INTRODUCTION
The power of molecular genetics has dramatically advanced our understanding of all aspects of gibberellin (GA) signaling. Many genes encoding GA response pathway components have been identified using Arabidopsis and cereal mutants, and more elaborate genetic screens are producing additional mutants that are providing new insights into this pathway. Now, a confluence of new approaches, such as functional genomics and proteomics, promises even more rapid progress in unraveling the physiology and biochemistry of GA-regulated processes. New levels of regulation are being identified that indicate that the GA response pathway is linked tightly to its biosynthesis and catabolism. Furthermore, the GA metabolism and response pathways integrate with other signaling pathways to regulate plant growth and development, and the interaction between these response pathways is likely to be extremely complex.
Several recent reviews have described GA biosynthesis and catabolism (Lange, 1998; Kamiya and García-Martínez, 1999; Hedden and Phillips, 2000; Yamaguchi and Kamiya, 2000) and the GA response pathway (Thornton et al., 1999b; Lovegrove and Hooley, 2000; Sun, 2000; Richards et al., 2001). This review focuses on recent results in the field of GA signaling. We begin by considering the aspects of the homeostatic and developmental control of GA biosynthesis and catabolism that relate to signaling. Then, we discuss components of the GA response pathway that have been identified primarily through the analysis of mutants exhibiting altered GA responses. We conclude with a summary of the cereal aleurone, which has proven to be an excellent model system for studies of GA signaling.
GA BIOSYNTHESIS AND CATABOLISM
GAs form a large family of diterpenoid compounds, some of which are bioactive growth regulators, that control such diverse developmental processes as seed germination, stem elongation, leaf expansion, trichome development, and flower and fruit development (Davies, 1995). There is much evidence that environmental stimuli, including light and temperature, can affect plant processes by either changing the GA concentrations and/or altering the responsiveness to GA (reviewed by Davies, 1995; Kamiya and García-Martínez, 1999).
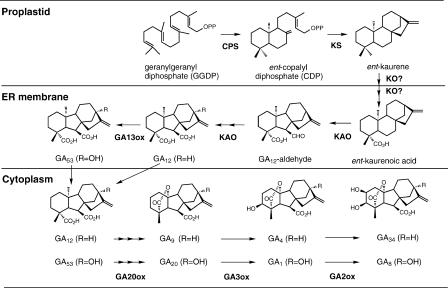
Studies of the regulation of GA concentration have been hindered by the low abundance of GAs, which necessitates the use of large samples and often precludes the analysis of single organs or portions of organs. Another way to gain insight into the regulation of GA concentrations is to examine the expression of genes encoding enzymes involved in GA biosynthesis and catabolism. Using reporter genes and in situ hybridization, it is possible to characterize expression at the level of single cells. Recently, the genes encoding most of the enzymes involved in GA biosynthesis (except the GA-13 hydroxylase; Figure 1) and the initial steps in catabolism were isolated and characterized. Examination of the expression patterns of these genes is revealing the sites of GA metabolism during development and the complex regulatory mechanisms by which endogenous developmental cues and light control the concentrations of bioactive GAs.
Figure 1.
Major GA Biosynthetic and Catabolic Pathways in Higher Plants.
The enzyme names are shown in boldface below or to the right of each arrow. GA9 and GA20 also can be converted to GA51 and GA29 by GA2ox. GA4 and GA1 are the bioactive GAs, and GA34 and GA8 are their inactive catabolites.
The biosynthesis of GA in higher plants can be divided into three stages: (1) biosynthesis of ent-kaurene in proplastids; (2) conversion of ent-kaurene to GA12 via microsomal cytochrome P450 monooxygenases; and (3) formation of C20- and C19-GAs in the cytoplasm (Figure 1). In the first stage, geranylgeranyl diphosphate, which serves as a common precursor for diterpenes (e.g., GAs and the phytol chain of chlorophyll) and tetraterpenes (carotenoids), is synthesized by either a mevalonate-dependent or a nonmevalonate pathway (reviewed by Lange, 1998; Hedden and Phillips, 2000). Geranylgeranyl diphosphate then is converted to ent-kaurene in a two-step cyclization reaction and catalyzed by ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS), with ent-copalyl diphosphate as the intermediate.
In the second stage of the pathway, stepwise oxidation followed by ring contraction catalyzed by ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO) produces GA12, which can be converted further to GA53 by 13-hydroxylation. In the third stage of GA biosynthesis, GA12 and GA53 are converted to various GA intermediates and bioactive GAs, including GA1 and GA4, by a series of oxidation steps catalyzed by 2-oxoglutarate–dependent dioxygenases, GA 20-oxidases (GA20ox), and GA 3-oxidases (GA3ox; Figure 1). Although 126 GAs have been identified in higher plants, fungi, and bacteria (http://www.plant-hormones.bbsrc.ac uk/gibberellin_information2.htm), only a small number are biologically active (e.g., GA1, GA3, GA4, and GA7) (Hedden and Phillips, 2000). Many of the other GAs are biosynthetic intermediates or catabolites of bioactive GAs. Figure 1 shows the two major pathways in plants that lead to the formation of GA1 and GA4. The amount of bioactive GAs can be affected by both the rate of their synthesis and the conversion to inactive forms by 2β-hydroxylation, which is catalyzed by GA 2-oxidases (GA2ox; Figure 1).
The synthesis of ent-kaurene, the first committed intermediate in the GA pathway, is catalyzed by CPS and KS. In Arabidopsis, extremely low amounts of CPS mRNA are found during plant development, and the expression pattern of this gene is cell type specific (Silverstone et al., 1997a). Rapidly growing tissues, including shoot apices, root tips, developing anthers, and seed, contain higher amounts of AtCPS mRNA. The expression pattern of AtKS is similar to that of AtCPS, but the overall amount of AtKS mRNA is much higher than that of AtCPS (Yamaguchi et al., 1998b; S. Yamaguchi and T.-p. Sun, unpublished results), suggesting that the synthesis of ent-kaurene is determined primarily by controlling the expression and location of CPS. The highly specific pattern of expression of the AtCPS gene and the greater expression of the AtKS gene both support previous speculations that AtCPS might act as a gatekeeper, controlling the location and activity of the early stages of GA biosynthesis.
CPS, KS, and KO each is encoded by a single gene in most plant species examined, and their expression is not under feedback regulation by the activity of the GA response pathway (reviewed by Hedden and Phillips, 2000). Two KAO genes are present in Arabidopsis, and both are expressed in all tissues examined (Helliwell et al., 2001a). In contrast, the cytosol-localized GA20ox, GA3ox, and GA2ox each is encoded by a small gene family, and each gene also shows tissue-specific expression patterns (reviewed by Hedden and Phillips, 2000). Overexpression of these genes in transgenic plants in some cases alters the concentrations of bioactive GA, indicating that the regulation of these genes is crucial in modulating flux in the late stages of the pathway (reviewed by Hedden and Phillips, 2000; Yamaguchi and Kamiya, 2000).
Another level of regulation of the GA biosynthetic pathway may reside in the subcellular compartmentalization of the pathway (Figure 1). Earlier biochemical studies suggested that ent-kaurene is synthesized in the proplastid because CPS and KS are localized in this organelle (Sun and Kamiya, 1994; Aach et al., 1997). KO is a cytochrome P450 enzyme that is thought to associate with the endoplasmic reticulum (ER) (reviewed by Graebe, 1987), and it is not clear how the hydrocarbon ent-kaurene is transported to the ER membrane. Recently, transient expression studies of AtKS–, AtKO–, AtKAO1–, and AtKAO2–green fluorescent protein (GFP) fusions in tobacco leaves demonstrated that AtKS-GFP and AtKO-GFP are targeted to the chloroplasts, whereas AtKAO1 and AtKAO2 fusion proteins are associated with the ER (Helliwell et al., 2001b). In vitro import experiments using isolated pea chloroplasts further confirmed that AtKS is imported into the chloroplast stroma, whereas the P450 enzyme AtKO is located on the outer surface of the chloroplast envelope.
These new findings provide a plausible mechanism for how AtKO gains access to its substrate ent-kaurene. However, these studies used the 35S promoter of Cauliflower mosaic virus to transiently overexpress the AtKO-GFP fusion protein in tobacco leaves, and some GFP signal also was detected in the cytoplasm (possibly in the ER) of epidermal cells (Helliwell et al., 2001b). Additionally, these authors' attempt to detect fluorescence in the transgenic plants carrying the same gene fusion was unsuccessful, most likely because of the strong fluorescence from the chlorophylls. Therefore, it needs to be verified whether AtKO is localized only on the plastid envelope in planta.
Previous analyses of the amounts of GA in GA response mutants indicate that the degree of tissue responsiveness to the bioactive GAs influences GA metabolism by a feedback mechanism (reviewed by Bethke and Jones, 1998; Phillips, 1998; Hedden and Phillips, 2000). With the few exceptions noted below, most of the GA20ox and GA3ox genes, whose products catalyze the penultimate and final steps, respectively, in the formation of bioactive GAs (GA1 and GA4), are downregulated by applied GA (reviewed by Hedden and Phillips, 2000; Yamaguchi and Kamiya, 2000). In contrast, the genes encoding 2-oxidases, which convert active GAs to inactive catabolites, are upregulated by GA treatment (Thomas et al., 1999; Elliott et al., 2001). This feedback regulation controlling the concentration of active GAs is discussed in more detail below.
DEVELOPMENTAL REGULATION OF GA BIOSYNTHESIS, CATABOLISM, AND TRANSPORT
In the last several years, a large number of genes encoding enzymes in all steps of the GA biosynthetic and catabolic pathways, except for 13-hydroxylation, have been isolated. Localization studies with these cloned genes using the techniques of in situ RNA hybridization and promoter-reporter genes are beginning to reveal the sites of GA metabolism. These studies also are revealing the complex mechanisms by which the amounts of bioactive GAs are modulated in different tissues to regulate plant growth and development.
During germination, GA promotes embryo growth and/or reduces the physical restraint imposed by the endosperm and testa that allows radicle protrusion. In Arabidopsis seed, the primary role of GA seems to be to facilitate the breakage of the seed coat. When the seed coat was damaged mechanically, ga1-3 mutant embryos were able to germinate (Silverstone et al., 1997b; Telfer et al., 1997). In tomato and tobacco, GA has been implicated in the induction of hydrolytic enzymes that degrade the endosperm (Groot and Karssen, 1987; Leubner-Metzger et al., 1996). Mutations that affect abscisic acid biosynthesis or seed coat structure in Arabidopsis or tomato were able to rescue the seed-germination defect of GA biosynthesis mutants, suggesting that GA is essential to overcome the inhibitory effects of the seed coat and abscisic acid–related embryo dormancy on seed germination (Groot and Karssen, 1987; Debeaujon and Koornneef, 2000).
In situ hybridization showed the presence of AtKO1, AtGA3ox1, and AtGA3ox2 mRNAs in the cortex and endodermis of embryo axes of germinating Arabidopsis seed, indicating that the synthesis of bioactive GAs from ent-kaurene occurs in these cells (Yamaguchi et al., 2001). Cortical cells in the embryo axis begin to expand before radicle protrusion. Therefore, the site of bioactive GA synthesis seems to occur in rapidly expanding cells, which are likely to be the GA-responding cells. Although AtGA3ox1 and AtGA3ox2 were expressed in the same cellular locations in the germinating embryo, only AtGA3ox1 expression was under feedback control by the GA response pathway. It was hypothesized that AtGA3ox2 may be required for the embryo to produce the higher level of bioactive GA needed to ensure efficient germination. Another surprising finding reported by Yamaguchi et al. (2001) is that the location of AtKO1, AtGA3ox1, and AtGA3ox2 expression appeared to be different from that of the early GA biosynthetic gene AtCPS.
Using an AtCPS promoter::β-glucuronidase (GUS) gene fusion in transgenic Arabidopsis, GUS activity was detected in the shoot apex and provasculature in both cotyledons and embryo axes, whereas AtKO1, AtGA3ox1, and AtGA3ox2 were expressed in the cortex and endodermis in embryo axes (as described above). Therefore, there is a physical separation of steps in the GA biosynthetic pathway in germinating Arabidopsis embryos, implying that the synthesis of active GAs in the germinating embryo requires intercellular transport (if only short range) of a pathway intermediate (CDP or ent-kaurene). Because earlier biochemical studies provided evidence that CPS and KS may form a complex in catalyzing the two consecutive cyclization reactions (West et al., 1982), it was suggested that ent-kaurene probably is the transported compound (Yamaguchi et al., 2001). Future analysis of AtKS mRNA localization in germinating seed will be crucial to determine the nature of the transported intermediate.
The physical separation of the early and late steps of GA biosynthesis during seed germination may be part of the mechanism that modulates the synthesis of bioactive GAs by regulating the transport of intermediate(s) between cells. Alternatively, this phenomenon may be simply the evolutionary consequence of this pathway. When detailed knowledge of the locations of the different steps of GA biosynthesis at later developmental stages becomes available, it will be interesting to discover whether physical separation of the early and late stages of the pathway occurs there as well. As described above, transiently expressed AtKO was shown to be targeted mainly to the plastids in tobacco leaf cells (Helliwell et al., 2001b). If ent-kaurene has to be transported between cells, presumably by unidentified carriers, the plastid envelope location of AtKO would seem unnecessary for the oxidation of ent-kaurene. This paradox could be attributable to differences between the germinating embryo and the leaf. Future studies are required to consolidate these results.
After germination, bioactive GAs promote stem elongation, leaf expansion, and root growth (Davies, 1995; Yaxley et al., 2001). In the plant species examined, bioactive GAs or ent-kaurene–synthesizing activities are present mainly in rapidly developing tissues, such as the shoot tips, expanding leaves and petioles near elongating internodes, and developing seed (Chung and Coolbaugh, 1986; Choi et al., 1995; Aach et al., 1997). Reverse transcriptase–mediated polymerase chain reaction or RNA gel blot analyses indicated that in Arabidopsis, CPS, KS, and KO mRNA accumulated to higher amounts in young seedlings compared with older vegetative tissues (Silverstone et al., 1997a; Helliwell et al., 1998; Yamaguchi et al., 1998b). Further examination of AtCPS expression using a promoter-GUS transgene showed that AtCPS is expressed not only in growing tissues, including shoot and root apices, but also in the vasculature of petioles and leaves (Silverstone et al., 1997a).
In the shoot apex of 2-day-old Arabidopsis seedlings, AtCPS is expressed in the two- to four-cell-layer-thick juvenile shoot apical meristem (SAM) and in the region below the SAM that is undergoing cell division, elongation, and differentiation (Silverstone et al., 1997a). Interestingly, AtCPS is no longer expressed in the adult vegetative SAM, with the L1/L2/L3 (corpus) arrangement of 5-day-old seedlings; rather, it continues to be expressed only in the region below the SAM. In the reproductive phase, the AtCPS promoter is active in the inflorescence meristem (IM) after bolting. These results suggest roles for GA in establishing the adult apical meristem, maintaining the IM, and establishing the floral primordia during the reproductive phase.
Recent studies of the expression of the GA20ox, GA3ox, and GA2ox genes in tobacco (Itoh et al., 1999; Sakamoto et al., 2001a) and rice (Sakamoto et al., 2001b) supported this hypothesis. In situ RNA hybridization showed that in tobacco, the GA20ox and GA3ox mRNAs were present in the leaf primordia and rib meristem near the SAM but not in the corpus cells of the SAM (Itoh et al., 1999; Sakamoto et al., 2001a). In rice, a high level of OsGA3ox2 mRNA was present in young leaves but not in the SAM, whereas OsGA2ox1 showed a ring-shaped expression pattern in the basal region of leaf primordia and young leaves near the vegetative SAM (Sakamoto et al., 2001b). The amount of the OsGA2ox1 mRNA was much reduced in the IM. These findings suggest that GA is synthesized in the region below the vegetative SAM and in young leaves and that the function of OsGA2ox1 might be to control the active GA level in the SAM by inactivating GAs synthesized in young leaves.
Recent studies provided evidence that the KNOX genes are involved in modulating GA biosynthesis in the SAM by inhibiting the expression of the GA20ox genes (Tanaka-Ueguchi et al., 1998; Sakamoto et al., 2001a). Most of the KNOX genes are expressed in the SAM but are not active in the lateral organ primordia (reviewed by Reiser et al., 2000). Based on the phenotype of loss-of-function knox mutants and KNOX overexpression lines, these genes function in the maintenance of the indeterminate cells of the SAM. Ectopic expression of KNOX genes (tobacco NTH15 or rice OSH1) in tobacco resulted in dwarfed and abnormal leaf phenotypes, which were rescued partially by exogenous GA applications (Kusaba et al., 1998; Tanaka-Ueguchi et al., 1998). GA measurements and RNA gel blot analysis showed that the amounts of GA1 and GA20ox mRNA in KNOX-overexpressing plants were reduced compared with those in the wild type (Tanaka-Ueguchi et al., 1998).
In vitro gel shift and DNase I protection assays provided evidence that the tobacco KNOX protein NTH15 binds to a 5′ upstream region and the first intron of a tobacco GA20ox gene (Sakamoto et al., 2001a). In contrast to the localization of the KNOX mRNA in the corpus of the SAM, the GA20ox mRNA was detected in the rib meristem and leaf primordia. Using a dexamethasone-inducible NTH15 system, Sakamoto et al. (2001a) showed elegantly that NTH15 suppresses the expression of the GA20ox gene in the corpus of the SAM and that deletion of the NTH15 binding site in the first intron of the GA20ox gene eliminated the effect of NTH15. These results suggest that the expression of KNOX in the corpus of the SAM in tobacco inhibits GA biosynthesis to maintain the indeterminate state of the corpus. On the other hand, GA production at the flank of the SAM may promote organized cell proliferation and consequently induce the determination of cell fate.
During Arabidopsis flower development, GA is essential for the development of stamens and petals (Koornneef and van der Veen, 1980). The AtCPS promoter is active in anther sac walls before dehiscence and in pollen (Silverstone et al., 1997a). It also is expressed in the funiculi, transmission tract, endosperm, and embryo axis of developing seed. Similarly, the transcripts of the GA3ox genes in tobacco and rice are localized in the pollen and tapetum of developing anthers (Itoh et al., 1999, 2001). In addition to acting in the anthers, GA synthesized in the anthers appears to control the development of other floral organs. In petunia, removal of the anthers blocked petal growth and the accumulation of pigments (Weiss and Halevy, 1989). The treatment of emasculated flowers with GA was sufficient to restore petal development.
CONTROL OF GA BIOSYNTHESIS AND CATABOLISM BY OTHER HORMONAL SIGNALS
In seedlings, brassinosteroid (BR) may affect GA biosynthesis (Bouquin et al., 2001). Mutants that are defective in BR response (bri1-201) or impaired in BR biosynthesis (cpd) have reduced amounts of the GA20ox1 mRNA, and exogenous BR treatment upregulated GA20ox1 expression. Because GA20ox expression is feedback inhibited by GA, BR and GA appear to have antagonistic effects on modulating the expression of this gene. Additionally, the effect of BR on GA20ox1 expression is independent of GA because induction of GA20ox1 mRNA by BR was observed in the cpd ga1-1 double mutant, which is deficient in both GA and BR biosynthesis.
The role of auxin in regulating GA biosynthesis is suggested by the findings that the GA1 content in stems of pea and tobacco was reduced by the application of an auxin transport inhibitor or decapitation (Ross, 1998; Wolbang and Ross, 2001). This reduction in the amount of GA1 in pea stems was accompanied by decreased GA3ox1 transcript in stems (Ross et al., 2000). The application of indoleacetic acid to decapitated pea stem was able to induce the accumulation of PsGA3ox1 mRNA and reduce the PsGA2ox1 transcript, implying that auxin transported from the shoot apex to the elongating stem is required to modulate the expression of both PsGA3ox and PsGA2ox1 to produce the bioactive GA1 (Ross et al., 2000).
LIGHT REGULATION OF GA BIOSYNTHESIS AND CATABOLISM
In addition to developmental regulation, the genes of GA biosynthesis and catabolism are subject to control by light quality and photoperiod, conditions that also modulate germination, flowering time, stem elongation, and/or tuberization of potato (reviewed by Davies, 1995; Kamiya and García-Martínez, 1999). Because of the feedback and feedforward control of GA metabolism, it often is difficult to determine whether the effects of light and photoperiod on GA synthesis and catabolism genes are direct or an indirect consequence of changes in the GA response pathway.
It is now clear that red light promotes seed germination by increasing GA biosynthesis (reviewed by Kamiya and García-Martínez, 1999; Yamaguchi and Kamiya, 2000) and by affecting tissue responsiveness to GA (Hilhorst and Karssen, 1988). Small-seeded plants such as Arabidopsis and lettuce require light for germination, and it was speculated that GA mediates the seed response to light because exogenous GAs mimicked the effects of red light in promoting seed germination (Hilhorst and Karssen, 1988). In Arabidopsis, both the de novo biosynthesis of GAs (Hilhorst and Karssen, 1988; Nambara et al., 1991) and appropriate light conditions (reviewed by Shinomura, 1997) are essential for seed germination. Red light treatment was able to decrease the concentration of GA4+7 that was needed to rescue the germination defect of the GA-deficient ga1 mutant (Hilhorst and Karssen, 1988), suggesting that red light increases the GA responsiveness of the tissue. In lettuce seed, a red light pulse specifically increased the amount of the bioactive GA1 (but not its precursors GA19 and GA20), and a subsequent far-red light pulse canceled the effect of the red light (Toyomasu et al., 1993). Derkx et al. (1994) also found that the amount of bioactive GA4 (but not its precursor GA9) in wild-type Arabidopsis seed irradiated with continuous white light was higher than the amount in dark-germinated seed.
These results suggest that 3β-hydroxylation of GA is controlled by light. RNA gel blot analyses further demonstrated that the expression of AtGA3ox1 and AtGA3ox2 in soaked seed of Arabidopsis and LsGA3ox1 in soaked seed of lettuce was induced within 1 hr after a brief red light treatment (Yamaguchi et al., 1998a; Toyomasu et al., 1998). The red light induction of expression of these genes was reversible by a far-red light pulse, indicating that these genes are controlled by phytochrome in Arabidopsis and lettuce. These results also suggested that phytochrome induces bioactive GA production by increasing the expression of GA3ox genes during light-induced seed germination.
Phytochrome also mediates the red light–induced inhibition of stem growth. The phyB mutants in garden pea (lv), cucumber (lh), and Arabidopsis (phyB) show increased responsiveness to the bioactive GAs (Weller et al., 1994, 1995; Lopez-Juez et al., 1995; Reed et al., 1996). These results demonstrate that the inhibition of stem elongation by light is associated with a decrease in tissue responsiveness to GAs. However, whether light also causes a reduction in the bioactive GA level in the stem is controversial (reviewed by Kamiya and García-Martínez, 1999). Several recent studies in pea have provided convincing evidence that during deetiolation of pea seedlings, the concentration of GA1 in the shoot tip decreases within 2 hr of light exposure, whereas the concentration of the inactive product of GA1 catabolism, GA8, increases (Ait-Ali et al., 1999; Gil and García-Martínez, 2000). O'Neill et al. (2000) further showed that after the initial reduction, the amount of GA1 increases after 24 hr of deetiolation. Some of the contradictory results from earlier studies could be attributable to GA measurements that were performed at later time points and/or the use of whole shoots instead of only shoot tips.
By analyzing gene expression using a more detailed time course during deetiolation, Reid et al. (2002) have demonstrated that rapid (0.5 to 1 hr) downregulation of PsGA3ox1 and upregulation of PsGA2ox2 are correlated with the reduction of GA1 level caused by red or blue light treatment. Interestingly, the amount of GA20ox1 transcript did not change during the first 0.5 to 1 hr of light, but it increased after 4 hr, probably as a result of feedback regulation. Reid et al. (2002) further showed that the reduction in GA1 concentration during deetiolation by red or far-red light is mediated by phyA, whereas the effect of blue light is mediated by an unidentified blue light receptor. These results suggest that the initial inhibition of stem growth by light is achieved by modulating GA3ox1 and GA2ox2 gene expression to reduce the endogenous GA1 concentration. But the long-term inhibitory effect of light on stem elongation probably is mediated, at least in part, by reducing the tissue responsiveness to GA.
Flowering in Arabidopsis is controlled by multiple pathways (Levy and Dean, 1998). One pathway mediates flowering under long day (LD) conditions. When Arabidopsis is grown under short day (SD) conditions, this pathway is inactive, but flowering eventually occurs because of induction by the daylength-independent autonomous pathway. The ga1-3 mutant never flowers under SD conditions (Wilson et al., 1992), indicating that GA is required for the induction of flowering by the autonomous pathway in Arabidopsis. The importance of GA in flower formation in a large number of LD plants also has been illustrated by inducing flowering in plants growing under noninductive SD conditions with exogenous GA (Metzger, 1995). However, it is technically very difficult to identify the bioactive GAs that control flower initiation. Recently, changes in the content of some GAs in the true shoot apex of the grass Lolium temulentum during LD induction of flowering were examined using very sensitive gas chromatography–mass spectrometry techniques (King et al., 2001b). This effort identified GA5 as one of the primary compounds involved in this process, because its concentration doubled in the shoot apex within 8 hr of LD treatment. However, it is unclear whether the increased GA5 is a result of de novo synthesis at the shoot apex or of transport of this compound from the leaves. In contrast, the amounts of GA1 and GA4 increased at later times after LD treatment, correlating with the role of these GAs in promoting stem elongation during bolting in dicots (Wu et al., 1996; Xu et al., 1997).
As described above, the AtCPS promoter is active in the IM and young floral primordia after bolting, but not in the adult vegetative SAM, suggesting a role for GA in the maintenance of the IM and in the establishment of the floral primordia during the reproductive phase (Silverstone et al., 1997a). It will be important to examine whether the expression of AtCPS resumes in the meristem before or after the transition from the SAM to the IM. Additionally, the expression pattern of later GA biosynthesis genes during floral induction needs to be determined.
Tuberization in potato is promoted by SD conditions and inhibited by GA applications (reviewed by Jackson, 1999; Kamiya and García-Martínez, 1999). Antisense phyB transgenic potato plants were able to form tubers under noninductive LD conditions. Grafting experiments using wild-type and antisense phyB plants further showed that phyB is involved in the production and/or transport of an inhibitor from leaves to the stolon in LD conditions (Jackson et al., 1998). Because GA is known to have an inhibitory effect on tuberization, several recent studies investigated the effect of light on GA biosynthesis under both SD and LD conditions (Carrera et al., 1999, 2000; Jackson et al., 2000). Three GA20ox genes were isolated from potato, all of them were expressed in stolons, and only GA20ox1 was expressed highly in leaves (Carrera et al., 1999). Overexpression of GA20ox1 under the control of either a leaf-specific promoter or the constitutive 35S promoter in transgenic potato resulted in taller stems, delayed tuberization, and reduced tuber dormancy in SD conditions (Carrera et al., 2000). Inhibition of GA20ox1 expression in transgenic plants carrying the 35S promoter::antisense GA20ox1 resulted in shorter stature and earlier tuber formation, but tuber dormancy was not altered (Carrera et al., 2000). A reduction in the amounts of both GA20 and GA1 was observed in the shoot tips and first leaves of the GA20ox1 antisense plants, indicating that downregulation of GA20ox1 expression contributes to reduced GA content in the leaves and earlier tuber formation. However, in addition to GA, another LD inhibitory signal is required for the light regulation of tuberization, because GA20ox1 antisense lines or severe GA biosynthetic potato mutants were unable to form tubers in LD conditions as readily as phyB antisense plants.
RNA gel blot analysis indicated that all three GA20ox genes in potato showed differential diurnal expression patterns under SD or LD conditions (Carrera et al., 1999). Additionally, GA20ox1 mRNA concentrations in leaves of phyB antisense plants were higher than in wild-type plants (Jackson et al., 2000). This finding supports the hypothesis that the more elongated stem of antisense phyB plants is caused by an increase in GA1 production in the leaves compared with the wild type. However, these results seem to contradict the notion that GA is one of the phyB-induced LD inhibitory signals in the suppression of tuberization. One possibility is that phyB induces the transport of GA(s) from the leaves to the stolons. Careful analysis of the effect of light on GA content and the expression of genes involved in GA metabolism in stolons and leaves will be required to elucidate the mechanism of light regulation of GA metabolism in tuber formation.
GA RESPONSE PATHWAY COMPONENTS
Mutants with defects in GA responses occur in several plant species. Here, we focus on the insights into the GA response pathway that have arisen from the analysis of the genes affected in these mutants.
Positively Acting Components
DWARF1
DWARF1 (D1) is the only gene in rice encoding a prototypical heterotrimeric G protein α-subunit (Gα) (Ashikari et al., 1999; Fujisawa et al., 1999). The d1 mutant exhibits phenotypes characteristic of GA deficiency, including semidwarfism and dark green coloration of leaves, but it has increased levels of active GA. Elongation of d1 internodes was 100-fold less sensitive to applied GA than elongation in the wild type. Surprisingly, the sensitivity of the second leaf to GA was indistinguishable from that in the wild type, but the total amount of growth induced by a saturating concentration of GA was reduced in d1 plants. Double mutants between d1 and slr1 exhibit a slender phenotype, suggesting that SLR1, a negative regulator of GA response (see below), functions downstream of D1 (Ueguchi-Tanaka et al., 2000). Although some phenotypes of d1 plants are consistent with heterotrimeric G proteins playing a positive role in the GA response pathway, the normal sensitivity of the second leaf to GA suggests that these proteins may not be involved in all GA responses. As in rice, only one gene encodes the prototypical Gα subunit of Arabidopsis. In contrast to the results from rice, however, loss-of-function mutations affecting Arabidopsis Gα, although they affect several signaling pathways, do not cause the dwarf phenotype that is typical of mutants with defective GA responses (Ullah et al., 2001; Wang et al., 2001). Therefore, the relative importance of the Gα protein in GA responses may vary between species.
PHOTOPERIOD RESPONSIVE 1
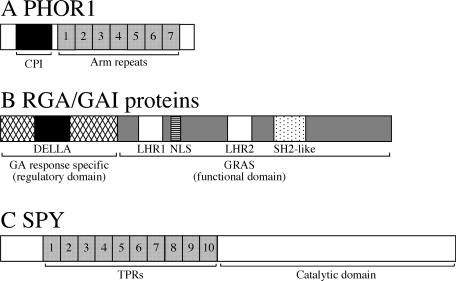
PHOTOPERIOD RESPONSIVE 1 (PHOR1) was identified in a screen for genes whose mRNA levels are increased in the leaves of potato during growth under short days (Amador et al., 2001). Antisense inhibition of PHOR1 expression caused a semidwarf phenotype, reduced response to GA, and increased levels of endogenous GAs. Overexpression of PHOR1 caused an overgrowth phenotype and an enhanced response to applied GA. GA treatment promoted nuclear localization of a PHOR1-GFP fusion protein in tobacco BY2 cells, and GA biosynthesis inhibitors caused the fusion protein to be localized to the cytosol. Analysis of deletion mutants has identified two domains that are important in the GA-regulated localization of PHOR1 (Figure 2A). Deletion of a conserved Cys-Pro-Ile motif (CPI) caused PHOR1-GFP to be localized constitutively to the nucleus, suggesting that CPI is a GA-inhibited cytosolic retention signal. PHOR1 also contains seven armadillo repeats. Deletion experiments suggest that the PHOR1 armadillo repeats function as a nuclear localization signal that can be overcome by the action of the CPI. The Drosophila armadillo protein and vertebrate β-catenin proteins, which are involved in the Wnt signaling pathway, contain armadillo repeats. During Wnt signaling, proteins with armadillo repeats move from the cytosol to the nucleus, associate with transcription factors, and induce gene expression (Dale, 1998; Willert and Nusse, 1998). These data suggest the following model for PHOR1 action: in the absence of GA signaling, CPI retains PHOR1 in the cytosol, in which it is inactive; during GA signaling, CPI is inhibited, allowing the armadillo repeats to localize PHOR1 to the nucleus and stimulate the transcription of genes encoding products with a positive role in the GA response.
Figure 2.
Schemes of PHOR1, RGA/GAI, and SPY.
(A) PHOR1 contains a CPI domain, which is a GA-repressible cytosolic retention signal, and the armadillo repeats, which function in nuclear localization and may allow it to interact with transcription factors.
(B) RGA/GAI proteins contain a highly conserved C-terminal region that is shared among all GRAS family members. The nuclear localization signal is absent in some of the GRAS members (Pysh et al., 1999; Schumacher et al., 1999; Bolle et al., 2000). RGA/GAI and other nuclear localization signal–containing GRAS family members are likely to function as transcriptional regulators. The N-terminal region of the RGA/GAI proteins is required for the inactivation of these proteins by the GA signal.
(C) SPY proteins contain two conserved domains: the TPR domain, which is believed to interact with other proteins; and the catalytic domain, which post-translationally modifies proteins with GlcNAc.
Arm, armadillo; LHR, Leu heptad repeat; NLS, nuclear localization signal.
MYB Transcription Factors
GAMYB is a GA-induced MYB transcription factor that was identified by its ability to activate the α-amylase promoter of barley (see below). Three GAMYB proteins from Arabidopsis have been shown to substitute functionally for barley GAMYB in aleurone cells (Gocal et al., 2001). One of these proteins, AtMYB33, is implicated in the induction of flowering by GA. Expression of AtMYB33 occurs in the shoot apex during the induction of flowering and is induced by GA. Expression of the floral meristem gene LEAFY (LFY) is induced by GA, and a specific promoter element, GOF9, has been shown to confer GA responsiveness (Blázquez and Weigel, 2000). AtGAMYB33 binds to GOF9, suggesting that, during the induction of flowering, GA-induced AtGAMYB33 binds to GOF9 and stimulates LFY expression. Because AtGAMYBs are expressed in seed and vegetative tissues, they also may participate in GA responses other than flowering.
GA regulation of another Arabidopsis MYB gene, GLABROUS1 (GL1), may play a role in the initiation and branching of trichomes (Perazza et al., 1998). The ga1 mutation causes Arabidopsis to have fewer trichomes, and treatment with GA reversed this effect (Chien and Sussex, 1996; Telfer et al., 1997; Perazza et al., 1998). Because GL1 mRNA is less abundant in ga1 and GA treatment increased the expression of a reporter gene that is driven by the GL1 promoter (Perazza et al., 1998), GA-induced expression of GL1 may promote both the initiation and branching of trichomes.
SLEEPY
A potential difficulty with genetic screens for positively acting components of the GA response pathway is that mutants affected in these components might not germinate. Germination of GA biosynthesis mutants, however, is restored by mutations that reduce abscisic acid synthesis or sensitivity (Koornneef et al., 1982; Nambara et al., 1992). The abi1-1 mutant is less sensitive to abscisic acid than the wild type and thus is able to germinate in the presence of 3 μM abscisic acid, a concentration that inhibits the germination of wild-type seed. An abi1-1 suppressor screen for mutants that do not germinate on 3 μM abscisic acid but do germinate when removed from abscisic acid has identified both new GA biosynthesis mutants and sleepy1 (sly1) (Steber et al., 1998). sly1 abi1-1 plants are dark-green dwarfs that are not rescued by the application of GA. In the absence of the abi1-1 allele, sly1 seed do not germinate.
PICKLE
The PICKLE (PKL) protein of Arabidopsis contains domains that are the hallmarks of CH3 chromatin-remodeling factors, and several of the phenotypes of pkl mutants suggest that it is involved in GA action (Ogas et al., 1997, 1999). Loss-of-function pkl mutants are GA-insensitive dwarfs that have increased amounts of GAs. With respect to flowering time, pkl and gai-1 exhibit a synergistic interaction, suggesting that both mutations affect the GA response pathway. Relative to the parental single mutants, the flowering of pkl gai-1 plants grown under SD conditions is delayed severely.
In addition to the phenotypes described above, pkl plants also exhibit phenotypes suggesting that PKL has a complex relationship with GA responses. Embryonic characters persist in pkl after germination, the most obvious being the accumulation of lipids and the expression of storage protein genes in the swollen tip of the primary root (Ogas et al., 1997). An intriguing aspect of these embryonic phenotypes is that they are not fully penetrant, and treatment with GA biosynthesis inhibitors increased the penetrance of the phenotypes, whereas treatment with GA reduced the penetrance. PKL also appears to play roles in responses beyond those mediated by GA. Acting in combination with the asymmetric1 or asymmetric2 mutation, pkl causes the formation of ectopic stipules and in rare cases ectopic meristems in the sinuses of leaves (Ori et al., 2000). In combination with crab claw, pkl causes the ectopic formation of adaxial carpel tissue (Eshed et al., 1999).
Although the role of PKL in GA responses is unclear, some insight into possible mechanisms is provided by studies of CH3 chromatin-remodeling factors. CH3 proteins are part of a large complex with histone deacetylase activity that inhibits transcription (Wade et al., 1998; Zhang et al., 1998). Therefore, pkl phenotypes may be caused by ectopic gene expression. It remains an open question whether the genes that are expressed ectopically in pkl play a direct or an indirect role in GA responses.
GIBBERELLIN-INSENSITIVE DWARF1
Recently, recessive GA-insensitive dwarf mutants of rice, gibberellin-insensitive dwarf1 (gid1), exhibiting phenotypes observed in GA-deficient plants, were identified (Sasaki et al., 2001). Expansion of the second leaf sheath and the induction of α-amylase did not occur in response to treatment with GA. In addition, GA20ox expression was increased in the mutant. GID1 has been cloned (M. Matsuoka, personal communication), and although the predicted GID1 protein has similarity to members of the Ser hydrolase family, which includes esterases, lipases, and proteases, the enzymatic function of GID1 has not been determined. Analysis of gid1 slr1 double mutants (Ueguchi-Tanaka et al., 2001) suggests that GID1 acts upstream of SLR1, a member of the GAI/RGA family of negative regulators of GA signaling (see below). Consistent with GID1 acting upstream of SLR1, an SLR1-GFP fusion protein became less abundant in response to GA treatment in wild-type plants but was unresponsive to GA treatment in gid1 (Ueguchi-Tanaka et al., 2001).
Negatively Acting Components
RGA/GAI Proteins
Many of the mutations that modify GA sensitivity affect genes encoding members of the RGA/GAI family. The RGA/GAI family is a subset of the larger GRAS family (Pysh et al., 1999). In addition to sharing a number of motifs with all members of the GRAS family, the N terminus of all RGA/GAI proteins contains the DELLA domain, which is absent from other GRAS proteins (Peng et al., 1997; Silverstone et al., 1998) (Figure 2B).
Mutations affecting RGA/GAI proteins have been identified in Arabidopsis (rga and gai), barley (sln1), maize (d8), rice (slr1), and wheat (reduced height [rht]) (Peng et al., 1997, 1999a; Silverstone et al., 1998; Ogawa et al., 2000; Ikeda et al., 2001; Chandler et al., 2002; Gubler et al., 2002). These mutations fall into two classes: semidominant mutations in Arabidopsis, maize, wheat, and barley causing dwarfism; and recessive loss-of-function mutations in Arabidopsis, barley, and rice causing increased growth (barley mutants representative of these classes are shown in Figure 3). The phenotypes of the recessive loss-of-function alleles indicate that these proteins are negative regulators of the response pathway. In barley and rice, the product of a single gene, SLN1 and SLR1, respectively, negatively regulates GA responses at all stages of development (Ogawa et al., 2000; Ikeda et al., 2001; Chandler et al., 2002; Gubler et al., 2002). In contrast, Arabidopsis contains a gene family encoding RGA, GAI, and three RGA-like (RGL) proteins that have overlapping functions (Sanchez-Fernandez et al., 1998; Dill and Sun, 2001; King et al., 2001a; Wen and Chang, 2002).
Figure 3.
Barley GA Mutants.
Plants are of equal age and are (from left to right) Himalaya (wild type), grd2 (GA-deficient putative GA3ox mutant [Chandler and Robertson, 1999]), Sln1d (dominant dwarf), and sln1c (slender).
RGA was identified in a screen for mutations suppressing the vegetative dwarfism of the GA-deficient ga1 mutant (Silverstone et al., 1997b). Loss-of-function rga alleles partially suppress most of the phenotypes of ga1 plants, including delayed abaxial trichome initiation, dwarfism of the rosette leaves, delayed flowering, dwarfism of the internodes of the floral shoot, and reduced apical dominance. In contrast to rga alleles, gai-t6, a loss-of-function allele, suppresses ga1 only weakly (Dill and Sun, 2001; King et al., 2001a). However, there is a synergistic interaction between rga and gai-t6 (Dill and Sun, 2001; King et al., 2001a). In combination, rga and gai-t6 double null mutations rescue the vegetative and delayed flowering phenotypes of ga1, even producing a GA-overdose phenotype, indicating that RGA and GAI are major repressors controlling these processes. rga gai-t6 ga1 triple mutants do not germinate and exhibit defects in petal and anther development, suggesting that the RGLs are involved in these processes. Recently, RGL1 was shown to be involved in germination (Wen and Chang, 2002). Transgenic Arabidopsis seed, in which the expression of RGL1 is silenced, were resistant to the inhibition of germination by the GA biosynthesis inhibitor paclobutrazol.
The GAI gene of Arabidopsis was identified as a semidominant mutation, gai-1, that greatly reduces GA responsiveness during vegetative development (Koornneef et al., 1985). The gai-1 mutant contains increased amounts of bioactive GAs, suggesting that RGA/GAI proteins are involved in the feedback regulation of GA biosynthesis (Talón et al., 1990). The gai-1 allele has a 51-nucleotide deletion that maintains the reading frame but deletes 17 amino acids from the DELLA domain (Peng et al., 1997). Interestingly, mutations similar to gai-1 affecting RGA/GAI family members from maize, wheat, and barley produce similar phenotypes (Peng et al., 1999a; Chandler et al., 2002; Gubler et al., 2002). The wheat Rht alleles were used to produce the wheat varieties that enabled the “green revolution.” Several recent experiments have demonstrated that it is possible to generate dwarf plants by expressing RGA/GAI proteins with a 17–amino acid deletion in the DELLA domain. Expression of DELLA domain deletion mutants of RGA and SLR1 in Arabidopsis and rice, respectively, caused semidwarfism (Dill et al., 2001; Ikeda et al., 2001). In addition, expression of the Arabidopsis gai-1 protein in rice caused dwarfism and abolished GA induction of α-amylase in the aleurone (Peng et al., 1999a; Fu et al., 2001), indicating that mutant RGA/GAI proteins function in heterologous species.
RGA/GAI proteins contain several protein motifs common to GRAS proteins, including Leu heptad repeats and nuclear localization signals (Figure 2B), that appear to have functional roles. Using GFP fusion proteins, Arabidopsis RGA and rice SLR1 have been shown to be localized to the nucleus (Silverstone et al., 1998, 2001; Ogawa et al., 2000; Itoh et al., 2002). Deletion analysis of rice SLR1 suggests that the Leu heptad domain functions in dimerization and also suggests that the C-terminal region is responsible for the repression of the GA response pathway (Itoh et al., 2002). The structural features and nuclear localization of RGA/GAI proteins suggest that they are transcription regulators, and rice SLR1 has been show to affect transcription when it is expressed in spinach (Ogawa et al., 2000). Analysis of the sequences of these proteins also has identified possible SH2 domains, suggesting that they may function like the STAT proteins (Peng et al., 1999a; Richards et al., 2000).
It is believed that active GA signaling inhibits the repressor action of the GAI/RGA proteins and that the DELLA domain, and perhaps surrounding regions, are required for this inhibition. GA overcomes the repressor activity of RGA, SLN1, and SLR1 by destabilizing them (Silverstone et al., 2001; Gubler et al., 2002; Itoh et al., 2002). Deletion of the DELLA domain caused the accumulation of RGA and SLR1 by rendering them insensitive to destabilization by GA (Dill et al., 2001; Itoh et al., 2002). The molecular mechanism by which GA affects the stability of the RGA/GAI proteins is not clear, but ubiquitin-mediated proteolysis is proposed to be involved (Dill et al., 2001; Gubler et al., 2002; Itoh et al., 2002). The model in which GA overcomes the repressive effects of RGA/GAI proteins by causing their destabilization predicts that their overexpression will inhibit GA responses. Overexpressing Arabidopsis GAI in rice caused dwarfism and reduced the induction of α-amylase by GA (Fu et al., 2001).
SPINDLY
Paclobutrazol, a GA biosynthesis inhibitor, blocks the germination of Arabidopsis seed and, if applied after germination, causes dwarfism. Screens for mutants that suppress both the germination and the dwarfing effects of paclobutrazol have identified recessive alleles of SPINDLY (SPY) (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996). Screens for suppressors of ga1 and gai-1 also have identified additional spy alleles (Wilson and Somerville, 1995; Silverstone et al., 1997b). Although in many cases the suppression was partial, all of the phenotypes of GA deficiency that have been examined are suppressed by spy (Jacobsen and Olszewski, 1993; Wilson and Somerville, 1995; Jacobsen et al., 1996, 1998; Peng et al., 1997; Silverstone et al., 1997b; Swain et al., 2001). In a wild-type background, spy causes several phenotypes that are observed when the wild type was treated repeatedly with GA3, including more erect rosette leaves with a pale green color, early flowering, and reduced seed set (Jacobsen and Olszewski, 1993). Overexpression of Arabidopsis SPY in Arabidopsis (Swain et al., 2001) and petunia (Izhaki et al., 2001) under the control of the 35S promoter of Cauliflower mosaic virus produced phenotypes consistent with reduced GA activity. Based on these results, SPY is believed to be a negative regulator of the GA response pathway. This hypothesis is supported by experiments in which barley SPY is overexpressed in barley aleurone (Robertson et al., 1998).
Experiments monitoring the expression of Arabidopsis SPY using a SPY::GUS reporter gene (Swain et al., 2002) and petunia SPY by reverse transcriptase–mediated polymerase chain reaction (Izhaki et al., 2001) have found that they are expressed constitutively at all stages of development and that their expression is not regulated by GA, other hormones, or light. Arabidopsis SPY is found in both the cytosol and the nucleus (Swain et al., 2002).
The SPY protein has overall similarity with UDP-GlcNAc protein transferase (OGT) from animals (Roos and Hanover, 2000) and has been shown to have OGT activity (Thornton et al., 1999a). Animal OGT is a nucleus- and cytosol-localized enzyme that transfers GlcNAc from UDP-GlcNAc to Ser and/or Thr of nuclear and cytosolic proteins (Comer and Hart, 2000; Hanover, 2001; Wells et al., 2001).
O-GlcNAc modification of proteins is a dynamic modification that regulates protein activity (Comer and Hart, 2000; Hanover, 2001; Wells et al., 2001). The extent of protein O-GlcNAc modification depends on metabolic and hormonal signals and the cell cycle. O-GlcNAc modification regulates protein activity by affecting localization, phosphorylation, stability, and interactions with other proteins. For a number of proteins, O-GlcNAc modification and phosphorylation have been shown to be reciprocal modifications occurring at the same Ser and Thr hydroxyl groups. Therefore, these proteins, which are modified reciprocally, can exist as three different isoforms: the unmodified, the O-GlcNAc modified, and the phosphorylated forms, each with a different activity. The proteins that are the targets of SPY activity have not been identified, although RGA/GAI proteins have been proposed to be targets (Silverstone et al., 1998; Thornton et al., 1999b).
The first half of SPY consists of 10 tetratricopeptide repeats (TPRs), and the C-terminal half is likely to be the catalytic domain (Figure 2C). TPRs act as scaffolds for the assembly of multiprotein complexes (Lamb et al., 1995; Das et al., 1998; Blatch and Lassle, 1999), suggesting that SPY is part of a multiprotein complex. Expression of the SPY TPR domain under the control of the 35S promoter of Cauliflower mosaic virus in both Arabidopsis (Tseng et al., 2001) and petunia (Izhaki et al., 2001) produced a SPY loss-of-function phenotype. The overexpressed TPR domain could interfere with SPY function either by interacting with SPY, preventing the formation of the active SPY enzyme, or by interacting with a SPY-interacting protein(s) and blocking the interaction of this protein with SPY. Animal OGT is a homotrimer, and deletion of the TPRs converted it to a monomer (Kreppel and Hart, 1999). The SPY TPR domain has been shown to interact with itself (Tseng et al., 2001), suggesting that SPY also is a homotrimer. Because TPR domains often mediate interactions with multiple proteins, it is likely that the SPY TPR domain interacts with additional proteins and that these interactions play a role in the GA response.
Animals contain a single OGT gene, and deletion of the mouse OGT gene results in embryo lethality (Shafi et al., 2000), indicating that O-GlcNAc modification of proteins is essential. In contrast to animals, Arabidopsis has two genes encoding OGTs. Although Arabidopsis mutants lacking the second OGT activity have no obvious phenotype, loss of both SPY and the second OGT causes male and female gamete lethality (L. Hartweck and N. Olszewski, unpublished data), indicating that OGT activity is essential and that SPY and the second OGT have partially redundant functions. Given the fact that OGT function is essential, it is not surprising that spy mutants exhibit phenotypes that are not explained by defects in GA response (Swain et al., 2001). Recent work suggests that SPY has a role in light signaling because spy mutants exhibit a long-hypocotyl phenotype when grown under far-red light, but hypocotyl length is similar to that in the wild type when grown under red light (T. Tseng and N. Olszewski, unpublished data).
SHORT INTERNODES
An overexpression allele of SHORT INTERNODES (SHI) caused by an activation tag Ds transposon produces a semidwarf phenotype that is not reversed by GA treatment and that increases the concentration of endogenous GAs (Fridborg et al., 1999). The wild-type SHI gene is expressed in young organs that are not undergoing rapid elongation growth (Fridborg et al., 2001), suggesting that SHI prevents young organs from initiating inappropriate elongation growth in response to GA. Because SHI is not expressed normally in cells that are undergoing rapid expansion, the semidwarfing effect of the SHI overexpression allele is likely attributable to ectopic expression. Expression of SHI in barley aleurone cells reduced the GA induction of α-amylase expression (Fridborg et al., 2001), indicating that overexpression of SHI can negatively regulate GA responses in a heterologous species. It remains an open question whether SHI functions in GA response or if overexpression somehow interferes with GA response. Although loss-of-function shi alleles do not produce a phenotype (Fridborg et al., 2001), SHI is part of a gene family with nine members, raising the possibility that it will be necessary to mutate several members before the loss-of-function phenotype can be determined. SHI proteins have a RING finger–class zinc finger motif, which is known to mediate protein–protein interactions involved in proteolysis or transcriptional regulation (Freemont, 2000; Peng et al., 2000). The IGGH domain is a motif that is unique to SHI family proteins, but the function of this domain has not been determined.
Additional Components with Less-Defined Function in GA Responses
GAR2
A dominant allele of GAR2, gar2-1, partially suppresses the dwarfism (Wilson and Somerville, 1995), GA overaccumulation, and delayed flowering phenotypes (Peng et al., 1999b) caused by gai-1. gar2-1 acts additively with spy-7, a weak spy allele, to completely suppress gai-1. The gar2-1 mutation confers resistance to paclobutrazol. In the absence of the gai-1 mutation, gar2-1 plants are indistinguishable from wild-type plants. Because of the dominant nature of the gar2-1 mutation, GAR2 could be either a negative or a positive regulator of GA response. gar2-1 could act as a dominant-negative mutation, indicating that GAR2 is a negative regulator of GA response; alternatively, it could act as a hypermorphic mutation, indicating that GAR2 is a positive-acting component of the GA response pathway.
Use of Reporter Genes to Identify GA Response Mutants
Recently, a screen for mutants exhibiting altered expression of a firefly luciferase (LUC) reporter gene that is driven by the GA-repressible GA20ox promoter was described (Meier et al., 2001). The screen identified both LUC super expression (lue) and LUC low expression (loe) mutants. Three recessive lue-class mutants, lue1, fpa1-3, and fpa1-4, have been characterized.
lue1 plants exhibit a number of phenotypes that, although consistent with defects in GA response, are not explained by a simple model. lue1 causes semidwarfism that is not reversed by GA treatment, but the plants are not dark green, another phenotype indicative of GA insensitivity. GA induction of GAI expression in lue1 was indistinguishable from that in the wild type. Consistent with the reduced sensitivity to GA, lue1 plants had increased GA20ox and GA3ox transcripts, but unlike its effect in the wild type, GA treatment of lue1 plants caused further accumulation of GA20ox and GA3ox transcripts.
Two lue mutants are allelic with the late-flowering mutant fpa1 and were named fpa1-3 and fpa1-4. FPA acts in the daylength-independent pathway to promote flowering. The response of several GA-regulated genes to GA in fpa1 mutants was quantitatively similar to that in the wild type, although the steady state expression of these mRNAs was altered in the mutant.
Feedback Regulation of GA Biosynthesis and Catabolism by the GA Response
The role for different GA response pathway components in the feedback regulation of GA biosynthesis has been demonstrated clearly by analyses of GA content and transcript levels of GA20ox and GA3ox genes in different GA response mutants. When the GA response pathway is derepressed as a result of loss-of-function mutations in the negative regulators of the pathway (e.g., barley sln, pea la crys, and Arabidopsis rga/gai-t6), bioactive GAs and/or GA20ox and GA3ox mRNAs are present at lower levels than in the wild type (Croker et al., 1990; Martin et al., 1996; Dill and Sun, 2001; Silverstone et al., 2001). Conversely, gain-of-function mutations in the repressors (e.g., maize d8 and Arabidopsis gai-1) or loss-of-function mutations in the positive components (e.g., rice d1 and potato phor1) of the GA response pathway often result in higher levels of bioactive GAs and/or upregulation of GA20ox and GA3ox gene expression (Fujioka et al., 1988; Talón et al., 1990; Xu et al., 1995; Cowling et al., 1998; Ueguchi-Tanaka et al., 2000; Amador et al., 2001). Although GA2ox expression has not been examined in these GA response mutants, we predict that changes in GA2ox mRNA levels would be opposite those of the GA20ox and GA3ox genes because GA2ox genes are feedforward regulated by the amount of bioactive GAs (Thomas et al., 1999; Elliott et al., 2001). This tight link between the activity of the GA response pathway and GA metabolism, in coordination with the rapid degradation of repressor proteins (RGA/SLR/SLN) by the GA signal, may allow the plant to modulate precisely GA-regulated processes.
SIGNALING IN CEREAL ALEURONE
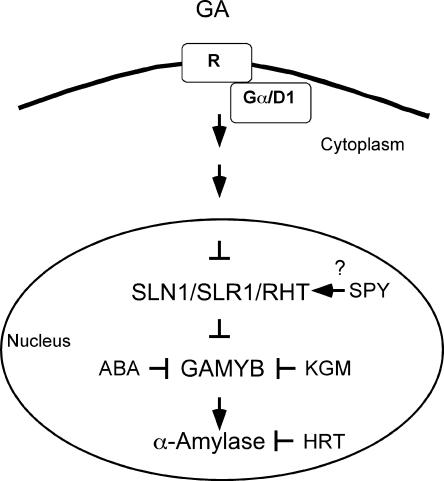
During the last four decades, the cereal aleurone has been a valuable system for studying GA regulation of gene expression. After germination, GAs are released from the embryo into the endosperm, triggering the expression of a number of genes encoding hydrolytic enzymes in aleurone cells. Many of these hydrolytic enzymes, which include α-amylase, proteases, and cell wall–degrading enzymes, are secreted and are responsible for digestion of the stored reserves in the starchy endosperm. Previous approaches to the investigation of the GA response in aleurone cells have relied largely on biochemical and molecular techniques, but recently, considerable progress has resulted from studies of cereal GA response mutants. Because the GA response is arguably best understood in this system, it is discussed by itself. Figure 4 presents a model of GA signaling in the aleurone.
Figure 4.
GA Response Pathway Leading to Gene Expression in Cereal Aleurone Cells.
ABA, abscisic acid.
GA Biosynthesis and Translocation
A number of studies have reported increases in GAs in cereal grains after imbibition (Yamada, 1982; Lenton and Appleford, 1991; Lenton et al., 1994); however, evidence demonstrating that the increase in GAs is necessary for germination remains elusive. A recent study has shown that the major increase in GA content in soaked barley grains occurs after the onset of radicle emergence, indicating that GA may not be required in the early germination steps (Jacobsen et al., 2002). However, it is clear that the synthesis of active GAs is required for the expression of hydrolytic enzymes in the endosperm (Lenton et al., 1994). The scutellum and the axis are the major sites of de novo GA synthesis in germinating grains, as judged by measuring ent-kaurene accumulation in paclobutrazol-treated wheat grains (Lenton et al., 1994). GA1, the major active GA, starts to accumulate rapidly in embryos of wheat grains after 24 hr of imbibition (Lenton and Appleford, 1991). This is followed by a concomitant increase in GA1 content in the endosperm of germinated wheat grains, which is thought to arise from passive diffusion of newly synthesized GA1 from the scutellum.
Upstream Components of the GA Response Pathway
Progress has been made in identifying components of the GA response pathway leading to the activation of early response genes such as GAMYB. Although the receptor has not been cloned, there is strong circumstantial evidence based on microinjection studies (Gilroy and Jones, 1994) and experiments with Sepharose beads coupled with GA suggesting that the GA receptor in aleurone cells is associated with the plasma membrane (Hooley et al., 1991).
Another component of the GA response pathway, heterotrimeric G protein, also is likely to be associated with the plasma membrane. The role of heterotrimeric G proteins in the GA response in aleurone cells has been suggested by experiments with Mas7, a constitutive stimulator of GDP/GTP exchange by Gα proteins (Jones et al., 1998). Aleurone cells from the rice dwarf mutant d1, which is defective in the α-subunit of the heterotrimeric G protein (Ashikari et al., 1999; Fujisawa et al., 1999), have reduced sensitivity to GA compared with the wild type (Ueguchi-Tanaka et al., 2000). However, the effect of the mutation was overcome by high GA concentrations, indicating that there may be Gα-dependent and Gα-independent response pathways leading to α-amylase expression. Genetic analysis indicated that the d1 mutation is hypostatic to the slr1 mutation of rice (Ikeda et al., 2001). Similarly, the gse1 mutant of barley, which reduces sensitivity to GA 100-fold, is hypostatic to sln1 and has reduced sensitivity to GA in aleurone cells (Chandler and Robertson, 1999). Further work is required to determine whether the GSE1 gene encodes a Gα protein.
In contrast to positive regulators such as D1, GAI/RGA-like proteins such as RHT, D8, SLR1, and SLN1 act as negative regulators of GA response in cereal aleurone (Peng et al., 1999a; Ikeda et al., 2001; Chandler et al., 2002; Gubler et al., 2002). Loss-of-function mutations (slr1-1 in rice and sln1b,sln1c in barley) in GAI/RGA-like genes result in constitutive α-amylase expression in aleurone cells. In contrast, mutants with dominant mutations in the DELLA region (Rht-B1b in wheat and Sln1d in barley) failed to produce GAMYB and α-amylase in the absence of applied GA and required 100-fold more GA than the wild type to induce the expression of these genes (Gale and Marshall, 1973; Ho et al., 1981; Ikeda et al., 2001; Chandler et al., 2002; Gubler et al., 2002).
Repressors of GA Responses Are Degraded Rapidly in Response to GA
Progress has been made recently in understanding how GA response regulates the SLN1 repression of gene expression in barley aleurone cells. SLN1 protein in barley aleurone cells is degraded within 5 to 10 min of treatment with GA (Gubler et al., 2002). This is similar to the observation in Arabidopsis, which showed rapid degradation of RGA in response to GA (Silverstone et al., 2001). Further work is required to determine whether the GA response involves targeting SLN1 for degradation via ubiquitin-mediated proteolysis. Mutations at both the N and C termini of SLN1 affected the stability of SLN1 in response to GA treatment (Gubler et al., 2002). The reduced GA insensitivity of Sln1d aleurone layers can be explained by increased SLN1d stability to GA. The loss-of-function sln1c mutant has a premature stop codon 18 amino acids before the C terminus that results in a polypeptide that also is resistant to GA-induced degradation.
The lag time (∼1 hr) between SLN1 disappearance and the expression of GAMYB, a transcription factor that induces α-amylase expression (see below), remains poorly understood (Gubler et al., 2002). Recent evidence indicating that SLR1 can act as a transcriptional activator of a repressor (Ogawa et al., 2000) may explain the lag time in GAMYB expression; however, further work is required to test this model in aleurone cells. Cyclic GMP may play an intermediary role between SLN1 and early response genes. Increases in cyclic GMP in response to GA correlate closely with increases in GAMYB protein in barley aleurone cells (Penson et al., 1996; Gubler et al., 2002). Studies with an inhibitor of guanylyl cyclase showed that the increase in cyclic GMP was necessary for GAMYB and α-amylase expression (Penson et al., 1996). There also is evidence that O-GlcNAc residues may play a role in the GA response in aleurone cells. Transient overexpression of HvSPY in GA-treated barley aleurone cells resulted in reduced α-amylase promoter activity (Robertson et al., 1998). Recent evidence indicates that a nucleoporin may be a target for HvSPY in barley aleurone cells (M. Robertson, personal communication).
GA Response Complexes and the Role of GAMYB
Various GA response complexes have been identified in the promoters of α-amylase genes (Gubler and Jacobsen, 1992; Lanahan et al., 1992; Rogers and Rogers, 1992). Functional analyses have shown that TAACAAA-like sequence motifs present in these GA response complexes play a key role in mediating the GA activation of transcription. The addition of the TAACAAA element to a minimal 35S promoter conferred GA responsiveness, indicating that the element can act as a GA response element (Skriver et al., 1991). Mutation of these elements caused a loss of GA responsiveness in α-amylase promoters, providing functional support for their role in regulating transcription (Gubler and Jacobsen, 1992; Rogers and Rogers, 1992). The TAACAAA element appears to function as a GA response element in GA-responsive promoters of other genes that are expressed in aleurone cells (Cercos et al., 1999).
Time-course studies have identified a number of early GA response genes that are expressed before α-amylase genes (Gubler et al., 1995; Chen et al., 1997). A MYB transcription factor, GAMYB, increased in amount in barley aleurone cells within 1 to 2 hr after GA application (Gubler et al., 1995, 2002). In vitro and transient expression experiments have shown that GAMYB binds specifically to the TAACAAA element of the GA response complex and is able to transactivate both α-amylase and a number of other GA-responsive promoters in transient expression experiments in the absence of GA (Gubler et al., 1995, 1999; Cercos et al., 1999). These results indicate that GAMYB may play an important role in activating the expression of GA-regulated genes.
Evidence is emerging that the GA regulation of GAMYB involves both transcriptional and post-transcriptional control. A GAMYB promoter:GUS gene responded weakly to GA in transient expression experiments with barley aleurone cells (Gomez-Cadenas et al., 2001). This is consistent with evidence from nuclear run-on experiments showing that GA caused a twofold increase in the rate of GAMYB transcription within 2 hr of application (Gubler et al., 2002). The presence of low amounts of GAMYB protein in aleurone layers treated without GA indicates the possible post-translational control of GAMYB function preventing transactivation of α-amylase genes (Gubler et al., 2002). A GAMYB binding protein, KGM, has been identified as a repressor of transcriptional activation of an α-amylase promoter by GAMYB (F. Woodger, personal communication). KGM, a mitogen-activated protein–like kinase, is expressed in aleurone cells in the absence of applied GA. Further work is required to show that phosphorylation of GAMYB by KGM is directly responsible for GAMYB inactivation. HRT is a repressor of α-amylase gene expression (Raventos et al., 1998). This nucleus-localized zinc-finger protein binds to a 21-bp sequence containing the TAACAAA element, but at present there is no evidence for GA control of its repressor function.
Interactions with Other Signaling Pathways
Along with GAMYB, other early GA responses in aleurone cells include increases in cytosolic calcium (Gilroy and Jones, 1992; Bush, 1996), calmodulin (Schuurink et al., 1996), and ER-localized Ca2+-ATPase (Chen et al., 1997). These changes highlight the importance of Ca2+ signaling in aleurone cells in mediating the GA response. A target for Ca2+/calmodulin signaling is an ER Ca2+-ATPase, which supplies Ca2+ for the synthesis and secretion of Ca2+-containing metalloenzymes such as α-amylase (Bush et al., 1993; Gilroy and Jones, 1993). Another target that is regulated positively by Ca2+ and calmodulin is a slow vacuolar channel in the protein storage vacuole (Bethke and Jones, 1994). This channel is believed to be involved with Ca2+ release from the protein storage vacuoles.
Many GA responses in cereal aleurone cells are inhibited by abscisic acid. Recent work has shown how these two signaling pathways interact in aleurone cells to regulate gene expression. α-Amylase expression in the sln1 loss-of-function mutant was blocked by abscisic acid, indicating that abscisic acid signaling acts downstream of SLN1 in barley aleurone cells (Gomez-Cadenas et al., 2001). This finding is consistent with the failure of abscisic acid to block GA-stimulated SLN1 degradation (Gubler et al., 2002). Evidence from nuclear run-on and transient expression experiments showed that abscisic acid prevents increases in GAMYB transcription elicited by GA (Gomez-Cadenas et al., 2001; Gubler et al., 2002). A protein kinase, PKABA1, is likely to play a role in the abscisic acid repression of GAMYB transcription (Gomez-Cadenas et al., 2001). It is highly likely, therefore, that abscisic acid blocks the GA response between SLN1 and GAMYB. There is evidence that Ca2+ signaling also may be a target for abscisic acid signaling. The GA-induced increase in cytosolic calcium is prevented by abscisic acid (Wang et al., 1991).
CONCLUSIONS
A conceptual overview illustrating the possible cellular locations and functions of the GA response components is shown in Figure 5. A close interaction between GA metabolism and GA response pathways has been demonstrated, but the molecular mechanism of feedback regulation needs to be elucidated. Investigations of early GA biosynthesis and gene expression have provided insight into how the ent-kaurene level is regulated to control the capacity for GA biosynthesis during plant development. Examination of the expression patterns of the later genes in GA biosynthesis and catabolism has revealed a fine-tuning mechanism that controls the levels of bioactive GAs in plants. These studies have revealed complex regulation of the GA metabolic pathway, by both developmental programs and the GA response pathway, that determines the concentration of active GA in different tissues. However, at present, we do not have a complete understanding of this regulation.
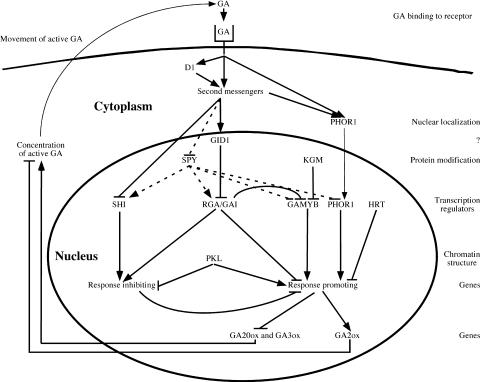
Figure 5.
Possible Roles of GA Response Components.
The cellular locations and functions of the GA response components discussed in this review are shown. A highly speculative order of action for these components also is shown. Although the diagram shows the components functioning in a single cell, there is no evidence that this occurs. Indeed, some aspects of the response pathway, such as the feedback regulation of GA levels, do not occur in all organs. Bioactive GA binds to an as yet unidentified GA receptor, directly or indirectly activates second messengers and G proteins (D1), and causes PHOR1 to localize to the nucleus and rapid degradation of RGA/GAI. GID1 is a positively acting component that is required for the degradation of GAI/RGA proteins. Although GID1 is shown in the cytosol, the cellular location has not been determined. In the nucleus, SPY and KGM inhibit GA responses by GlcNAc, modifying or phosphorylating component(s) of the pathway. Several putative transcription regulators (SHI, RGA/GAI proteins, GAMYB, PHOR1, and HRT) modify the transcription of genes encoding products that either promote or inhibit the GA response. Some of the products of these genes may be positive or negative regulators acting in a transcriptional cascade that leads to the GA response. The maintenance of chromatin structure by PKL is important for the proper expression of genes involved in the response. One outcome of the GA response is reduced transcription of GA20ox and GA3ox and enhanced expression of GA2ox, which leads to a reduction in the pool of bioactive GA.
To achieve this understanding, it will be necessary to conduct detailed localization studies of all genes for the entire pathway in the same species by in situ RNA hybridization, by promoter-reporter fusion, and, if possible, by immunostaining using specific antibodies. The list of GA response pathway components identified by molecular genetic analyses is impressive and expanding, and it is clear that these approaches will continue to be productive. As the components identified genetically are cloned, the analysis of genetic interactions, biochemical and cell biological studies, and gene profiling by microarray analyses will be needed to decipher the detailed mechanisms by which these components function. These future studies also will help to identify the interactors and/or targets of these components of the GA response pathway. The use of multiple plant systems has accelerated the isolation of new GA signaling components and will be essential in revealing differences in the relative importance of these components among species. However, to fully understand the relationships between the different components, it will be necessary to study them in model organisms.
Acknowledgments
We thank Peter Chandler, Caren Chang, Rod King, Nick Harberd, Chris Helliwell, Makoto Matsuoka, Jim Reid, John Ross, and Eva Sundberg for sharing data before publication. We also thank Peter Chandler, Chris Helliwell, Russell Jones, Rod King, Masumi Robertson, and Aron Silverstone for helpful comments on the manuscript, and Peter Chandler for providing Figure 3. This work was supported by grants from the National Science Foundation (MCB-0112826 to N.O. and IBN-0078003 to T.-p.S.), the U.S.–Israel Binational Agricultural Research and Development Fund (IS-2837-97 to N.O.), and the U.S. Department of Agriculture (99-35304-8061 and 01-35304-10892 to T.-p.S.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010476.
References
- Aach, H., Bode, H., Robinson, D.G., and Graebe, J.E. (1997). ent-Kaurene synthase is located in proplastids of meristematic shoot tissues. Planta 202 211–219. [Google Scholar]
- Ait-Ali, T., Frances, S., Weller, J., Reid, J.B., Kendrick, R.E., and Kamiya, Y. (1999). Regulation of gibberellin 20-oxidase and gibberellin 3β-hydroxylase transcript accumulation during de-etiolation of pea seedlings. Plant Physiol. 121 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador, V., Monte, E., García-Martínez, J.-L., and Prat, S. (2001). Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106 343–354. [DOI] [PubMed] [Google Scholar]
- Ashikari, M., Wu, J., Yano, M., Sasaki, T., and Yoshimura, A. (1999). Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc. Natl. Acad. Sci. USA 96 10284–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke, P.C., and Jones, R.L. (1994). Ca2+-calmodulin modulates ion channel activity in storage protein vacuoles of barley aleurone cells. Plant Cell 6 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke, P.C., and Jones, R.L. (1998). Gibberellin signaling. Curr. Opin. Plant Biol. 1 440–446. [DOI] [PubMed] [Google Scholar]
- Blatch, G.L., and Lassle, M. (1999). The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays 21 932–939. [DOI] [PubMed] [Google Scholar]
- Blázquez, M.A., and Weigel, D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404 889–892. [DOI] [PubMed] [Google Scholar]
- Bolle, C., Koncz, C., and Chua, N.-H. (2000). PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 14 1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Bouquin, T., Meier, C., Foster, R., Nielsen, M.E., and Mundy, J. (2001). Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 127 450–458. [PMC free article] [PubMed] [Google Scholar]
- Bush, D.S. (1996). Effects of gibberellic acid and environmental factors on cytosolic calcium in wheat aleurone cells. Planta 199 89–99. [Google Scholar]
- Bush, D.S., Biswas, A.K., and Jones, R.L. (1993). Hormonal regulation of Ca2+ transport in the endomembrane system of the barley aleurone. Planta 189 507–515. [Google Scholar]
- Carrera, E., Jackson, S.D., and Prat, S. (1999). Feedback control and diurnal regulation of gibberellin 20-oxidase transcript levels in potato. Plant Physiol. 119 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera, E., Bou, J., García-Martínez, J.L., and Prat, S. (2000). Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant J. 22 247–256. [DOI] [PubMed] [Google Scholar]
- Cercos, M., Gomez-Cadenas, A., and Ho, T.-H.D. (1999). Hormonal regulation of a cysteine proteinase gene, EBP1, in barley aleurone layers: Cis- and trans-acting elements involved in the co-ordinated gene expression regulated by gibberellins and abscisic acid. Plant J. 19 107–118. [DOI] [PubMed] [Google Scholar]
- Chandler, P.M., and Robertson, M. (1999). Gibberellin dose-response curves and the characterization of dwarf mutants of barley. Plant Physiol. 120 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, P.M., Marion-Poll, A., Ellis, M., and Gubler, F. (2002). Mutants at the Slender1 locus of ‘Himalaya’ barley: Molecular and physiological characterization. Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- Chen, X., Chang, M., Wang, B., and Wu, R. (1997). Cloning of a Ca2+-ATPase gene and the role of cytosolic Ca2+ in the gibberellin-dependent signaling pathway in aleurone cells. Plant J. 11 363–371. [DOI] [PubMed] [Google Scholar]
- Chien, J.C., and Sussex, I.M. (1996). Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 111 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.-H., Yoshizawa, K., Kobayashi, M., and Sakurai, A. (1995). Distribution of endogenous gibberellin in vegetative shoots of rice. Plant Cell Physiol. 36 997–1001. [Google Scholar]
- Chung, C.H., and Coolbaugh, R.C. (1986). ent-Kaurene biosynthesis in cell-free extracts of excised parts of tall and dwarf pea seedlings. Plant Physiol. 80 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer, F.I., and Hart, G.W. (2000). O-Glycosylation of nuclear and cytosolic proteins: Dynamic interplay between O-GlcNAc and O-phosphate. J. Biol. Chem. 275 29179–29182. [DOI] [PubMed] [Google Scholar]
- Cowling, R.J., Kamiya, Y., Seto, H., and Harberd, N.P. (1998). Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol. 117 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croker, S.J., Hedden, P., Lenton, J.R., and Stoddart, J.L. (1990). Comparison of gibberellins in normal and slender barley seedlings. Plant Physiol. 94 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, T.C. (1998). Signal transduction by the Wnt family of ligands. Biochem. J. 329 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A.K., Cohen, P.T.W., and Barford, D. (1998). The structure of the tetratricopeptide repeats of protein phosphatase 5: Implications for TPR-mediated protein-protein interactions. EMBO J. 17 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, P.J., ed (1995). Plant Hormones: Physiology, Biochemistry and Molecular Biology. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Debeaujon, I., and Koornneef, M. (2000). Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 122 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkx, M.P.M., Vermeer, E., and Karssen, C.M. (1994). Gibberellins in seeds of Arabidopsis thaliana: Biological activities, identification and effects of light and chilling on endogenous levels. Plant Growth Regul. 15 223–234. [Google Scholar]
- Dill, A., and Sun, T.-p. (2001). Synergistic de-repression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., Jung, H.-S., and Sun, T.-p. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, R.C., Smith, J.L., Lester, D.R., and Reid, J.B. (2001). Feed-forward regulation of gibberellin deactivation in pea. J. Plant Growth Regul. 20 87–94. [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209. [DOI] [PubMed] [Google Scholar]
- Freemont, P.S. (2000). Ubiquitination: RING for destruction? Curr. Biol. 10 R84–R87. [DOI] [PubMed] [Google Scholar]
- Fridborg, I., Kuusk, S., Moritz, T., and Sundberg, E. (1999). The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell 11 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridborg, I., Kuusk, S., Robertson, M., and Sundberg, E. (2001). The Arabidopsis protein SHI represses gibberellin responses in Arabidopsis and barley. Plant Physiol. 127 937–948. [PMC free article] [PubMed] [Google Scholar]
- Fu, X., Sudhakar, D., Peng, J., Richards, D.E., Christou, P., and Harberd, N.P. (2001). Expression of Arabidopsis GAI in transgenic rice represses multiple gibberellin responses. Plant Cell 13 1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, S., Yamane, H., Spray, C.R., Gaskin, P., MacMillan, J., Phinney, B.O., and Takahashi, N. (1988). Qualitative and quantitative analyses of gibberellins in vegetative shoots of normal, dwarf-1, dwarf-2, dwarf-3, and dwarf-5 seedlings of Zea mays L. Plant Physiol. 88 1367–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa, Y., Kato, T., Ohki, S., Ishikawa, A., Kitano, H., Sasaki, T., Asahi, T., and Iwasaki, Y. (1999). Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc. Natl. Acad. Sci. USA 96 7575–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D., and Marshall, G.A. (1973). Insensitivity to gibberellin in dwarf wheats. Ann. Bot. 37 729–735. [Google Scholar]
- Gil, J., and García-Martínez, J.L. (2000). Light regulation of gibberellin A1 content and expression of genes coding for GA 20-oxidase and GA 3β-hydroxylase in etiolated pea seedlings. Physiol. Plant. 108 223–229. [Google Scholar]
- Gilroy, S., and Jones, R.L. (1992). Gibberellic acid and abscisic acid coordinately regulate cytoplasmic calcium and secretory activity in barley aleurone protoplasts. Proc. Natl. Acad. Sci. USA 89 3591–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy, S., and Jones, R.L. (1993). Calmodulin stimulation of unidirectional calcium uptake by the endoplasmic reticulum of barley aleurone. Planta 190 289–296. [Google Scholar]
- Gilroy, S., and Jones, R.L. (1994). Perception of gibberellin and abscisic acid at the external face of the plasma membrane of barley (Hordeum vulgare L.) aleurone protoplasts. Plant Physiol. 104 1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal, G.F.W., Sheldon, C.C., Gubler, F., Moritz, T., Bagnall, D.B., MacMillan, C.P., Li, S.F., Parish, R.W., Dennis, E.S., Weigel, D., and King, R.W. (2001). GAMYB-like genes, flowering and gibberellin signaling in Arabidopsis. Plant Physiol. 127 1682–1693. [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadenas, A., Zentalla, R., Walker-Simmons, M., and Ho, T.-H.D. (2001). Gibberellin/abscisic acid antagonism in barley aleurone cells: Site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13 667–679. [PMC free article] [PubMed] [Google Scholar]
- Graebe, J.E. (1987). Gibberellin biosynthesis and control. Annu. Rev. Plant Physiol. 38 419–465. [Google Scholar]
- Groot, S.P.C., and Karssen, C.M. (1987). Gibberellins regulate seed germination in tomato by endosperm weakening: A study with gibberellin-deficient mutants. Planta 171 525–531. [DOI] [PubMed] [Google Scholar]
- Gubler, F., and Jacobsen, J.V. (1992). Gibberellin-responsive elements in the promoter of a barley high-pI α-amylase gene. Plant Cell 4 1435–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Kalla, R., Roberts, J., and Jacobsen, J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Raventos, D., Keys, M., Watts, R., Mundy, J., and Jacobsen, J.V. (1999). Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 17 1–9. [DOI] [PubMed] [Google Scholar]
- Gubler, F., Chandler, P., White, R., Llewellyn, D., and Jacobsen, J. (2002). GA signaling in barley aleurone cells: Control of SLN1 and GAMYB expression. Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- Hanover, J.A. (2001). Glycan-dependent signaling: O-Linked N-acetylglucosamine. FASEB J. 15 1865–1876. [DOI] [PubMed] [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5 523–530. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Sheldon, C.C., Olive, M.R., Walker, A.R., Zeevaart, J.A.D., Peacock, W.J., and Dennis, E.S. (1998). Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. USA 95 9019–9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A., Chandler, P.M., Poole, A., Olive, M.R., Dennis, E.S., and Peacock, W.J. (2001. a). The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 98 2065–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A., Sullivan, J.A., Mould, R.M., Gray, J.C., Peacock, J., and Dennis, E.S. (2001. b). A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. 28 201–208. [DOI] [PubMed] [Google Scholar]
- Hilhorst, H.W.M., and Karssen, C.M. (1988). Dual effect of light on the gibberellin- and nitrate-stimulated seed germination of Sisymbrium officinale and Arabidopsis thaliana. Plant Physiol. 86 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, T.-H.D., Nolan, R.C., and Shute, D.E. (1981). Characterization of a gibberellin-insensitive dwarf wheat, D6899. Plant Physiol. 67 1026–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooley, R., Beale, M.H., and Smith, S.J. (1991). Gibberellin perception at the plasma membrane of Avena fatua aleurone protoplasts. Planta 183 274–280. [DOI] [PubMed] [Google Scholar]
- Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Kawaide, H., Chen, X., Kamiya, Y., and Matsuoka, M. (1999). The gene encoding tobacco gibberellin 3β-hydroxylase is expressed at the site of GA action during stem elongation and flower organ development. Plant J. 20 15–24. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sentoku, N., Kitano, H., Matsuoka, M., and Kobayashi, M. (2001). Cloning and functional analysis of two gibberellin 3β-hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA 98 8909–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhaki, A., Swain, S.M., Tseng, T.-s., Borochov, A., Olszewski, N.E., and Weiss, D. (2001). The role of SPY and its TPR domain in the regulation of gibberellin action throughout the life cycle of Petunia hybrida plants. Plant J. 28 1–11. [DOI] [PubMed] [Google Scholar]
- Jackson, S.D. (1999). Multiple signaling pathways control tuber induction in potato. Plant Physiol. 119 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.D., James, P., Prat, S., and Thomas, B. (1998). Phytochrome B affects the levels of a graft-transmissible signal involved in tuberization. Plant Physiol. 117 29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.D., James, P.E., Carrera, E., Prat, S., and Thomas, B. (2000). Regulation of transcript levels of a potato gibberellin 20-oxidase gene by light and phytochrome B. Plant Physiol. 124 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, J.V., Pearce, D.W., Poole, A.T., Pharis, R.P., and Mander, L.N. (2002). Abscisic, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant., in press. [DOI] [PubMed]
- Jacobsen, S.E., and Olszewski, N.E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., Binkowski, K.A., and Olszewski, N.E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 93 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., Olszewski, N.E., and Meyerowitz, E.M. (1998). SPINDLY's role in the gibberellin response pathway. Symp. Soc. Exp. Biol. 51 73–78. [PubMed] [Google Scholar]
- Jones, H.D., Smith, S.J., Desikan, R., Plakidou-Dymock, S., Lovegrove, A., and Hooley, R. (1998). Heterotrimeric G proteins are implicated in gibberellin induction of α-amylase gene expression in wild oat aleurone. Plant Cell 10 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, Y., and García-Martínez, J. (1999). Regulation of gibberellin biosynthesis by light. Curr. Opin. Plant Biol. 2 398–403. [DOI] [PubMed] [Google Scholar]
- King, K., Moritz, T., and Harberd, N. (2001. a). Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, R.W., Moritz, T., Evans, L.T., Junttila, O., and Herlt, A.J. (2001. b). Long-day induction of flowering in Lolium temulentum involves sequential increases in specific gibberellins at the shoot apex. Plant Physiol. 127 624–632. [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., and van der Veen, J.H. (1980). Induction and analysis of gibberellin-sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 58 257–263. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Jorna, M.L., Brinkhorst-van der Swan, D.L.C., and Karssen, C.M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 61 385–393. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Elgersma, A., Hanhart, C.J., van Loenen, M.E.P., van Rijn, L., and Zeevaart, J.A.D. (1985). A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant. 65 33–39. [Google Scholar]
- Kreppel, L.K., and Hart, G.W. (1999). Regulation of a cytosolic and nuclear O-GlcNAc transferase: Role of the tetratricopeptide repeats. J. Biol. Chem. 274 32015–32022. [DOI] [PubMed] [Google Scholar]
- Kusaba, S., Kano-Murakami, Y., Matsuoka, M., Tamaoki, M., Sakamoto, T., Yamaguchi, I., and Fukumoto, M. (1998). Alteration of hormone levels in transgenic tobacco plants overexpressing the rice homeobox gene OSH1. Plant Physiol. 116 471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J.R., Tugendreich, S., and Hieter, P. (1995). Tetratricopeptide repeat interactions: To TPR or not to TPR? Trends Biochem. Sci. 20 257–259. [DOI] [PubMed] [Google Scholar]
- Lanahan, M.B., Ho, T.-H.D., Rogers, S.W., and Rogers, J.C. (1992). A gibberellin response complex in cereal alpha-amylase gene promoters. Plant Cell 4 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, T. (1998). Molecular biology of gibberellin synthesis. Planta 204 409–419. [DOI] [PubMed] [Google Scholar]
- Lenton, J.R., and Appleford, N.E.J.G. (1991). Gibberellin production and action during germination of wheat. In Gibberellin, N. Takahashi, B.O. Phinney, and J. MacMillan, eds (New York: Springer-Verlag), pp. 125–135.
- Lenton, J.R., Appleford, N.E.J., and Croker, S.J. (1994). Gibberellins and α-amylase gene expression in germinating wheat grains. Plant Growth Regul. 15 261–270. [Google Scholar]
- Leubner-Metzger, G., Frundt, C., and Meins, F.J. (1996). Effects of gibberellins, darkness and osmotica on endosperm rupture and class I β-1,3 glucanase induction in tobacco seed germination. Planta 199 282–288. [Google Scholar]
- Levy, Y.Y., and Dean, C. (1998). The transition to flowering. Plant Cell 10 1973–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Juez, E., Kobayashi, M., Sakurai, A., Kamiya, Y., and Kendrick, R.E. (1995). Phytochrome, gibberellins, and hypocotyl growth. Plant Physiol. 107 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovegrove, A., and Hooley, R. (2000). Gibberellin and abscisic acid signalling in aleurone. Trends Plant Sci. 5 102–110. [DOI] [PubMed] [Google Scholar]
- Martin, D.N., Proebsting, W.M., Parks, T.D., Dougherty, W.G., Lange, T., Lewis, M.J., Gaskin, P., and Hedden, P. (1996). Feed-back regulation of gibberellin biosynthesis and gene expression in Pisum sativum L. Planta 200 159–166. [DOI] [PubMed] [Google Scholar]
- Meier, C., Bouquin, T., Nielsen, M.E., Raventos, D., Mattsson, O., Rocher, A., Schomburg, F., Amasino, R.M., and Mundy, J. (2001). Gibberellin response mutants identified by luciferase imaging. Plant J. 25 509–519. [DOI] [PubMed] [Google Scholar]
- Metzger, J.D. (1995). Hormones and reproductive development. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 617–648.
- Nambara, E., Akazawa, T., and McCourt, P. (1991). Effects of the gibberellin biosynthetic inhibitor uniconazol on mutants of Arabidopsis. Plant Physiol. 97 736–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara, E., Naito, S., and McCourt, P. (1992). A mutant of Arabidopsis which is defective in seed development and storage protein accumulations is a new abi3 allele. Plant J. 2 435–441. [Google Scholar]
- Ogas, J., Cheng, J.-C., Sung, Z.R., and Somerville, C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277 91–94. [DOI] [PubMed] [Google Scholar]
- Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, M., Kusano, T., Katsumi, M., and Sano, H. (2000). Rice gibberellin-insensitive gene homolog, OsGAI, encodes a nuclear-localized protein capable of gene activation at transcriptional level. Gene 245 21–29. [DOI] [PubMed] [Google Scholar]
- O'Neill, D.P., Ross, J.J., and Reid, J.B. (2000). Changes in gibberellin A1 levels and response during de-etiolation of pea seedlings. Plant Physiol. 124 805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Peng, H., Begg, G.E., Schultz, D.C., Friedman, J.R., Jensen, D.E., Speicher, D.W., and Rauscher, F.J., III (2000). Reconstitution of the KRAB-KAP-1 repressor complex: A model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J. Mol. Biol. 295 1139–1162. [DOI] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signalling pathway that negatively regulates gibberellin responses. Genes Dev. 11 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., et al. (1999. a). “Green Revolution” genes encode mutant gibberellin response modulators. Nature 400 256–261. [DOI] [PubMed] [Google Scholar]
- Peng, J., Richards, D.E., Moritz, T., Cano-Delgado, A., and Harberd, N.P. (1999. b). Extragenic suppressors of the Arabidopsis gai mutation alter the dose-response relationship of diverse gibberellin responses. Plant Physiol. 119 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penson, S.P., Schuurink, R.C., Fath, A., Gubler, F., Jacobsen, J.V., and Jones, R.L. (1996). cGMP is required for gibberellic acid-induced gene expression in barley aleurone. Plant Cell 8 2325–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza, D., Vachon, G., and Herzog, M. (1998). Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol. 117 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, A. (1998). Gibberellins in Arabidopsis. Plant Physiol. Biochem. 36 115–124. [Google Scholar]
- Pysh, L.D., Wysocka-Diller, J.W., Camilleri, C., Bouchez, D., and Benfey, P.N. (1999). The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18 111–119. [DOI] [PubMed] [Google Scholar]
- Raventos, D., Skriver, K., Schlein, M., Karnahl, K., Rogers, S.W., Rogers, J.C., and Mundy, J. (1998). HRT, a novel zinc finger, transcriptional repressor from barley. J. Biol. Chem. 273 23313–23320. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Foster, K.R., Morgan, P.W., and Chory, J. (1996). Phytochrome B affects responsiveness to gibberellins in Arabidopsis. Plant Physiol. 112 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, J.B., Botwright, N.A., Smith, J.J., O'Neill, D.P., and Kerckhoffs, L.H.J. (2002). Control of gibberellin levels and gene expression during de-etiolation in pea. Plant Physiol. 128 734-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, L., Sánchez-Baracaldo, P., and Hake, S. (2000). Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol. Biol. 42 151–166. [PubMed] [Google Scholar]
- Richards, D.E., Peng, J., and Harberd, N.P. (2000). Plant GRAS and metazoan STATs: One family? Bioessays 22 573–577. [DOI] [PubMed] [Google Scholar]
- Richards, D.E., King, K.E., Ait-Ali, T., and Harberd, N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 67–88. [DOI] [PubMed] [Google Scholar]
- Robertson, M., Swain, S.M., Chandler, P.M., and Olszewski, N.E. (1998). Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell 10 995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, J.C., and Rogers, S.W. (1992). Definition and functional implications of gibberellin and abscisic acid cis-acting hormone response complexes. Plant Cell 4 1443–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos, M.D., and Hanover, J.A. (2000). Structure of O-linked GlcNAc transferase: Mediator of glycan-dependent signaling. Biochem. Biophys. Res. Commun. 271 275–280. [DOI] [PubMed] [Google Scholar]
- Ross, J.J. (1998). Effects of auxin transport inhibitors on gibberellins in pea. J. Plant Growth Regul. 17 141–146. [Google Scholar]
- Ross, J.J., O'Neill, D.P., Smith, J.J., Kerckhoffs, L.H.J., and Elliott, R.C. (2000). Evidence that auxin promotes gibberellin A1 biosynthesis in pea. Plant J. 21 547–552. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., Kamiya, N., Ueguchi-Tanaka, M., Iwahori, S., and Matsuoka, M. (2001. a). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T., Kobayashi, M., Itoh, H., Tagiri, A., Kayano, T., Tanaka, H., Iwahori, S., and Marsuoka, M. (2001. b). Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 125 1508–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Fernandez, R., Ardiles-Diaz, W., van Montagu, M., Inze, D., and May, M.J. (1998). Cloning of a novel Arabidopsis thaliana RGA-like gene, a putative member of the VHIID-domain transcription factor family. J. Exp. Bot. 49 1609–1610. [Google Scholar]
- Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Ito, H., Kobayashi, M., Kitano, H., and Matsuoka, M. (2001). Screening of rice GIBBERELLIN-INSENSITIVE DWARF 1 mutants (GID1). In 17th International Conference on Plant Growth Substances; July 1–6, 2001; Brno, Czech Republic.
- Schumacher, K., Schmitt, T., Rossberg, M., Schmitz, G., and Theres, K. (1999). The Lateral Suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 96 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurink, R.C., Chan, P.V., and Jones, R.L. (1996). Modulation of calmodulin mRNA and protein levels in barley aleurone. Plant Physiol. 111 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafi, R., Iyer, S.P., Ellies, L.G., O'Donnell, N., Marek, K.W., Chui, D., Hart, G.W., and Marth, J.D. (2000). The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 97 5735–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T. (1997). Phytochrome regulation of seed germination. J. Plant Res. 110 151–161. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Chang, C.-w., Krol, E., and Sun, T.-p. (1997. a). Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 12 9–19. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Mak, P.Y.A., Casamitjana Martínez, E., and Sun, T.-p. (1997. b). The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T.-p. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Jung, H.-S., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T.-p. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriver, K., Olsen, F.L., Rogers, J.C., and Mundy, J. (1991). cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc. Natl. Acad. Sci. USA 88 7266–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber, C.M., Cooney, S., and McCourt, P. (1998). Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.-p. (2000). Gibberellin signal transduction. Curr. Opin. Plant Biol. 3 374–380. [DOI] [PubMed] [Google Scholar]
- Sun, T.-p., and Kamiya, Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, S.M., Tseng, T.S., and Olszewski, N.E. (2001). Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol. 126 1174–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain, S.M., Tseng, T.-s., Thornton, T., Gopalraj, M., and Olszewski, N.E. (2002). SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant. Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- Talón, M., Koornneef, M., and Zeevaart, J.A.D. (1990). Accumulation of C-19-gibberellins in the gibberellin-insensitive dwarf mutant gai of Arabidopsis thaliana (L.) Heynh. Planta 182 501–505. [DOI] [PubMed] [Google Scholar]
- Tanaka-Ueguchi, M., Itoh, H., Oyama, N., Koshioka, M., and Matsuoka, M. (1998). Over-expression of a tobacco homeobox gene, NTH15, decreases the expression of a gibberellin biosynthetic gene encoding GA 20-oxidase. Plant J. 15 391–400. [DOI] [PubMed] [Google Scholar]
- Telfer, A., Bollman, K.M., and Poethig, R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124 645–654. [DOI] [PubMed] [Google Scholar]
- Thomas, S.G., Phillips, A.L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, T., Kreppel, L., Hart, G., and Olszewski, N. (1999a). Genetic and biochemical analysis of Arabidopsis SPY. In Plant Biotechnology and in Vitro Biology in the 21st Century, A. Altman, M. Ziv, and S. Izhar, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 445–448.
- Thornton, T.M., Swain, S.M., and Olszewski, N.E. (1999. b). Gibberellin signal transduction presents … the SPY who O-GlcNAc'd me. Trends Plant Sci. 4 424–428. [DOI] [PubMed] [Google Scholar]
- Toyomasu, T., Tsuji, H., Yamane, H., Nakayama, M., Yamaguchi, I., Murofushi, N., Takahashi, N., and Inoue, Y. (1993). Light effects on endogenous levels of gibberellins in photoblastic lettuce seeds. J. Plant Growth Regul. 12 85–90. [Google Scholar]
- Toyomasu, T., Kawaide, H., Mitsuhashi, W., Inoue, Y., and Kamiya, Y. (1998). Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol. 118 1517–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, T.-s., Swain, S.M., and Olszewski, N.E. (2001). Ectopic expression of the tetratricopeptide repeat domain of SPINDLY causes defects in gibberellin response. Plant Physiol. 126 1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Fujisawa, Y., Kobayashi, M., Ashikari, M., Iwasaki, Y., Kitano, H., and Matsuoka, M. (2000). Rice dwarf mutant d1, which is defective in the α subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. USA 97 11638–11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Itoh, H., Kobayashi, M., Kitona, H., and Matsuoka, M. (2001). Characterization of rice dwarf mutant, GIBBERELLIN-INSENSITIVE DWARF 1 (GID1). In 17th International Conference on Plant Growth Substances; July 1–6, 2001; Brno, Czech Republic.
- Ullah, H., Chen, J.G., Young, J.C., Im, K.H., Sussman, M.R., and Jones, A.M. (2001). Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292 2066–2069. [DOI] [PubMed] [Google Scholar]
- Wade, P.A., Jones, P.L., Vermaak, D., and Wolffe, A.P. (1998). A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 8 843–846. [DOI] [PubMed] [Google Scholar]
- Wang, M., Duijn, B.V., Schram, A.W., and Van, D.U. (1991). Abscisic acid induces a cytosolic calcium decrease in barley aleurone protoplasts. FEBS Lett. 278 69–74. [DOI] [PubMed] [Google Scholar]
- Wang, X.Q., Ullah, H., Jones, A.M., and Assmann, S.M. (2001). G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292 2070–2072. [DOI] [PubMed] [Google Scholar]
- Weiss, D., and Halevy, A.H. (1989). Stamens and gibberellin in the regulation of corolla pigmentation and growth in Petunia hybrida. Planta 179 89–96. [DOI] [PubMed] [Google Scholar]
- Weller, J.L., Ross, J.J., and Reid, J.B. (1994). Gibberellins and phytochrome regulation of stem elongation in pea. Planta 192 489–496. [Google Scholar]
- Weller, J.L., Nagatani, A., Kendrick, R.E., Murfet, I.C., and Reid, J.B. (1995). New lv mutants of pea are deficient in phytochrome B. Plant Physiol. 108 525–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, L., Vosseller, K., and Hart, G.W. (2001). Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNAc. Science 291 2376–2378. [DOI] [PubMed] [Google Scholar]
- Wen, C.-K., and Chang, C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, C.A., Shen-Miller, J., and Railton, I.D. (1982). Regulation of kaurene synthetase. In Plant Growth Substances 1982, P.F. Wareing, ed (London: Academic Press), pp. 81–90.
- Willert, K., and Nusse, R. (1998). Beta-catenin: A key mediator of Wnt signaling. Curr. Opin. Genet. Dev. 8 95–102. [DOI] [PubMed] [Google Scholar]
- Wilson, R.N., and Somerville, C.R. (1995). Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 108 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.N., Heckman, J.W., and Somerville, C.R. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang, C.M., and Ross, J.J. (2001). Auxin promotes gibberellin biosynthesis in decapitated tobacco plants. Planta 214 153–157. [DOI] [PubMed] [Google Scholar]
- Wu, K., Li, L., Gage, D.A., and Zeevaart, J.A.D. (1996). Molecular cloning and photoperiod-regulated expression of gibberellin 20-oxidase from the long-day plant spinach. Plant Physiol. 110 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.-L., Li, L., Wu, K., Peeters, A.J.M., Gage, D.A., and Zeevaart, J.A.D. (1995). The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: Molecular cloning and functional expression. Proc. Natl. Acad. Sci. USA 92 6640–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.-L., Gage, D.A., and Zeevaart, J.A.D. (1997). Gibberellins and stem growth in Arabidopsis thaliana. Plant Physiol. 114 1471–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K. (1982). Determination of endogenous gibberellins in germinating barley by combined gas chromatography-mass spectrometry. J. Am. Soc. Brew. 40 18–25. [Google Scholar]
- Yamaguchi, S., and Kamiya, Y. (2000). Gibberellin biosynthesis: Its regulation by endogenous and environmental signals. Plant Cell Physiol. 41 251–257. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S., Smith, M.W., Brown, R.G.S., Kamiya, Y., and Sun, T.-p. (1998. a). Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seed. Plant Cell 10 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S., Sun, T.-p., Kawaide, H., and Kamiya, Y. (1998. b). The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol. 116 1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S., Kamiya, Y., and Sun, T.-p. (2001). Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 28 443–453. [DOI] [PubMed] [Google Scholar]
- Yaxley, J.R., Ross, J.J., Sherriff, L.J., and Reid, J.B. (2001). Gibberellin biosynthesis mutations and root development in pea. Plant Physiol. 125 627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., LeRoy, G., Seelig, H.P., Lane, W.S., and Reinberg, D. (1998). The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95 279–289. [DOI] [PubMed] [Google Scholar]