Abstract
Significant progress has been made in elucidating the mechanism of abscisic acid (ABA)–regulated gene expression, including the characterization of an ABA-responsive element (ABRE), which is regulated by basic domain/Leu zipper transcription factors. In addition to the ABRE, a coupling element (CE1) has been demonstrated to be involved in ABA-induced expression. However, a trans factor that interacts with CE1 has yet to be characterized. We report the isolation of a seed-specific maize ABI4 homolog and demonstrate, using a PCR-based in vitro selection procedure, that the maize ABI4 protein binds to the CE-1 like sequence CACCG. Using electrophoretic mobility shift assays, we demonstrate that recombinant ZmABI4 protein binds to the CE1 element in a number of ABA-related genes. ZmABI4 also binds to the promoter of the sugar-responsive ADH1 gene, demonstrating the ability of this protein to regulate both ABA- and sugar-regulated pathways. ZmABI4 complements Arabidopsis ABI4 function, because abi4 mutant plants transformed with the ZmABI4 gene have an ABA- and sugar-sensitive phenotype. Identification of the maize ABI4 ortholog and the demonstration of its binding to a known ABA response element provide a link between ABA-mediated kernel development and the regulation of ABA response genes.
INTRODUCTION
Abscisic acid (ABA) plays an important role in a number of agronomically important traits, including the plant's response to abiotic stress and the deposition of storage protein and starch during seed development (Bewley and Black, 1994; Davies, 1995; Wobus and Weber, 1999). A greater understanding of the mechanism of ABA-regulated gene expression has been achieved in genetic and molecular studies (Grill and Himmelbach, 1998; McCourt, 1999). The dissection of the signal transduction pathway from ABA to gene activation has led to the identification of cis-acting elements important to ABA-induced gene expression. Most of these studies have identified ABA-responsive element (ABRE) or G-box–like elements as the important cis element for ABA-induced gene expression (Shen and Ho, 1995; Shen et al., 1996; Busk et al., 1997; Hobo et al., 1999a).
A substantial body of biochemical work has identified basic domain/Leu zipper (bZIP)–type transcription factors in the regulation of ABA-responsive genes because they have high binding affinity for ABRE/G-box (Guiltinan et al., 1990; Choi et al., 2000; Uno et al., 2000). Identification of ABI5 as a bZIP protein using a map-based cloning approach (Finkelstein and Lynch, 2000) confirms the role that this class of transcription factor plays in ABA-regulated gene expression. Other cis elements important for ABA-induced gene expression have been identified as coupling elements (Shen and Ho, 1995). Although Coupling Element3 (CE3) has been reported to be an ABRE/G-box equivalent (Hobo et al., 1999a), CE1 is distinct. The CE1-like element DRE2 also has been demonstrated to be important for the ABA induction of maize rab17 gene expression, and in vivo footprinting has demonstrated that an unidentified regulatory protein binds to this element during seed maturation and in ABA-treated embryos (Busk et al., 1997).
In addition to ABI5/bZIP transcription factors that interact with ABREs, two other transcription factors (ABI3/VP1 and ABI4) have been characterized that play a role in regulating the ABA response during seed development (Finkelstein and Somerville, 1990; Finkelstein, 1994). The ABI3 and VP1 proteins, which are thought to be orthologous proteins from Arabidopsis and maize, respectively, control key regulatory steps during the maturation stage of seed development (Finkelstein and Somerville, 1990; McCarty et al., 1991). The abi4 mutant from Arabidopsis was isolated from a similar ABA insensitivity screen used for abi3 and abi5 and was demonstrated to have similar effects on seed dormancy and the expression of maturation-specific seed proteins (Finkelstein, 1994). In addition, the abi4 mutant has been isolated from Arabidopsis in independent screens for Glc and Suc insensitivity (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rook et al., 2001), salt tolerance (Quesada et al., 2000), and regulation of the sugar-responsive gene ApL3, which encodes the large subunit of ADP-Glc pyrophosphorylase (Rook et al., 2001). Together, these results suggest that ABI4 is a focal point in the signal transduction pathways of ABA and sugar signaling.
Cloning of ABI4 from Arabidopsis revealed an AP2 domain characteristic of a class of transcription factors known to regulate plant development (Finkelstein and Goodman, 1998). Several genes have been shown to have altered expression in the abi4 mutant, including ABA response genes, such as Em6 and other late embryogenesis genes (Arenas-Huertero et al., 2000; Soderman et al., 2000), genes modulated by sugars, such as ApL3 and ADH (Arenas-Huertero et al., 2000; Rook et al., 2001), and photosynthesis-related genes, such as plastocyanin (Arenas-Huertero et al., 2000; Rook et al., 2001).
In this article, we describe a seed-specific gene from maize that has homology with the Arabidopsis ABI4 gene. Using an in vitro selection technique, we demonstrate that this protein binds to the sequence CACCG, which corresponds to the previously characterized CE1 element found in ABA-responsive genes. Transformation of the maize protein into the Arabidopsis abi4 mutant rescues the ABA- and sugar-sensitive phenotype, demonstrating that maize ABI4 can complement the Arabidopsis gene.
RESULTS
Isolation and Sequence Analysis of the ZmABI4 Gene
Three ESTs with a deduced protein sequence highly homologous with that of the AP2 domain of the Arabidopsis ABI4 protein were identified from the Pioneer/Dupont database. All three ESTs are from 20-days after pollination (DAP) maize embryo libraries and represent transcripts of the same gene. After sequencing of the full insert from the longest clone, an open reading frame of 747 bp was detected that, when translated, included the AP2 domain in frame. This sequence was used subsequently to isolate an upstream fragment of 2521 bp 5′ of the ATG start codon by genome walking. The sequence upstream of the open reading frame contains several potential TATA boxes, including one located 71 bp upstream of the longest cDNA (Figure 1). Genomic sequence analysis by PCR also indicated that the maize ABI4 homolog has no intron (data not shown). Based on sequence homology, we designated the maize homolog ZmABI4.
Figure 1.
Nucleotide and Deduced Amino Acid Sequences of the ZmABI4 Gene, Including the Upstream Region.
The deduced amino acid sequence is presented under the DNA sequence, and the stop codon is marked with an asterisk. A putative TATA sequence is underlined, and the start of the longest cDNA is double underlined.
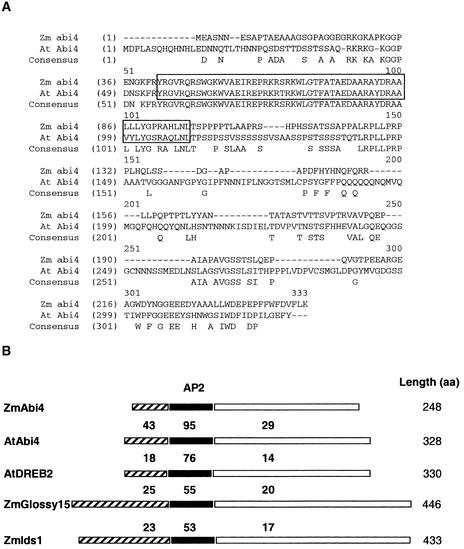
The deduced amino acid sequence revealed a significant degree of identity between the ZmABI4 and AtABI4 proteins in the AP2 domain (50 of 55 residues identical). However, the ZmABI4 protein is smaller, and outside of the AP2 domain, the sequence is divergent (Figure 2A). The overall structural features of AtABI4 are largely present in ZmABI4. For example, a Ser/Thr-rich sequence is present (9 of 24 residues, compared with 19 of 28 for Arabidopsis), as is a small Gln-rich domain (3 of 14 residues, compared with 13 of 21). The C-terminal half of the ZmABI4 protein is Pro rich, but a defined Pro domain is missing from the maize sequence. Lastly, the C-terminal 40 residues are high in acidic residues, similar to the Arabidopsis sequence. The identity between ZmABI4 and AtABI4 in the AP2 domain is significantly higher than that between ZmABI4 and other AP2 domain proteins in maize or Arabidopsis, and the overall size and structure are more similar between the ABI4 proteins than between the ABI4 proteins other AP2 proteins (Figure 2B).
Figure 2.
Comparison of ZmABI4 with AtABI4 and Other AP2 Proteins.
(A) Alignment of the deduced amino acid sequence of AtABI4 (At) and ZmABI4 (Zm). Single-letter amino acid residues were aligned, and the consensus sequence is presented underneath. The conserved AP2 domain is boxed.
(B) Comparison of the ZmABI4 protein sequence with other AP2 domain–containing protein sequences in maize and Arabidopsis. A scheme of the predicted ZmABI4 protein, divided into the N-terminal, AP2, and C-terminal regions, is aligned with AtABI4, OsABI4 (contigs 4094 and 1079; draft rice sequence), AtDREB2, ZmGlossy15, and ZmIds1. The percentage identity with ZmABI4 is listed above the scheme for each sequence.
Recently, a draft sequence of the rice genome was reported (Yu et al., 2002), and within the rice database, a single sequence shares significant homology with the AtABI4 AP2 domain, indicating that a single ABI4 homolog exists in rice. The rice ABI4 (OsABI4) also is highly homologous with ZmABI4, sharing 100% identity in the AP2 domain and the same structural features described above. Therefore, within available databases, the deduced maize protein sequence presented here represents the closest structural homolog of the Arabidopsis ABI4 protein.
Expression of the ZmABI4 Gene
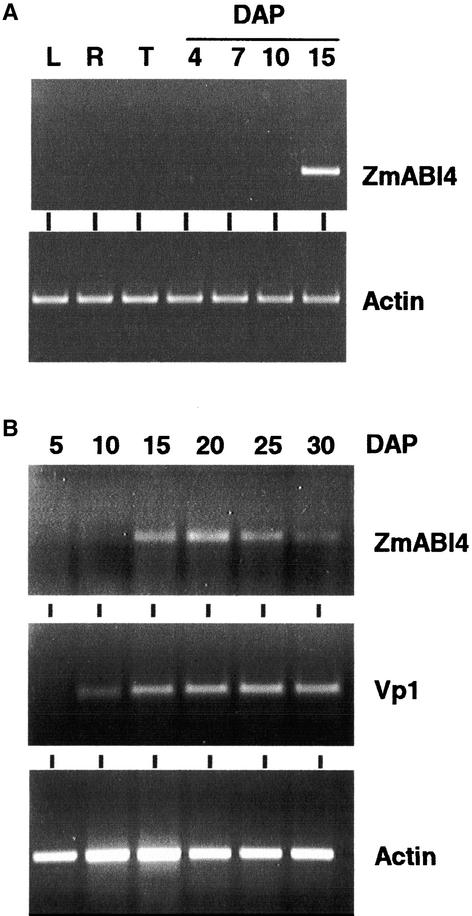
The ZmABI4 sequence is present only in libraries from 20-DAP kernels, and reverse transcriptase–mediated (RT) PCR analysis of vegetative tissues demonstrated that the ZmABI4 message was undetectable in leaf, root, and tassel tissues (Figure 3A), in contrast to Arabidopsis, in which AtABI4 was present at low levels in vegetative tissues (Soderman et al., 2000). In addition, treatment of vegetative tissues or 10-DAP kernels with ABA or high sugars did not induce transcript accumulation to detectable levels (data not shown). Collectively, the expression data indicate that ZmABI4 is kernel specific. During kernel development, ZmABI4 mRNA was detectable as early as 10 DAP, accumulated between 10 and 15 DAP (Figure 3B) at the transition to the storage phase of kernel development (starting after ∼12 DAP), and peaked at ∼20 DAP (Figure 3B). By contrast, mRNA levels for VP1, a transcription factor correlated with the storage and maturation phases of seed development, began to accumulate earlier and did not peak in expression; transcript continued to be present later in seed development (Figure 3B).
Figure 3.
Expression of ZmABI4 and VP1 during Seed Development.
(A) Expression of ZmABI4 in vegetative tissues and during early kernel development. RT-PCR analysis of ZmABI4 expression in leaf (L), root (R), tassel (T), and kernel tissue collected at 5, 7, 10, and 15 DAP.
(B) Transcript accumulation in kernel tissue collected throughout seed development (from 5 to 30 DAP) was measured by RT-PCR. Actin expression was included as a constitutive control.
Determination of the ZmABI4 Binding Sites
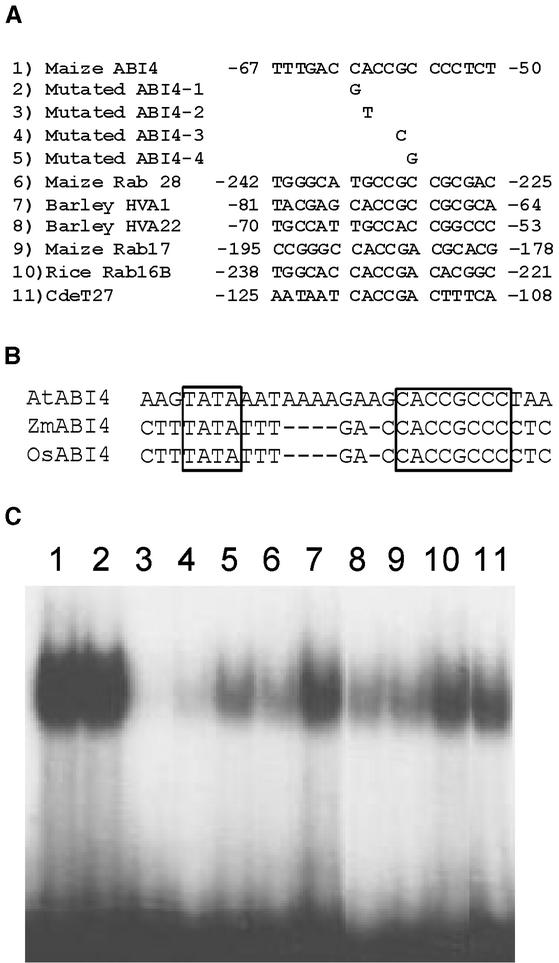
To determine the sequences to which the ABI4 protein may bind, and to gain a greater understanding of the function of the ABI4 protein, we used a PCR-assisted method to determine the binding site for this putative DNA binding protein. Recombinant ZmABI4 protein was incubated with a library of random double-stranded oligonucleotides in a binding reaction. After gel electrophoresis, the shifted protein/DNA complex was excised from the gel, purified, and subjected to PCR amplification. This procedure was repeated five additional times to enrich the sequences bound by the ZmABI4 protein. The enriched sequences from the sixth round of selection were cloned, and 52 independent clones were sequenced. The selected sequences are shown in Figure 4.
Figure 4.
ZmABI4 Binding Sequences Determined by PCR-Assisted Binding Site Selection.
Selected sequences are aligned around the consensus CACCG. Reverse complementary stands are presented for the sequences in group II. The clone number for each independent sequence is indicated at left. The two primer sites for the oligonucleotide used in the binding site selection are underlined. The selected sequences that correspond to ZmABI4, rab17, and HVA22 gene promoter elements are indicated at right.
Of 52 sequences, the majority (42) have the 5-bp consensus sequence CACCG, suggesting that the ZmABI4 protein has affinity for this pentamer sequence (Figure 4). We found that the CE1 site, a cis element shown previously to be functionally important to ABA-regulated gene expression (Shen and Ho, 1995), resembles the ZmABI4 binding sites. The CE1 element in the HVA22 promoter and the DRE2 element in the rab17 promoter both were identified in the selected sequences (Figure 4), and the ABI4 selected binding site also is present in the promoters of a number of other ABA-regulated genes (Figure 5A). Interestingly, a putative binding site was present in the maize ABI4 upstream region (Figures 4 and 5A), and a highly conserved ABI4 binding site also was present in the upstream sequence of the Arabidopsis and rice ABI4, immediately downstream from a putative TATA box, as observed in the maize upstream region (Figure 5B).
Figure 5.
Binding of ZmABI4 Protein to Upstream Sequences in ABA-Related Genes.
(A) Sequence alignment of upstream regions of ZmABI4 and ABA-regulated genes containing CE1-like sequences as well as mutated ZmABI4 binding sites used in electrophoretic mobility shift assay analysis. Numbering of sequence location is from the transcription start site for each gene except ZmABI4, which is numbered from the translation start. Sequences of maize ABI4, maize rab28 (Busk et al., 1999), barley HVA1 (Shen et al., 1996), barley HVA22 (Shen and Ho, 1995), maize rab17 (Busk et al., 1997), rice rab16B (Ono et al., 1996), and Craterostigma plantagineum CdeT27 (Michel et al., 1993) are presented.
(B) Alignment of the putative TATA box and the ABI4 binding site of Arabidopsis (AtABI4), maize (ZmABI4), and rice (OsABI4) ABI4 genes. The TATA box and the ABI4 binding site are boxed.
(C) Electrophoretic mobility shift assay analysis of ZmABI4 binding to the CE1-like sequences presented in (A).
ZmABI4 Binds to CE1-Like Elements in the Promoters of ABA- and Sugar-Responsive Genes and to Its Own Upstream Region
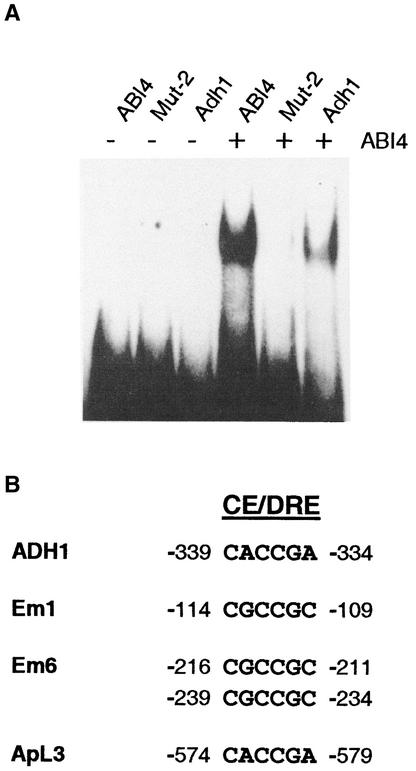
To confirm the binding of ZmABI4 to the selected site, double-stranded DNA probes were created to correspond to the CE1-like sequences found in ABA-related genes from a range of species. Figure 5C shows that recombinant ZmABI4 protein binds to a range of promoter elements, including those from the ZmABI4, maize rab28, maize rab17, barley HVA22, barley HVA1, and rice rab16b genes. Among these promoter elements, HVA22 CE1 and rab17 DRE2 elements have been shown to be important for ABA induction (Shen and Ho, 1995; Busk et al., 1997).
To determine the role of ZmABI4 in regulating sugar-sensitive genes, we studied the sugar-regulated maize ADH1 gene (Koch et al., 2000; X. Niu and N.J. Bate, unpublished data). Analysis of the maize ADH1 promoter revealed the presence of an ABI4 binding site (Tikhonov et al., 1999) 347 bp upstream of the translational start. Figure 6A shows that recombinant ABI4 binds to the ADH1 promoter with high affinity. Further analysis of the upstream sequences and promoter regions of a number of Arabidopsis sugar-responsive genes revealed the presence of ABI4 binding sites (Figure 6B). Expression of these genes in the Arabidopsis abi4 mutant also has been shown to be modified, relative to that of the wild type (Arenas-Huertero et al., 2000; Huijser et al., 2000; Rook et al., 2001).
Figure 6.
Electrophoretic Mobility Shift Assay Analysis of ZmABI4 Binding to the Maize ADH1 Promoter and Alignment of Promoter Elements of Sugar-Regulated Genes in Arabidopsis.
(A) Binding of maize ABI4 protein to the ABI4 binding site in the maize ADH1 gene (AF123535). The ADH1 sequence (5′-CGCCGCCACCGCTTGGCG-3′ [−246 to −229]; the ABI4 binding site is shown in boldface) was used for gel shift reactions (lanes 1 and 4). ABI4 (lanes 2 and 5) and Mut-2 (lanes 3 and 6) sequences were used as controls (see Figure 4A). Lanes 1 to 3 and 4 to 6 show reactions with and without ZmABI4, respectively.
(B) Potential ABI4 binding sites (designated CE/DRE) in Arabidopsis sugar-regulated genes (ADH1, Em6, PC, and ApL3). Numbering is from the transcription start site, except for ApL3, which is numbered relative to the start of translation.
Rescue of the Arabidopsis abi4 Mutant with ZmABI4
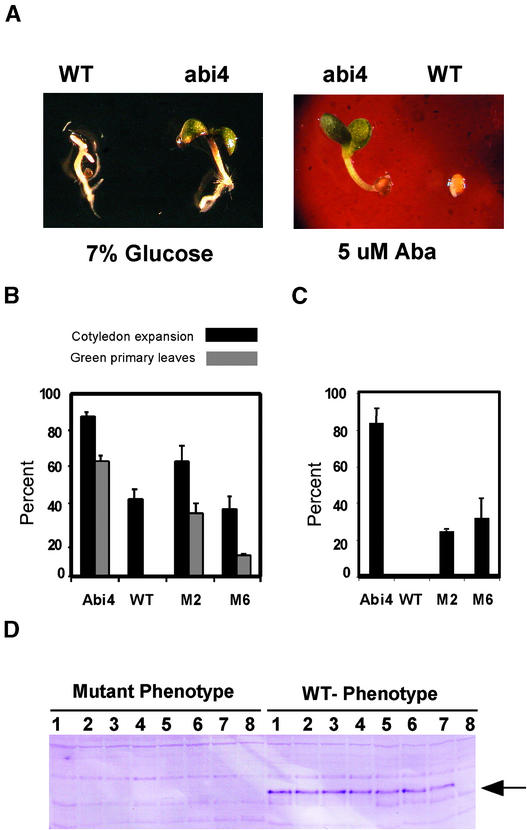
To demonstrate functional equivalence between AtABI4 and ZmABI4, we transferred the ZmABI4 gene, under the control of a constitutive promoter (the maize ubiquitin promoter, with glufosinate resistance as the selectable marker), into the abi4-1 mutant of Arabidopsis (obtained from the ABRC Stock Center, Columbus, OH). Glufosinate resistance was used to select a number of primary transformants, and seeds from these plants were used to establish copy number. Two independent single-insert lines of abi4/Ubi:ZmABI4 were selected for further characterization. To establish the ability of the ZmABI4 gene to rescue the Arabidopsis abi4 mutation, we used an abi4-1 line homozygous for the mutation and segregating for the presence of the Ubi::ZmABI4 transgene. Seeds from these lines were grown in the presence of repressive concentrations of ABA (5 μM) or Glc (7%).
When germinated in the presence of 5 μM ABA, wild-type plants produced a radicle but did not develop further (Figure 7A, right). Similarly, wild-type plants germinated in the presence of 7% Glc did germinate but had a reduced frequency of cotyledon expansion and did not produce green true leaves (Figure 7A, left). Therefore, early development of seedlings under these repressive conditions provides an effective screen to assess the ability of the ZmABI4 gene to rescue the phenotype of the Arabidopsis abi4 mutation. Surface-sterilized mutant seeds segregating for the presence of the transgene were placed onto minimal medium plates containing 5 μM ABA or 7% Glc. Single-insert lines segregating for the transgene have a 3:1 ratio of wild-type to mutant phenotype if ZmABI4 rescues the Arabidopsis abi4 mutation. If the transgene does not rescue the Arabidopsis mutant, the seedlings all have the mutant phenotype.
Figure 7.
The Maize ZmABI4 Gene Can Rescue Wild-Type Sensitivity in the abi4 Mutant Background of Arabidopsis Grown in the Presence of 5 μM ABA or 7% Glc.
(A) Wild-type (WT) and abi4-1 mutant phenotypes of plants germinated in the presence of 7% Glc or 5 μM ABA.
(B) Rescue of the abi4-1 mutant phenotype by ectopic ZmABI4 expression in plants germinated in the presence of 7% Glc. Comparison of wild-type (Columbia), abi4-1, and two segregating ZmABI4 transgenic lines in an abi4-1 background. The mean frequencies of cotyledon expansion and green primary leaves are expressed as the mean percentage and standard deviation of three plates containing >100 seeds each.
(C) Rescue of the sensitivity to ABA phenotype by ectopic ZmABI4 expression. Imbibed seeds were spread onto plates containing 5 μM ABA as in Figure 6B and scored after 5 days of constant illumination for their ability to develop expanded green cotyledons.
(D) Phenotype of segregating plants correlates with the presence of ZmABI4. Segregating seedlings grown on 7% Glc medium (from [B]) were grouped into wild-type or mutant phenotypes and placed onto medium without selection. After 10 days of recovery, proteins were extracted, and the presence of the ZmABI4 protein was detected by protein gel blot analysis with a ZmABI4-specific antibody.
Figure 7B indicates that in two independent lines of abi4 mutant Arabidopsis, ectopic expression of the ZmABI4 gene compensated for the genetic lesion in Arabidopsis abi4. Approximately 75% of the seedlings had the wild-type phenotype when grown on 5 μM ABA, in keeping with a 3:1 segregation of the transgene.
Screening of the same segregating population for the ability to germinate and form true green leaves in the presence of inhibitory concentrations of Glc gave similar results (Figure 7C). Mutant seed lines containing a segregating transgene were treated as described above, except that 7% Glc was the inhibitory reagent. Plants were allowed to imbibe for 5 days to ensure that germination was initiated before plating. Both transgenic lines rescued the mutant phenotype in a ratio close to the expected 3:1 (Figure 7C).
To further demonstrate cosegregation of the wild-type phenotype and the transgene, seedlings with the wild-type and mutant phenotypes were removed from the 7% Glc plates and placed onto nonselective minimal medium plates containing 2% Glc. After 1 week of recovery, the plants were assayed for the presence of the transgene by protein gel blot analysis with a ZmABI4-specific antibody. None of the plants (eight of eight surveyed) with a mutant phenotype contained the transgene, whereas seven of eight seedlings with the wild-type phenotype possessed a functional transgene when grown on 7% Glc (Figure 7D).
DISCUSSION
In this study, we present the characterization of the maize homolog of Arabidopsis ABI4 and demonstrate that it binds to a cis-regulatory site shown previously to be essential for the induction of genes by ABA. Although the protein sequence for the maize and Arabidopsis genes are divergent outside of the AP2 domain, the overall similarity is in line with orthologous proteins in maize and Arabidopsis, such as VP1 and ABI3. No EST is present in the Pioneer/Dupont or public maize databases that has greater homology with the Arabidopsis ABI4 gene, and there is significant structural divergence between the ABI4 proteins and other AP2 domain proteins from either species. Furthermore, screening of the Arabidopsis, maize, and rice databases with ABI4 sequences did not identify any other AP2 proteins with greater identity than AtABI4, ZmABI4, and OsABI4. All three ABI4 sequences share a stretch of significant homology, including an ABI4 binding site, immediately downstream of the TATA box (Figure 5B). Together, these results suggest that the AP2 protein presented here is the maize ortholog of the Arabidopsis ABI4 gene.
The expression pattern of ZmABI4 is consistent with a transcription factor that plays a role in the initiation or maintenance of the storage phase of maize seed development. The prestorage-to-storage phase transition occurs at ∼12 to 15 DAP, continuing until the maturation phase begins at ∼30 DAP. Expression of ZmABI4 appears to be under strict developmental control, because no expression was detectable in vegetative tissues and ZmABI4 transcript was not induced by ABA or sugars in leaf, root, or early kernel tissue. The timing of ZmABI4 gene expression also was consistent with the induction of the CE1 element containing genes such as rab17 and ADH1. Rab17 message was low in abundance during early seed development and began to accumulate rapidly in the embryo between 15 and 20 DAP (data not shown), coincident with ZmABI4 expression. Similarly, ADH1 expression was induced approximately fourfold between 8 and 20 DAP (data not shown). However, in vegetative tissues, rab17 was induced in response to abiotic stress, indicating that a distinct regulatory protein binds to the CE1 element outside of seed development, as has been suggested by footprinting data (Busk et al., 1997).
Mapping results provide further evidence to link ZmABI4 with seed development and yield. ZmABI4 was mapped on the maize genome and found to be located on chromosome 6 (bin 5) (data not shown), which is a similar map location as that of rab17 (Frova et al., 1998). This region of chromosome 6 has been shown to be the location of significant quantitative trait loci in previous studies of yield under drought stress conditions (Ribaut et al., 1997; Quarrie et al., 1999), but it is not associated with leaf ABA content (Tuberosa et al., 1998).
A model for the role of ABI4 in the regulation of ABA-responsive genes is presented in Figure 8. Two hybrid screens using VP1 as bait isolated TRAB1, a bZIP protein (Hobo et al., 1999b), but ABI3 (VP1 ortholog) and ABI4 do not interact physically (Nakamura et al., 2001). These physical interaction results are consistent with functional studies in which it has been shown that ABRC3 (bZIP complex) can be activated by VP1, whereas ABRC1 (bZIP plus the ABI4 configuration) is not activated by VP1 (Shen et al., 1996). From these results, it is clear that different regulatory mechanisms exist to control ABA-responsive genes, and other regulatory factors that interact with ABI4 are required to complete our understanding of ABI4-mediated gene expression.
Figure 8.
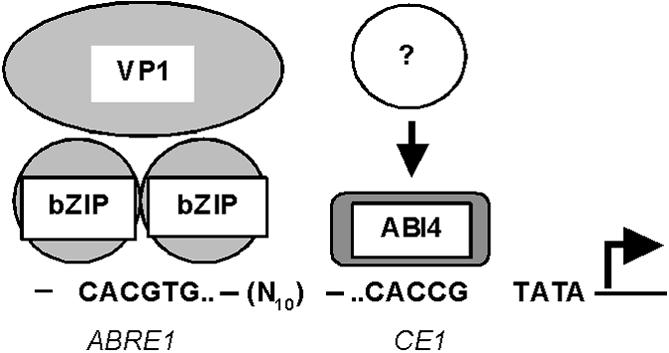
Model for the Developmental Expression of ABA-Responsive Genes.
In storage-phase embryos, ABI4 binds to the CE1 element, and in conjunction with bZIP, VP1, and other unidentified components, activates storage- and maturation-phase genes, such as rab17.
CE1-like elements also are present in the promoters of Arabidopsis sugar-responsive genes, including Em6, ADH1, PC, and ApL3, consistent with the fact that these genes have altered expression in the abi4 mutant. Significantly, PC and ApL3 promoter mutant screens identified ABI4 as a crucial regulatory factor in Arabidopsis (Huijser et al., 2000; Rook et al., 2001). The ability of ZmABI4 to rescue sugar sensitivity when transformed into mutant abi4-1 plants provides further evidence that ABI4-mediated transcriptional regulation of sugar-responsive genes through the CE1-like element is an important control point. In Arabidopsis, ABI4 is thought to be an important regulatory component in the progression of early seedling development, responding to sugar and ABA signals that act together or separately (Gibson, 2000). Although it is clear that ZmABI4 can rescue sugar response functions of the Arabidopsis abi4 mutant, the role of ZmABI4 in maize appears to be strictly developmental (i.e., responding to a kernel development program to regulate storage-related genes). Whether or not ZmABI4 function is mediated by ABA and sugars remains to be determined.
The presence of a strong binding site for ABI4 in its own promoter in maize, rice, and Arabidopsis is interesting in light of their conserved locations downstream of the putative TATA box. Binding of regulatory proteins to sites downstream of the TATA box has been reported to be important to the transcriptional regulation of Shrunken (Bellmann and Werr, 1992) and NPR1 (Yu et al., 2001). Binding of ABI4 to its own promoter may serve a function distinct from binding to downstream ABA/sugar-responsive genes. One possibility is that binding to a site immediately downstream suppresses ABI4 transcription in an autoinhibitory manner, because protein binding to TATA-proximal regions has been shown to repress transcription (Heins et al., 1992).
Collectively, the data presented here provide strong evidence for the conservation of regulatory mechanisms that control seed development in plants. By establishing a link between mutant phenotype, gene expression, and DNA binding, the characterization of ZmABI4 provides further insight into the role of ABI4 in ABA and sugar signal transduction pathways and into the regulatory framework that controls seed development.
METHODS
cDNA and Gene Isolation
A sequence with a deduced AP2 domain homologous with the Arabidopsis thaliana ABI4 gene was identified from the Pioneer/Dupont EST databases. Genomic DNA upstream of the coding sequence was isolated using the Universal GenomeWalker Kit from Clontech (Palo Alto, CA). Additional sequence information was obtained by PCR analysis of genomic DNA and cDNA. Mapping of the maize (Zea mays) ABI4 gene (ZmABI4) was performed as described previously (Tarchini et al., 2000).
Expression Analysis by Reverse Transcriptase–Mediated PCR
For reverse transcriptase–mediated (RT) PCR analysis, oligonucleotides were designed to the 3′ end of the maize ABI4 gene (5′-GGCGAGGAGGAGGATTATGC-3′ and 5′-CCTCAATCACACCCTTAG-GG-3′) to amplify a 307-bp gene-specific fragment. RNA was isolated from maize tissues using the Plant RNeasy kit from Qiagen (Valencia, CA) or the Trizol method (Invitrogen, Carlsbad, CA). Purified RNA from each sample was subjected to DNase treatment according to the manufacturer's instructions (amplification-grade DNase; Invitrogen). RT-PCR was performed with 1 μg of total RNA using the ThermoScript kit (Invitrogen), and the cDNA was used in PCR to detect ZmABI4 transcript. The same reactions were used to detect actin message as a positive control. For the detection of VP1 transcript, a 328-bp fragment was amplified using the gene-specific primers 5′-TGCAAGGGGAGGTCTCCGGA-3′ and 5′-GGGCCCAAC-TCACAAACCAAACC-3′.
Gel-Shift Assays and DNA Binding Site Selection
Nearly full-length maize ABI4 protein (amino acids 6 to 248) was overexpressed in Escherichia coli as a glutathione S-transferase fusion and purified according to established protocols (Pharmacia glutathione S-transferase fusion purification manual). The purified protein was used for gel-shift assays as described (Niu et al., 1999). For DNA probes, 18-bp complementary oligonucleotides with four base overhangs were synthesized and filled with Klenow in the presence of 32P.
DNA binding site selection for maize ABI4 was essentially as described (Niu et al., 1999) except that the random oligonucleotide 5′-CAGCTGAATTCGTAGGAC(N)10GAACGGATCCAGATCTCA-3′ was used. After six rounds of selection, the selected sequences were cloned into pCR2.1 (Invitrogen) and sequenced.
Transgenic Construct and Plant Transformation
A DNA fragment encoding amino acids 6 to 248 of maize ABI4 was cloned into a superbinary transgenic vector by incorporating a translation start site ATG into an NcoI site. Ectopic overexpression of maize ABI4 was engineered by placing it under the control of the maize ubiquitin promoter.
The floral dip method was used for Arabidopsis transformation essentially as described (Clough and Bent, 1998). abi4-1 mutant plants (stock number CS8104; obtained from the ABRC Stock Center at Ohio State University, Columbus) were used for transformation. Subsequently, seeds from dipped plants were grown on soil and sprayed four times with glufosinate (1 mg/mL) to select for recombinant plants, and the seeds were harvested. Copy number was established by segregation of glufosinate resistance in the subsequent generation.
Arabidopsis Growth Conditions and Rescue Experiments
For rescue experiments, a segregating single-insert line was used. Seeds were sterilized in 30% bleach and rinsed and allowed to imbibe in 0.1% agar for 5 days before plating onto minimal medium plates containing 2% Suc, 2% Glc, 7% Glc, or 2% Suc plus 5 μM abscisic acid. For abscisic acid rescue experiments, the plants were scored at 5 days after sowing, and successful germination was defined as the development of green cotyledons (Finkelstein, 1994). For Glc rescue experiments, plants were scored at 9 days after sowing for the presence of expanded cotyledons and green primary leaf formation (Arenas-Huertero et al., 2000). In each experiment, three plates with ∼100 seeds were scored, and means and standard deviations were calculated. Protein gel blot analysis was performed using the manufacturer's protocols (Bio-Rad) using a rabbit polyclonal antibody raised against recombinant ZmABI4 protein (amino acid residues 144 to 248).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are AAAA01003042 (AtABI4 AP2 domain), AY125490 (ZmABI4), AAD25937 (AtABI4), BAA33795 (AtDREB2), TO3981 (ZmGlossy15), ACC05206 (ZmIds1), X77943 (Arabidopsis ADH1), Z11157 (Em6), S67901 (PC), and Y18432 (ApL3).
Acknowledgments
The authors thank Michael Muszynski, Norbert Brugière, and Steven Rothstein for critical reading of the manuscript and members of the Yield Stability Group for useful discussion. We also thank the Arabidopsis Biological Resource Center for providing abi4-1 seeds. The Pioneer Hi-Bred facilities also are gratefully acknowledged for mapping and protein production.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003400.
References
- Arenas-Huertero, F., Arroyo, A., Zhou, L., Sheen, J., and Leon, P. (2000). Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant vegetative development by sugar. Genes Dev. 14, 2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Bellmann, R., and Werr, W. (1992). Zmhox1a, the product of a novel maize homeobox gene, interacts with the Shrunken 26 bp feedback control element. EMBO J. 11, 3367–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley, J.D., and Black, M. (1994). Seeds: Physiology of Development and Germination. (New York: Plenum Press).
- Busk, P., Jensen, A.B., and Pages, M. (1997). Regulatory elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize. Plant J. 11, 1285–1295. [DOI] [PubMed] [Google Scholar]
- Busk, P.K., Pujal, J., Jessop, A., Lumbreras, V., and Pages, M. (1999). Constitutive protein-DNA interactions on the abscisic acid-responsive element before and after developmental activation of the rab28 gene. Plant Mol. Biol. 41, 529–536. [DOI] [PubMed] [Google Scholar]
- Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (1995). The plant hormones: Their nature, occurrence and functions. In Plant Hormones: Physiology, Biochemistry and Molecular Biology, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–12.
- Finkelstein, R., and Goodman, H. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 10, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R., and Lynch, T. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5, 765–771. [Google Scholar]
- Finkelstein, R.R., and Somerville, C.R. (1990). Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol. 94, 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frova, C., Caffulli, A., and Pallavera, E. (1998). Mapping quantitative trait loci for tolerance to abiotic stresses in maize. J. Exp. Zool. 282, 164–170. [Google Scholar]
- Gibson, S.I. (2000). Plant sugar-response pathways: Part of a complex regulatory web. Plant Physiol. 124, 1532–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill, E., and Himmelbach, A. (1998). ABA signal transduction. Curr. Opin. Plant Biol. 1, 412–418. [DOI] [PubMed] [Google Scholar]
- Guiltinan, M.J., Marcotte, W.R., Jr., and Quatrano, R.S. (1990). A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250, 267–271. [DOI] [PubMed] [Google Scholar]
- Heins, L., Frohberg, C., and Gatz, C. (1992). The Tn10-encoded Tet repressor blocks early but not late steps of assembly of the RNA polymerase II initiation complex in vivo. Mol. Gen. Genet. 232, 328–331. [DOI] [PubMed] [Google Scholar]
- Hobo, T., Asada, M., Kowyama, Y., and Hattori, T. (1999. a). ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 19, 679–689. [DOI] [PubMed] [Google Scholar]
- Hobo, T., Kowyama, Y., and Hattori, T. (1999. b). A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 96, 15348–15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser, C., Kortstee, A., Pego, J., Weisbeek, P., Wisman, E., and Smeekens, S. (2000). The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: Involvement of abscisic acid in sugar responses. Plant J. 23, 577–585. [DOI] [PubMed] [Google Scholar]
- Koch, K., Ying, Z., Wu, Y., and Avigne, W.T. (2000). Multiple paths of sugar-sensing and a sugar/oxygen overlap for genes of sucrose and ethanol metabolism. J. Exp. Bot. 51, 417–427. [DOI] [PubMed] [Google Scholar]
- Laby, R.J., Kincaid, M.S., Kim, D., and Gibson, S.I. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23, 587–596. [DOI] [PubMed] [Google Scholar]
- McCarty, D., Hattori, T., Carson, C.B., Vasil, V., Lazar, M., and Vasil, I.K. (1991). The Viviparous-1 development gene of maize encodes a novel transcriptional activator. Cell 66, 895–905. [DOI] [PubMed] [Google Scholar]
- McCourt, P. (1999). Genetic analysis of hormone signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 219–243. [DOI] [PubMed] [Google Scholar]
- Michel, D., Salamini, F., Bartels, D., Dale, P., Baga, M., and Szalay, A. (1993). Analysis of a desiccation and ABA-responsive promoter isolated from the resurrection plant Craterostigma plantagineum. Plant J. 4, 29–40. [DOI] [PubMed] [Google Scholar]
- Nakamura, S., Lynch, T.J., and Finkelstein, R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26, 627–635. [DOI] [PubMed] [Google Scholar]
- Niu, X., Renshaw-Gegg, L., Miller, L., and Guiltinan, M.J. (1999). Bipartite determinants of DNA-binding specificity of plant basic leucine zipper proteins. Plant Mol. Biol. 41, 1–13. [DOI] [PubMed] [Google Scholar]
- Ono, A., Izawa, T., Chua, N.H., and Shimamoto, K. (1996). The rab16B promoter of rice contains two distinct abscisic acid-responsive elements. Plant Physiol. 112, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrie, S.A., Lazic-Jancic, V., Kovacevic, D., Steed, A., and Pekic, S. (1999). Bulk segregant analysis with molecular markers and its use for improving drought resistance in maize. J. Exp. Bot. 50, 1299–1306. [Google Scholar]
- Quesada, V., Ponce, M.R., and Micol, J. (2000). Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154, 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribaut, J.-M., Jiang, C., Gonzalez-de-Leon, D., Edmeades, G.O., and Hoisington, D.A. (1997). Identification of quantitative trait loci under drought conditions in tropical maize. 2. Yield components and marker-assisted selection strategies. Theor. Appl. Genet. 94, 887–896. [Google Scholar]
- Rook, F., Corke, F., Card, R., Munz, G., Smith, C., and Bevan, M.W. (2001). Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signaling. Plant J. 26, 421–433. [DOI] [PubMed] [Google Scholar]
- Shen, Q., and Ho, T.-H.D. (1995). Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q., Zhang, P., and Ho, T.-H. (1996). Modular nature of abscisic acid (ABA) response complexes: Composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8, 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderman, E.M., Brocard, I.M., Lynch, T.J., and Finkelstein, R.R. (2000). Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 124, 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini, R., Biddle, P., Wineland, R., Tingey, S., and Rafalski, A. (2000). The complete sequence of 340 kb of DNA around the rice Adh1-Adh2 region reveals interrupted colinearity with maize chromosome 4. Plant Cell 12, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A.P., SanMiguel, P.J., Nakajima, Y., Gorenstein, N.M., Bennetzen, J.L., and Avramova, Z. (1999). Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96, 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuberosa, R., Sanguineti, M.C., Landi, P., Salvi, S., Casarini, E., and Conti, S. (1998). RFLP mapping of quantitative trait loci controlling abscisic acid concentration in leaves of drought-stressed maize (Zea mays L.). Theor. Appl. Genet. 97, 744–755. [Google Scholar]
- Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus, U., and Weber, H. (1999). Seed maturation: Genetic programs and control signals. Curr. Opin. Plant Biol. 2, 33–38. [DOI] [PubMed] [Google Scholar]
- Yu, D., Chen, C., and Chen, Z. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13, 1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 76, 79–92. [DOI] [PubMed] [Google Scholar]