated auxin response, we also tested whether AXR1 and CSN can confer resistance to JA (del Pozo and Estelle, 1999; Schwechheimer et al., 2001). We found that root growth inhibition and VSP transcript induction are impaired in CSN5 antisense plants as well as in axr1-3 mutants (Figures 4A and 4B). Together, these findings suggest that JA-mediated responses require CSN, AXR1, and AtRBX1 function, the latter likely functioning as a subunit of the SCFCOI1 complex.
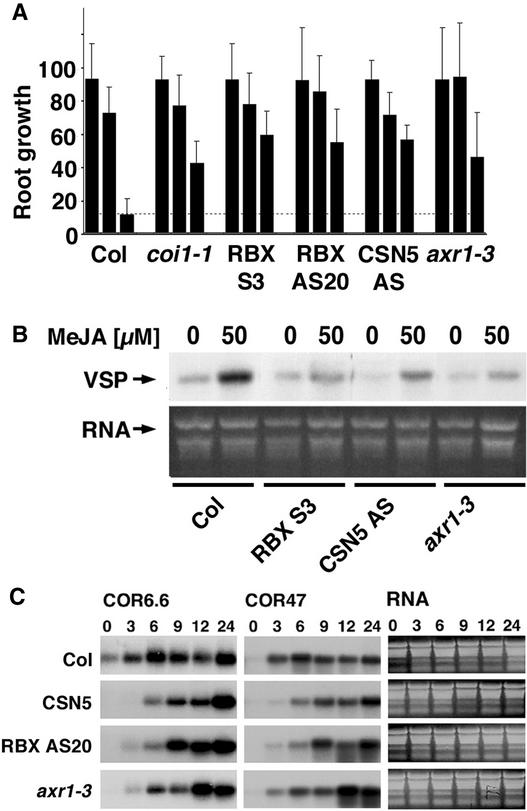
Figure 4.
Plants with Reduced AtRBX1, AXR1, and CSN Function Show Reduced Response to MeJA and Cold.
(A) Root growth inhibition by MeJA. Relative root growth was measured using NIH Image software. Root growth in the absence of MeJA was set at 100%. Bars from left to right represent root growth in the absence of MeJA, with 0.01 μM MeJA, and with 0.1 μM MeJA. Col, Columbia wild type.
(B) VSP transcript induction by MeJA. Expression of the VSP gene was induced with 50 μM MeJA. A total of 5 μg of total RNA was loaded in each lane.
(C) Plants with reduced AtRBX1, AXR1, and CSN function are delayed in their response to cold stress. The RNA gel blots show the induction kinetics of the cold-induced genes COR6.6 and COR47 after transfer of plants from standard growth conditions to 0°C. Measurements were taken at 0, 3, 6, 9, 12, and 24 h. A total of 5 μg of total RNA was loaded per lane.
Cold stress rapidly induces the expression of a series of cold-regulated (COR) genes in Arabidopsis (Stockinger et al., 1997). By analogy to the auxin induction of AUX/IAA expression and the JA induction of VSP expression, cold-regulated gene expression also may be controlled by a transcriptional repressor that represses COR expression in the absence of the cold stimulus. Therefore, induction by cold treatment would require the activity of an E3 that induces the degradation of this transcriptional repressor. To test this hypothesis, we assayed the induction of the cold-induced genes COR6.6 and COR47 in cold-treated wild-type and AtRBX1 transgenic plants. The induction of the two COR genes was delayed in the AtRBX1 transgenic plants compared with the wild type, suggesting that an AtRBX1-containing SCF complex may play a role in cold induction (Figure 4C). Because our previous results had suggested that CSN and AXR1 are essential for AtRBX1-mediated processes, we assayed cold-induced gene expression in transgenic plants with reduced CSN function and in the axr1-3 mutant. We found that CSN and AXR1 also are required for the timely induction of COR genes after the onset of cold treatment (Figure 4C). Together, these data suggest that the timely cold induction of gene expression requires the activities of CSN, AXR1, and the SCF complex subunit AtRBX1.
AXR1 Is Required for the COP1/COP10/CSN-Mediated Repression of Photomorphogenesis in the Dark
The repression of photomorphogenesis in the dark is mediated by COP1, COP10, and CSN (Wei et al., 1994; Chamovitz et al., 1996; Kwok et al., 1996; Osterlund et al., 2000; Suzuki et al., 2002). COP1 has characteristics of an E3 ubiquitin ligase and interacts with COP10, an E2 ubiquitin–conjugating enzyme variant (Osterlund et al., 2000; Suzuki et al., 2002). COP1 interacts physically with HY5 and HYH and mediates the dark-dependent degradation of these transcription factors (Ang et al., 1998; Osterlund et al., 2000; Holm et al., 2002). Together, these results provide substantial evidence that COP1 and COP10 represent components of an E2/E3 protein degradation pathway that also requires CSN function (Osterlund et al., 2000; Suzuki et al., 2002).
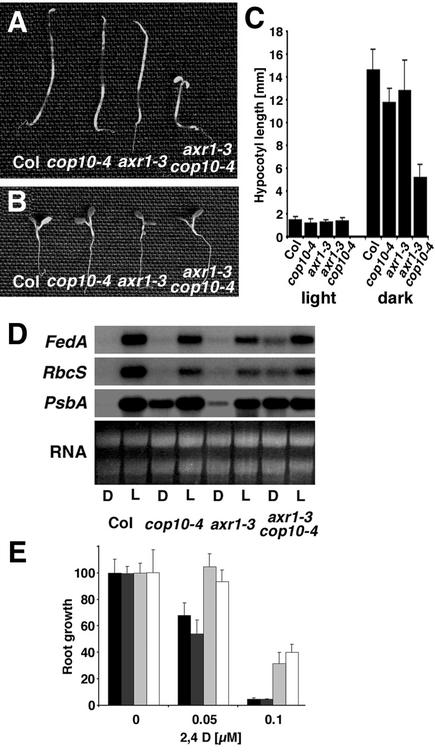
Because we observed the involvement of both CSN and AXR1 in several E3-mediated processes, we were curious to determine whether AXR1 is involved in the repression of photomorphogenesis in the dark. To this end, we generated double mutants between axr1-3 and cop10-4. cop10-4 is the weakest allele of all cop1, cop10, and CSN mutants and displays only a subtle photomorphogenic phenotype when grown in the dark (Figure 5A) (Suzuki et al., 2002). We found that the axr1-3 cop10-4 double mutant had a strong photomorphogenic phenotype compared with either of the parental lines or the wild type (Figures 5A to 5C). This photomorphogenic phenotype could represent an enhancement of the photomorphogenic pathway mediated by cop10, implying a role of AXR1 in photomorphogenesis or, alternatively, representing an enhancement AXR1-mediated auxin response. The misregulation of light-induced genes, such as the genes for type A ferredoxin (FedA), the small subunit of the ribulose-1,5-bisphosphate carboxylase gene (RbcS), and the 32-kD protein of photosystem II gene (PsbA), is the hallmark of the strong cop1, cop10, and CSN mutant alleles (Kwok et al., 1996). Therefore, we reasoned that misregulation of these genes in the axr1-3 cop10-4 double mutant compared with the single mutants could give an indication of the role of AXR1 in COP10-mediated photomorphogenesis. Interestingly, we observed that the expression of these light-induced genes in the dark was enhanced strongly in the axr1-3 cop10-4 double mutants compared with the single mutants and the wild type (Figure 5D). To determine whether COP10 also functions in AXR1-mediated auxin responses, we assayed the auxin sensitivity of the axr1-3 cop10-4 seedlings. We found that the level of auxin insensitivity of the axr1-3 cop10-4 double mutants compared with the auxin-resistant axr1-3 parental line was not altered in a root growth inhibition assay, suggesting that COP10 function is not required for the proper auxin response (Figure 5E). Therefore, we concluded that AXR1 is an important component of the COP1/COP10/CSN-mediated repression of photomorphogenesis in the dark.
Figure 5.
AXR1 Function Is Required for the Repression of Photomorphogenesis in the Dark.
(A) and (B) Six-day-old dark-grown (A) and light-grown (B) seedlings of the wild type (Col), axr1-3, cop10-4, and the axr1-3 cop10-4 double mutant.
(C) Statistical analysis of hypocotyl length of dark- and light-grown single and double mutant seedlings. Although the single mutants showed a minor reduction in hypocotyl length in dark-grown seedlings, hypocotyl length was reduced dramatically in the axr1-3 cop10-4 double mutant. Cotyledons were open and expanded in the double mutant throughout dark-grown development.
(D) RNA gel blot with total RNA prepared from 6-day-old Columbia wild-type and mutant seedlings grown in the dark (D) and in continuous white light (L). The blots were probed with cDNA fragments for type A ferredoxin (FedA), the small subunit of the ribulose-1,5-bisphosphate carboxylase gene (RbcS), or the 32-kD protein of photosystem II gene (PsbA). A total of 5 μg of total RNA was loaded per lane.
(E) The axr1-3 cop10-4 double mutant is not altered in its reduced response to auxin. The auxin root growth inhibition assay was as described for Figure 3 with wild-type (black bars), cop10-4 (dark gray bars), axr1-3 (light gray bars), and axr1-3 cop10-4 double mutant (white bars) seedlings.
DISCUSSION
We have provided molecular, physiological, and genetic evidence that AtRBX1 functions as an SCF-type E3 complex component in Arabidopsis. Transgenic plants with reduced AtRBX1 function showed a number of morphological defects and exhibited reduced responses to auxin, JA, and cold. We predict that AtRBX1 functions as a subunit of SCF-type E3s in Arabidopsis; thus, the observed phenotypes most likely reflect defects in processes mediated by these E3s. In the case of reduced auxin response, this could be an impairment of the activity of SCFTIR1, whereas reduced JA response may be attributable to an uncharacterized SCFCOI1 complex. Loss of SCFTIR1 or SCFCOI1 function can explain only some of the phenotypes that we observed in the AtRBX1 reduced-expression plants. The AtRBX1-containing E3 complexes that mediate the other phenotypes remain to be elucidated. The Arabidopsis genome contains possibly >500 F-box domain proteins (Bachmair et al., 2001). Many of these F-box proteins are related structurally to the bona fide SCF complex F-box protein TIR1 (Xiao and Jang, 2000). These proteins represent good candidates for SCF complex subunits, and it is expected that Arabidopsis harbors a considerable number of unidentified SCF complexes that could mediate the morphological and physiological processes that are impaired in the AtRBX1 reduced-expression plants.
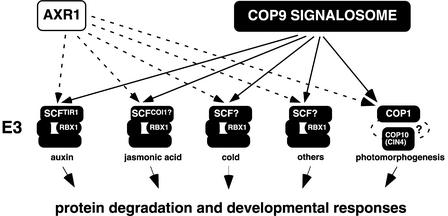
We have shown that CSN and AXR1 are required for several likely E3-mediated processes in addition to the previously reported SCFTIR1-mediated auxin response (Figure 6). The sum of the individual defects evoked by the loss of various E3-mediated responses may be sufficient to explain the strong pleiotropic phenotype of CSN loss-of-function mutants (Figure 2E) (Kwok et al., 1996). Most interestingly, we found that AXR1 also participates in the repression of photomorphogenesis in the dark, a process that requires the activity of a non-SCF-type E3 consisting of the RING finger protein COP1 and its interactors CIP8 and COP10. AXR1 mediates the neddylation of cullins, and these proteins represent a protein family in many eukaryotes, including Arabidopsis (Bachmair et al., 2001). Cullins have been found in E3 ubiquitin ligase complexes other than SCF-type E3s (Deshaies, 1999). Therefore, AXR1 may mediate photomorphogenesis by controlling the activity of the COP1/COP10 ubiquitination complex through the neddylation of an associated, unidentified cullin homolog.
Figure 6.
The Roles of CSN and AXR1, as Supported by Data Obtained in This Study.
A role for AXR1 and CSN was shown for the responses to auxin, JA, cold, and light (photomorphogenesis) and for other undefined processes.
The finding that AXR1 mutants are impaired in multiple developmental responses implies that axr1 loss-of-function mutants have severe developmental defects that are similar to those observed in the CSN loss-of-function mutants or the severe AtRBX1 transgenic lines. Compared with these mutants and transgenic lines, however, the apparent developmental defects observed in the axr1 loss-of-function mutants are rather subtle (Lincoln et al., 1990). This inconsistency could be explained by the presence of at least one additional AXR1 homologous gene in the Arabidopsis genome, At2g32410. AXR1 shares 80% amino acid identity with its homolog. Thus, the relatively weak morphological phenotype of the axr1 mutants could be explained if these two proteins share redundant functions such that the AXR1 homolog replaces most of the AXR1 function in the axr1 mutants. This hypothesis also finds further support in the recent finding that transgenic Arabidopsis plants with reduced function of ECR1, the second subunit of the heterodimeric Nedd8/RUB1-activating enzyme, have phenotypes that are more severe and that cover a wider range of phenotypes than those reported for the AXR1 loss-of-function mutants (del Pozo et al., 2002).
METHODS
Biological Material and Plant Transformation Constructs
Arabidopsis thaliana var Columbia plants were used in all experiments. The plants were grown using standard growth conditions (16-h-day/8-h-night) on soil or on solid 1 × GM/1% Suc (Gibco Life Technologies) in a controlled-environment chamber at 22°C. EST 110C19T7 was identified as a clone containing the full-length AtRBX1 clone. The AtRBX1 open reading frame was amplified by PCR, and the PCR product was subcloned into vector pCR2.1 (Invitrogen, Carlsbad, CA). The AtRBX1 open reading frame was inserted as an EcoRI fragment into the EcoRI-digested vector pGreen 35S(3) in the sense and antisense orientations to generate RBX sense and antisense lines by Agrobacterium tumefaciens–mediated transformation (Hellens et al., 1999). Expression of the AtRBX1 transgene was examined by RNA gel blot analysis and by semiquantitative reverse transcriptase–mediated PCR, as described previously (Frohman et al., 1988). Twenty amplification cycles were used for amplification of the sense transcript, and 25 cycles were used for amplification of the antisense transcript. The COP9 signalosome (CSN5) antisense and cosuppression plants were described previously (Kwok et al., 1998; Schwechheimer et al., 2001).
Antibody Production and Immunoblot Analysis
A fragment corresponding to amino acids 1 to 320 of AtCUL1 was amplified by PCR and expressed as a glutathione S-transferase fusion in vector pGEX-4T-1. The PCR-amplified full-length AtRBX1 open reading frame was expressed in the same vector. Gel-eluted recombinant protein was used for the production of polyclonal antibodies in rabbits. The CSN5 and ASK2 antibodies have been described previously (Schwechheimer et al., 2001). Gel-filtration and immunoblot analyses were performed as described previously (Serino et al., 1999; Schwechheimer et al., 2001).
Physiological Experiments
For the auxin root growth inhibition assay, seedlings were grown on 1 × GM/1% Suc on vertically positioned plates at 22°C. At day 4, seedlings were transferred to medium containing 2,4-D and grown for another 5 days. The root length of 20 seedlings was measured using NIH Image software. Root length increase was set relative to that of seedlings grown in the absence of 2,4-D (100%). The number of lateral roots was determined from 20 seedlings grown for 10 days on vertically positioned 1 × GM/1% Suc plates. For RNA gel blot analysis, leaf material from adult RBX S3 plants (severe phenotype) and from wild-type plants was induced for 1 h with 5 μM 2,4-D in 40 mL of liquid 1 × GM. RNA was prepared using the RNeasy kit (Qiagen, Valencia, CA). A total of 25 μg of total RNA was loaded and hybridized with a probe for IAA3 (EST 36C1T7). Root growth inhibition experiments with methyl jasmonate (MeJA; Bedoukian, Danbury, CT) were performed in a similar manner to the 2,4-D experiments except that root length was determined at 7 days after transfer to MeJA-containing medium. MeJA induction experiments were performed in a similar manner except that induction was performed at 7 h with 5 μM MeJA. The blot was probed with EST 36C1T7, which encodes VSP. For cold induction experiments, adult tissue was placed in precooled GM medium (1% Suc) at 0°C for the time indicated in Figure 4. RNA gel blots were probed with EST probes corresponding to COR6.6 (103L16T7) and COR47 (110K23T7).
Double Mutant Analysis
axr1-3 mutant pollen was used to pollinate cop10-4 homozygous mutant plants. Homozygous double mutant lines were identified by preselecting the seedlings for the axr1-3 phenotype on 2,4-D–containing medium. cop10-4 homozygous mutants were identified using an EcoRI polymorphism (Suzuki et al., 2002). The axr1-3 mutation was confirmed by sequencing of the genomic axr1 locus. Hypocotyl measurements were taken and averaged from 20 seedlings (7 days old), and the same material was used to prepare RNA for RNA gel blot analysis.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Acknowledgments
We thank Kathrin Schrick and Jirì Friml for their helpful comments on the manuscript. We thank the Arabidopsis Stock Center at Ohio State University for providing ESTs and seed stocks. This work was supported by National Science Foundation Grant MCD-0077217 and Binational Agricultural Research and Development Fund Grant IS-3123-99 to X.-W.D. C.S. was the recipient of a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (Schw751/1-1).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003434.
References
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. [DOI] [PubMed] [Google Scholar]
- Abel, S., Oeller, P.W., and Theologis, A. (1994). Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 91, 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang, L.-H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.-W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. [DOI] [PubMed] [Google Scholar]
- Bachmair, A., Novatchkova, M., Potuschak, T., and Eisenhaber, F. (2001). Ubiquitylation in plants: A post-genomic look at a post-translational modification. Trends Plant Sci. 6, 463–470. [DOI] [PubMed] [Google Scholar]
- Chamovitz, D.A., Wei, N., Osterlund, M.T., von Arnim, A.G., Staub, J.M., Matsui, M., and Deng, X.-W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86, 115–121. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., Dharmasiri, S., Hellmann, H., Walker, L., Gray, W.M., and Estelle, M. (2002). AXR1-ECR1–dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., and Estelle, M. (2000). Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem. Sci. 25, 133–138. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., Timpte, C., Tan, S., Callis, J., and Estselle, M. (1998). The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280, 1760–1763. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., and Estelle, M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96, 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and Cullin/RING H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., Okamoto, H., Stoop-Myer, C., and Deng, X.W. (2002). Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J. 30, 385–394. [DOI] [PubMed] [Google Scholar]
- Hellens, R., Joyce, N., and Mullineaux, P. (1999). A new and versatile Agrobacterium-based plant transformation vector. In Plant Biotechnology and in Vitro Biology in the 21st Century, A. Altman, M. Ziv, and S. Izhar, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 155–158.
- Hershko, A., and Chiechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (2000). All in the ubiquitin family. Science 5479, 563–564. [DOI] [PubMed] [Google Scholar]
- Holm, M., Ma, L.G., Qu, L.J., and Deng, X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura, T., Koepp, D.M., Conrad, M.N., Skowyra, D., Moreland, R.J., Iliopoulos, O., Lane, W.S., Kaelin, W.G.J., Elledge, S.J., Conaway, R.C., Harper, J.W., and Conaway, J.W. (1999). RBX1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284, 657–661. [DOI] [PubMed] [Google Scholar]
- Kwok, S.F., Piekos, B., Miséra, S., and Deng, X.-W. (1996). A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 110, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, S.F., Solano, R., Tsuge, T., Chamovitz, D.A., Ecker, J.R., Matsui, M., and Deng, X.-W. (1998). Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, H.M., Lincoln, C.A., Timpte, C., Lammer, D., Turner, J., and Estelle, M. (1993). Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364, 161–164. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Zhou, C., Wolf, D.A., Wei, N., Shevchenko, A., and Deshaies, R.J. (2001). COP9 signalosome promotes cleavage of NEDD8-CUL1 conjugates. Science 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.-W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.-W. (2001. a). Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted development processes in Arabidopsis. Plant Cell 13, 2393–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.-W. (2001. b). A role of Arabidopsis COP9 signalosome in multifaceted developmental processes revealed by the characterization of its subunit 3. Development 128, 4277–4288. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.-W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Serino, G., Tsuge, T., Kwok, S., Matsui, M., Wei, N., and Deng, X.-W. (1999). Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11, 1967–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra, D., Koepp, D.M., Kamura, T., Conrad, M.N., Conaway, R.C., Conaway, J.W., Elledge, S.J., and Harper, J.W. (1999). Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284, 662–665. [DOI] [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, G., Yanagawa, Y., Kwok, S., Matsui, M., and Deng, X.W. (2002). Arabidopsis COP10 is an ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 16, 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., Chamovitz, D.A., and Deng, X.-W. (1994). Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 78, 117–124. [DOI] [PubMed] [Google Scholar]
- Xiao, W., and Jang, J.-C. (2000). F-box proteins in Arabidopsis. Trends Plant Sci. 5, 454–457. [DOI] [PubMed] [Google Scholar]
- Xie, D.-X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W.L., Ma, H., Peng, W., Huang, D., and Xie, D. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]