INTRODUCTION
Steroids play a role as essential hormones in plants as well as in animals. Plants produce numerous steroids and sterols, some of which are recognized as hormones in animals (Geuns, 1978; Jones and Roddick, 1988). Brassinolide (BL) is the most bioactive form of the growth-promoting plant steroids termed brassinosteroids (BRs). Grove et al. (1979) purified 4 mg of BL from ∼40 kg of bee-collected rape pollen to determine its structure, which shows similarity to animal steroid hormones (Figure 1). Due to its low concentration, the identification of BL took 10 years of dedicated work on the part of U.S. Department of Agriculture researchers at a cost of over one million U.S. dollars (Mandava, 1988). Today, over 40 naturally occurring BRs are known; these carry an oxygen moiety at the C-3 position in combination with others at the C-2, C-6, C-22 or C-23 positions (Bishop and Yokota, 2001; Figure 1). Initial interest in brassinosteroids was based on the growth-promoting properties of pollen extracts studied by Mitchell et al. (1970). Although results from these early experiments revealed potential hormonal activities, many of the observed BR-induced effects appeared to be similar to those of other known plant hormones. The definition of BRs as hormones therefore did not gain wide acceptance, and the difficulty in obtaining and quantifying BRs restricted the related research. However, through the dedication and perseverance of a select group of researchers, numerous BRs were identified and metabolism studies performed, enabling the elucidation of the biochemical pathway leading to the production of BL (see Figure 2). Many reviews have covered this early research period (Adam and Marquardt, 1986; Mandava, 1988; Sakurai and Fujioka, 1993; Clouse, 1997; Sasse, 1997; Yokota, 1997) and an excellent book provides intriguing details on related subjects (see Sakurai et al., 1999). The observation that Arabidopsis mutations abolishing the biosynthesis of BL result in a dwarf phenotype that can be restored to a wild-type phenotype by externally provided BL and intermediates of BR biosynthesis (Li et al., 1996; Szekeres et al., 1996) resulted in a wider acceptance that BRs are essential plant hormones. Characterization of BR-insensitive mutants showing a similar dwarf phenotype has subsequently identified key genes in BR signaling (Clouse et al., 1996; Li and Chory, 1997; Li and Nam, 2002). Recent advances in the study of plant steroid hormone biosynthesis and signaling are highlighted by several reviews (Clouse and Sasse, 1998; Szekeres and Koncz, 1998; Altmann, 1999; Li and Chory, 1999; Schumacher and Chory, 2000; Bishop and Yokota, 2001; Friedrichsen and Chory, 2001; Mussig and Altmann, 2001).
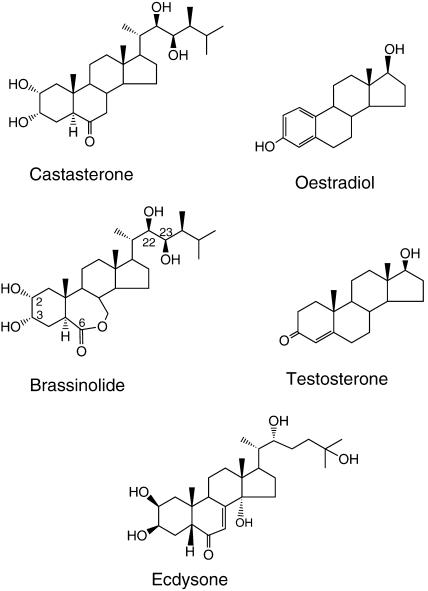
Figure 1.
Structures of Steroid Hormones.
Chemical structure of brassinolide and castasterone plant steroid hormones, in comparison with the mammalian sex steroid hormones testosterone and oestradiol, and the insect steroid hormone ecdysone. Highlighted are carbon numbers of BL having oxygen moieties that are important for BR activity.
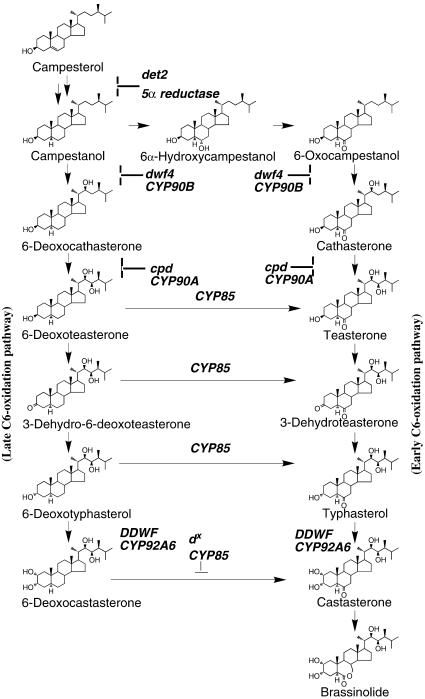
Figure 2.
BR Biosynthesis Pathway.
A simplified scheme of the BR biosynthesis pathway including steps of early and late C-6 oxidation that lead to the synthesis of BL (Noguchi et al., 2000; Shimada et al., 2001). Mutations in genes coding for key biosynthetic enzymes are in bold. det-2, DEETIOLATED-2, steroid 5α reductase (Li et al., 1996); dwf4, DWARF4 cytochrome P450 CYP90B, C-22 hydroxylase (Choe et al., 1998); cpd, CONSTITUTIVE PHOTOMORPHOGENIC and DWARFISM cytochrome P450 CYP90A, C-23 hydroxylase (Szekeres et al., 1996); dx, DWARF cytochrome P450 CYP85, C-6 oxidase (Bishop et al., 1996, 1999; Shimada et al., 2001); DDWF, DARK-INDUCED dwf-LIKE-1, cytochrome P450 CYP92A6, C-2 hydroxylase (Kang et al., 2001).
In animal cells, steroid hormones are perceived by binding to their cognate nuclear steroid receptors, which are present in the cytoplasm. Once activated, the receptor complex is transferred to the nucleus to promote or repress transcription of hormone-responsive genes (Beato et al., 1995). This is called genomic signaling because it involves the genome in generating a physiological response. Steroid hormones can also elicit cellular responses in the presence of inhibitors of transcription or protein synthesis, and in enucleated cells. This mechanism is often referred to as nongenomic steroid signaling because it is not mediated by the genome. Some of the nongenomic signaling mechanisms imply a direct interaction between nuclear steroid receptors and other signaling molecules. Nongenomic steroid signaling pathways can also stimulate alterations in second messenger levels, ion fluxes, and protein kinase activities via as-yet uncharacterized steroid carrier proteins and plasma membrane–associated receptors (for reviews, see Wehling, 1997; Falkenstein et al., 2000). The annotation of the Arabidopsis genome sequence (Arabidopsis Genome Initiative, 2000) has confirmed the previous notion (Li and Chory, 1997; McCarty and Chory, 2000) that plants lack close homologs of animal nuclear steroid receptors. Steroid signaling in plants that also leads to alteration in transcription or a physiological response is therefore probably mediated by alternative mechanisms that may be similar to nongenomic steroid signaling in animals.
This review highlights the characterization of genes and mutations that is beginning to reveal the mode of steroid hormone action in plants, but at the same time is mindful that this was preceded by the characterization of BR's effects on plant growth and development. To identify regulatory functions in plant steroid signaling, a genetic approach was exploited that aimed at the isolation of brassinosteroid insensitive (bri) Arabidopsis mutants (Clouse et al., 1996). Intriguingly, initial characterization of these bri mutants led only to the identification of multiple mutant alleles of the BRI1 gene, which was subsequently cloned by chromosome walking (Li and Chory, 1997). BRI1 encodes a leucine-rich repeat receptor-like kinase (LRR-RLK) that provides an important key to deciphering primary events in BR signaling. In addition to summarizing our current knowledge on the regulatory role of BRI1, this review provides a brief look at the characterization of further genes that is beginning to provide an insight into some intricate signaling functions controlling steroid hormone action in plants.
Physiological Activity of BRs
The leaf bending caused by extracts from Distylium racemosum in the rice lamina inclination assay (Marumo et al., 1968) is one of the earliest reports of BR activity in plants. In this assay, BRs caused dose-dependent swelling of the adaxial cells in the joint between the leaf blade and sheath of etiolated rice seedlings. The angle of the leaf bending caused by the swelling is used as a sensitive bioassay during BR isolation and quantification procedures (Crozier et al. 2000). An early report on growth-promoting activity of “brassin,” from which BL was isolated, was based on BL-mediated stimulation of elongation of bean 2nd and 3rd internode, with higher concentrations causing splitting of the stem (Mitchell et al., 1970; Grove et al., 1979). BL and synthetic analogs were subsequently used in various bioassays, which indicated that in addition to leaf bending, cell elongation, and cell division, BL also effects source/sink relationships (Krizek and Mandava, 1983b), proton pumping and membrane polarization (Cerana et al., 1983; Romani et al., 1983), photosynthesis (Braun and Wild, 1984), and stress responses including thermotolerance (Wilen et al., 1995; Dhaubhadel et al., 1999) and senescence (Mandava et al., 1981). In addition to these effects, BRs promote vascular differentiation (Iwasaki and Shibaoka, 1991; Yamamoto et al., 1997) and reorientation of microtubules (Mayumi and Shibaoka, 1995). Key observations in BR's interaction with other hormones in stem elongation include a synergistic response with auxins and an additive effect with gibberellins (Mandava et al., 1981; Yopp et al., 1981). However, BRs inhibit root elongation (Guan and Roddick, 1988a, 1988b) that may be the consequence of BR-induced ethylene synthesis (Arteca et al., 1983; Arteca and Bachman, 1987).
BR-Related Dwarf Mutants
Because BRs have such strong growth-promoting properties, it is not surprising that mutants involved in BR biosynthesis and signaling display a dramatic phenotype, dark green dwarfs (Figure 3). When grown in the dark, the BR-deficient dwarf mutants show a de-etiolated phenotype and lack of normal skotomorphogenesis, which suggests a potential interaction between BR and light signaling pathways. Such interactions between light and BR responses had been previously reported (Krizek and Mandava, 1983a, 1983b; Arteca and Bachman, 1987), and the availability of mutants now allows their more detailed analysis. Other phenotypes exhibited by BR-deficient dwarfs that correlate well with preceding physiological experiments include delayed senescence, lack of xylem formation, and in some cases, sterility problems. Certain BR-deficient mutants were identified in screens for sensitivity to other hormones, for example, abscisic acid (Ephritikhine et al., 1999), which led to the observation that BRs enhance seed germination (Leubner-Metzger, 2001; Steber and McCourt, 2001). BR signaling mutants also show altered responses to ABA (Li et al., 2001b) and auxins (Koka et al., 2000), although their primary phenotype is BR related. In addition to genetic approaches yielding BR-deficient and -insensitive dwarfs, a chemical approach has been developed that uses specific inhibitors of BR biosynthesis. Brassinazole is such an inhibitor and, when applied at micromolar concentrations, generates phenotypes mimicking those of BR-deficient dwarf mutants (Asami et al., 2000; Nagata et al., 2001) and thus provides a useful tool for discerning the effect of BRs in plant species for which BR mutants have not been identified.
Figure 3.
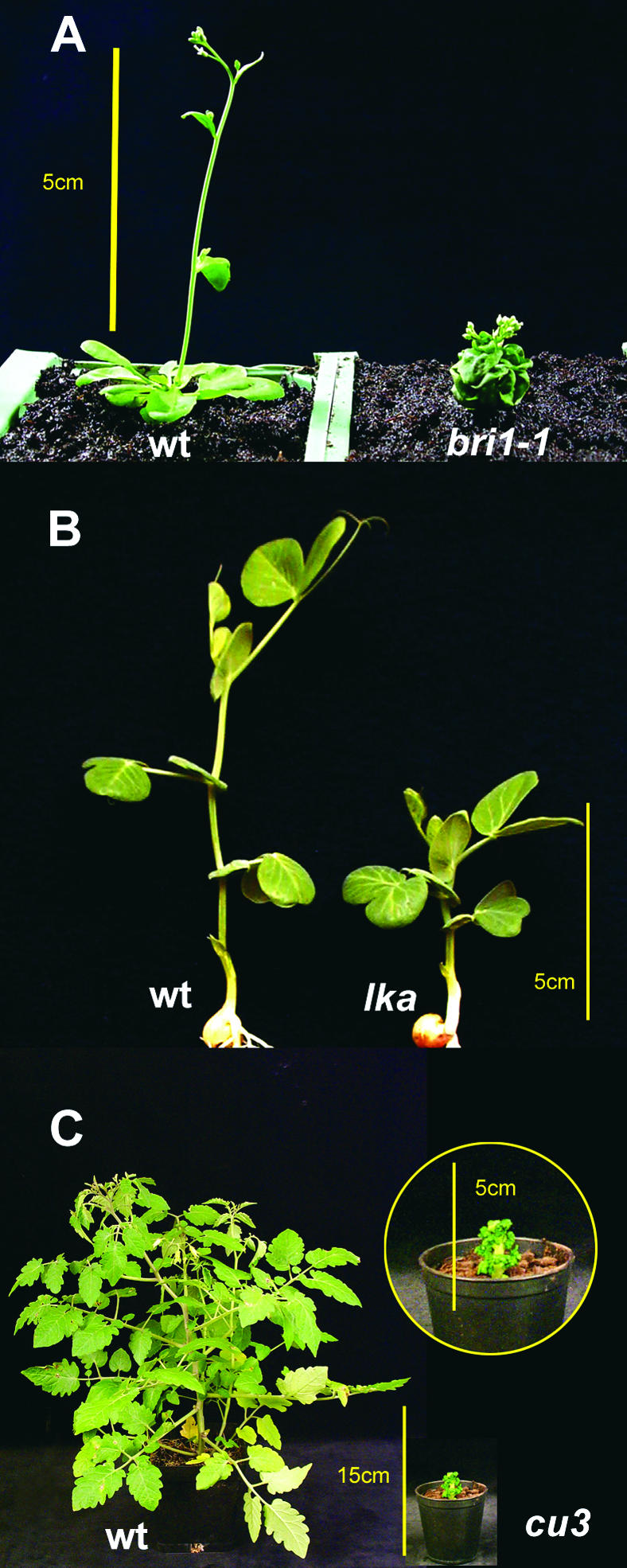
Dwarf Phenotype of bri1 Mutants in Different Plant Species.
(A) Six-week-old wild-type and bri1-1 mutant Arabidopsis plants.
(B) Twelve-day-old wild-type and lka mutant pea plants.
(C) Seven-week-old wild-type and cu-3 mutant tomato plants.
WT, wild type. Bars in (A), (B), and enlarged inset photograph of cu-3 in (C), 5 cm; bar in (C), 15 cm.
BRI1, a Conserved Function in BR Perception
The Arabidopsis bri1 mutant was originally identified by its normal root elongation in the presence of BL (Clouse et al., 1996). Repeated screens for BL-insensitive mutations first resulted only in the identification of numerous mutant alleles of BRI1 (Kauschmann et al., 1996; Li and Chory, 1997; Noguchi et al., 1999; Friedrichsen et al., 2000), indicating that BRI1 represents a major nonredundant component of BR perception. Nonetheless, careful re-examination of dwarf mutants from the original collection used to clone BRI1 identified a gain-of-function mutation in the BRASSINOSTEROID INSENSITIVE-2 (BIN2) locus, which results in reduced BL signaling accompanied by abscisic acid hypersensitivity (Li et al., 2001b). BR-insensitive mutants have also been characterized in rice (Yamamuro et al., 2000), pea (lka; Nomura et al., 1997, 1999) and tomato (cu-3 [curl-3]; Koka et al., 2000) (see Figure 3). Recent cloning experiments indicate that these mutants are defective in BRI1 homologs (Yamamuro et al., 2000; T. Nomura and T. Yokota, personal communication; G.J. Bishop, unpublished data), highlighting the conservation of BRI1 function in BR perception of higher plants. It is also interesting to note that the tomato cu-3 mutant displays hypersensitivity to the auxin 2,4 dichlorophenoxyacetic acid (2,4D) but a normal response to indole acetic acid (Koka et al., 2000). Thus, this mutant may be helpful in the study of potential cross talk between auxin and BR signaling.
BRI1 Is a Member of the LRR-RLK Family
Receptor-like kinases (RLKs) show considerable structural diversity. Classification of RLKs based on the comparison of their domain structures (Torii, 2000; Torii and Clark, 2000) shows that LRR-RLKs represent one of the largest groups, with over 170 genes in the Arabidopsis genome (Arabidopsis Genome Initiative, 2000). LRR domains are involved in protein–protein interactions and consist of repeating units of ∼24 amino acids rich in leucine (Kobe and Deisenhofer, 1994, 1995). Each repeat is composed of an α-helix and β-sheet hairpin with the β-sheet forming the surface for protein–protein interaction. LRR-RLKs are involved in a variety of plant processes, including pathogen resistance (Xa21; Song et al., 1995), flagellin sensing (FLS2; Gomez-Gomez and Boller, 2000), meristem proliferation (CLAVATA1; Clark et al., 1997), abscission (HAESA-RLK5; Jinn et al., 2000), and regulation of organ size (ERECTA; Torii et al., 1996).
BRI1 is a typical plasma membrane–associated LRR-RLK, which carries an N-terminal signal peptide and an extracellular domain of 25 imperfect leucine-rich repeats (LRRs; Li and Chory, 1997). The presence of a 70-amino-acid loop-out “island” found between repeats 21 and 22 of BRI1 is characteristic of a specific family of RLKs. Such “islands” are observed in the Cf-9 (Jones et al., 1994), CLAVATA2 (Jeong et al., 1999), and TOLL LRR receptors (Hashimoto et al., 1988), which lack a cytosolic kinase domain (Figure 4A). At either end of the LRR region in BRI1, pairs of cysteine residues are found that, together with a putative leucine-zipper motif at the N terminus, may facilitate the dimerization of BRI1.
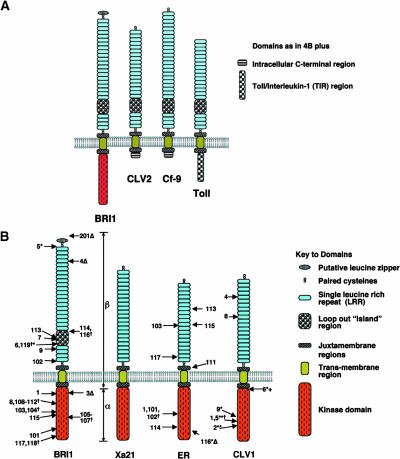
Figure 4.
Schematic Structure of BRI1 and Similar LRR-RLKs and LRR receptor–like Proteins.
(A) Similarities in LRR-domain topologies highlighting a similar location of loop-out island region (for details see main text). Source of primary sequence information: BRI1, BL signaling (Li and Chory, 1997); CLV2, LRR-R involved in control of plant meristem size and differentiation (Jeong et al., 1999); Cf-9, involved in plant pathogen signaling (Jones et al., 1994); and Toll, involved in Drosophila embryo development and immunity pathways (Hashimoto et al., 1988). Note: domains are not to scale and the N-terminal signal peptides are not shown.
(B) Domain topology of selected LRR-RLKs highlighting the location of sequenced mutations. BRI1, location of brassinolide insensitive 1 (Li and Chory, 1997) and additional alleles (Noguchi et al., 1999; Friedrichsen et al., 2000; Bouquin et al., 2001); Xa21, (Song et al., 1995); ER, erecta-1 (Torii et al., 1996) and additional alleles (Lease et al., 2001); CLV1, clavata-1 alleles (Clark et al., 1997). Missense mutations indicated by arrows are shown on the left, whereas nonsense, deletion (Δ) and insertion (+) mutations are indicated on the right. (†), independent mutations at the same position. All alleles are strong (null alleles) unless indicated. (*), weak alleles; (**), intermediate alleles. α and β indicate the approximate regions used in domain swaps between BRI1 and Xa21 (He et al., 2000). Note: domains are not to scale, and the N-terminal signal peptide is not shown. Also note: allele bri-201 is a deletion mutant in the N-terminal region that is not shown.
The relative functional importance of different LRR-RLK domains can be inferred from the location of mutations identified in the different sequenced alleles. Analysis of the erecta alleles indicates that mutations are located in both kinase and LRR regions (Lease et al., 2001; Figure 4B). Similarly, mutations in the CLV-1 (CLAVATA1) gene have been found in all regions (Clark et al., 1997; Figure 4B). By contrast, mutations in the bri1 alleles are prevalent in the kinase and 70-amino-acid island regions (Li and Chory, 1997; Noguchi et al., 1999; Friedrichsen et al., 2000; Bouquin et al., 2001; Figure 4B). The lack of missense mutations in the LRR region of BRI1 suggests that conservation of LRR sequences is not as necessary as in the ERECTA and CLV1 LRRs, indicating the importance of the island region in BRI1. Nonetheless, sequence comparison of the Arabidopsis and rice BRI1 homologs reveals a much higher conservation of kinase than LRR domains, including divergence in the 70-amino-acid island (Yamamuro et al., 2000).
Evolutionary Links between RLKs
BRI1 has been shown to function as a serine/threonine kinase lacking tyrosine kinase activity (Friedrichsen et al., 2000; Oh et al., 2000), similar to other members of the plant LRR-RLK family, including ERECTA (Lease et al., 2001) and CLV1 (Williams et al., 1997). By contrast, with the exception of the transforming growth factor β receptors, most animal RLKs display tyrosine kinase activity, which suggests a potential split in the evolution of RLKs prior to the separation of animals and plants. The analysis of the Arabidopsis genome sequence indicates that all plant RLKs share a common ancestry and form a monophyletic group with Pelle, a cytoplasmic serine/threonine kinase from Drosophila (Shiu and Bleecker, 2001). Based on homology between the kinase domains, the animal receptor tyrosine kinases and the Pelle RLK clade share a common ancestry, suggesting that diversification arose from an early duplication event. Overall, these studies indicate that plant RLKs evolved by sequential recruitment and fusion of various domains to an ancestral kinase and further expansion of certain classes through duplication events.
Modular Activities of RLK Domains
One of the key characteristics of animal steroid nuclear receptors is the discrete modular nature of the functional domains mediating steroid and DNA binding (Beato et al., 1995). In comparison, a modular domain organization in the BRI1 protein was revealed by an intriguing and informative experiment aimed at determining whether the extracellular LRR region is essential for BL sensing. The extracellular BRI1 LRR domain plus the transmembrane domain and juxtamembrane region of 65 amino acids were fused to the kinase domain of rice RLK Xa-21 and expressed in rice cells (He et al., 2000; Figure 4B). Addition of BL to the rice cultures expressing the chimeric BRI1-Xa21 receptor elicited downstream activation of disease resistance responses, including hydrogen peroxide production, defense gene expression, and cell death. The observed cellular responses were thus similar to those obtained by stimulation of the rice culture with Xa-21–specific elicitor. The specificity of BL induction was demonstrated by the absence of activation of pathogenic signaling when using mutant proteins that either had a mutation in the 70-amino-acid island of the BRI1 LRR domain or carried a mutant Xa21 kinase domain (He et al., 2000). These results indicated that the 70-amino-acid island in the extracellular domain of BRI1 is essential either for direct binding of steroid hormone or for proper folding and interaction of the LRR domains of dimerized BRI1 with some accessory factor(s) that may be required for BL perception. The latter possibility is more likely, considering a relatively lower homology of LRR and 70-amino-acid-island sequences between rice OsBRI1 and Arabidopsis BRI1 kinases. Hence, utilization of chimeric RLKs may provide a general means of identifying ligands of as-yet uncharacterized LRR-RLKs, as well as enabling the dissection of downstream signaling responses. The identification of the ligands for BRI1 and other LRRs is definitely a high priority for future research.
BRI1 Is Located in the Plasma Membrane
Using a C-terminal BRI1 translational fusion to the green fluorescent protein (GFP), BRI1 was found to be localized to the plasma membrane, suggesting that it may in fact function as a receptor for an extracellular ligand (Friedrichsen et al., 2000). Plants overexpressing BRI1-GFP display elongated petioles similar to those of plants that overproduce the DWARF4-encoded CYP90B C-22 steroid hydroxylase (see BR biosynthesis pathway in Figure 2), and show a reduced inhibition of growth in the presence of the BR biosynthesis inhibitor brassinazole (Wang et al., 2001). These phenotypes suggest that the BRI1 protein may be limiting, although the BRI1 gene is abundantly expressed in all tissues (Li and Chory, 1997; Friedrichsen et al., 2000).
In analogy to binding studies of radiolabeled steroids with animal nuclear steroid receptors, tritiated BL with high specific activity was used as a ligand in plasma membrane binding assays (Wang et al., 2001). The binding constant for BL in membrane samples of wild-type plants was 10.8 ± 3.2 nM (Wang et al., 2001). Unlike the nonspecific competitor ecdysone, castasterone, the immediate precursor of BL (CS, Figure 1), reduced BL binding by a factor of ∼5-fold, which is consistent with the previous suggestion (Yokota, 1997; Bishop et al., 1999) that CS is a bioactive compound that triggers steroid signaling through BRI1. Immunoblotting of protein extracts prepared from BRI1-GFP–expressing plants with antibodies recognizing either the N-terminal region of BRI1 or GFP revealed a shift in size suggesting autophosphorylation of BRI1 in the presence of BL. In fact, no change in the electrophoretic mobility of BRI1 was observed when the extracts were incubated with protein phosphatases or were isolated from BL-treated bri1 mutants containing mutations in the BRI1 kinase domain. Most importantly, BL binding activity could be co-immunoprecipitated with the BRI1 protein, indicating that BRI is a criticial component of BR signaling. Overall, these experiments confirmed that binding of BL, either directly or through an accessory factor (e.g., a steroid binding protein), to the LRR domain results in autophosphorylation of BRI1 (Wang et al., 2001).
BRS1, an Upstream Component in BL Signaling
To gain further insights into BL signaling, a gain-of-function suppressor screen was performed with a weak bri1 allele (bri1-5; Li et al., 2001a). Plants carrying the bri1-5 allele with a missense mutation affecting the first cysteine pair of BRI1 are fertile (Noguchi et al., 1999), which enabled their transformation with an activation-tagging T-DNA vector. Screening of 2500 tagged lines led to the identification of a single dominant bri1 suppressor dominant (brs1-1D) mutation that resulted in overexpression of a serine carboxypeptidase–like protein (Li et al., 2001a). The bri1 suppression phenotype was reproduced by overexpression of the BRS cDNA using the promoter of cauliflower mosaic virus 35S RNA. However, the brs1-1D mutations suppressed only those bri1 alleles that carried a mutation in the extracellular LRR domain and not in the intracellular kinase domain (Li et al., 2001a). Furthermore, brs1-1D did not suppress the dwarf phenotype of BR biosynthesis mutants, indicating that steroid hormone synthesis is required for alleviation of the bri1 phenotype by overexpression of BRS1. Specificity to the BL signaling pathway was observed through the inability of BRS to suppress the clavata1 and erecta mutant phenotypes.
To explain these observations, Li et al. (2001a) suggested that BRS1 might act on a protein that is required for BL perception. For example, BRS1 might convert from inactive to active form one or more putative sterol binding proteins (SBPs) that are represented by several genes in the Arabidopsis genome (Arabidopsis Genome Initiative, 2000). Alternatively, BRS1 may use BRI1 as potential substrate to generate an activated BL receptor. This latter possibility is suggested by the observation of a potentially processed BRI1 product (Wang et al., 2001). This scenario implies that other proteases in Arabidopsis might perform a similar function, because the knockout mutation of the BRS1 locus does not lead to a dwarf BL-insensitive phenotype (Li et al., 2001a). Nonetheless, BRS1 does not appear to process the chimeric BRI1-Xa21 receptor (He et al., 2000), and further investigation is therefore necessary to define its role in BR perception (Li et al., 2001a).
BRI1 and Regulation of BR Biosynthesis
In recent years many mutants have been recovered that are defective in BR biosynthesis, and the corresponding genes were cloned (Bishop and Yokota, 2001). BL, the end product of BR biosynthesis pathways, is synthesized by sequential hydroxylation reactions (Figure 2) catalyzed by cytochrome P450 enzymes (P450s). In analogy to animal steroid hydroxylases, the transcription of the CPD gene, encoding the C-23 steroid hydroxylase CYP90A1 (Szekeres et al., 1996), was found to be inhibited by BL and numerous bioactive BR biosynthesis intermediates (Mathur et al., 1998). Steroid hormone–mediated downregulation of CPD is inhibited by the protein biosynthesis inhibitor cycloheximide, indicating a need for de novo protein synthesis in genomic effects of BR signaling (Mathur et al., 1998). This model is also supported by the observations that both CPD and DWF4 show derepressed expression in the bri1-5 mutant that is impaired in BR perception (Choe et al., 2001), and that BR intermediates accumulate in bri1 mutants of Arabidopsis and other plant species (Noguchi et al., 1999; Nomura et al., 1999; Yamamuro et al., 2000; T. Nomura, T. Yokota, and G.J. Bishop, unpublished data). BRI1 thus appears to play a key role in activation of a signaling pathway controlling the repression of P450 genes involved in BR biosynthesis.
A surprising finding indicated that BL biosynthesis is probably also controlled by direct interaction of P450s with signaling proteins. Kang et al. (2001) found that the CYP92A6, a C-2 BR hydroxylase encoded by the DDWF (DARK-INDUCED dwf-LIKE-1) (Figure 2) gene, interacts in the yeast two-hybrid system with the PRA2 light-repressible/dark-inducible small G protein. PRA2 is proposed to act as a connection between the light-signaling and BR-biosynthesis pathways by stimulating DDWF activity in the endoplasmic reticulum, leading to greater BL production and etiolation in the dark. As the level of (6-deoxo)castasterone, the product of the DDWF enzyme, is increased in light-grown bri1 mutants, it will be intriguing to see how BL signaling through BRI1 controls C-2 hydroxylation, in addition to potentially downregulating the DDWF gene. Nevertheless, lack of observable mutant phenotype in pra2 antisense lines (Kang et al., 2001) suggests that post-translational regulation of DDWF may be confined to early development.
BR Signaling and Regulation of Gene Expression
In addition to genes in BR biosynthesis, BL appears to negatively regulate the transcription of a large battery of genes that consequently show derepressed expression in the BR-biosynthesis mutants, such as cpd (Szekeres et al., 1996). However, only a few BR-induced genes have been identified so far, despite considerable efforts. Initially, one BR-induced gene, BRU1, was isolated from soybean and found to encode a xyloglucan endotransferase (XET) (Zurek and Clouse, 1994). Subsequently, similar BL-induced XETs have been characterized in Arabidopsis (Xu et al., 1996; Klahre et al., 1998), rice (Uozu et al., 2000), and tomato (Koka et al., 2000), demonstrating that the induction of XETs correlates with cell wall loosening during BL-induced growth responses. Among other potentially BR-induced genes, the expression of CDC2b cyclin-dependent kinase is upregulated by BRs in the absence of light, but unaffected in the light (Yoshizumi et al., 1999). The fact that the cyclin gene CycD3 is also inducible by BRs (Hu et al., 2000) suggests that BR signaling may also play a role in the regulation of the cell cycle, although further research is required to support this intriguing possibility. In addition, the BR induction of an extracellular invertase (Goetz et al., 2000) provides a potential link to previous findings that suggests alteration of source-sink relationships in response to BRs.
More recently, another BL-induced gene was found to encode an Arabidopsis homolog of TRIP, an interacting partner of transforming growth factor β type II receptor kinase, that also represents a subunit of the eukaryotic translation initiation factor eIF3 in animals (Jiang and Clouse, 2001). Transgenic plants expressing antisense TRIP-1 RNA display developmental defects that resemble the phenotype of BR-deficient and -insensitive mutants. This suggests that the TRIP-1 WD-domain protein might establish a link between BL signaling and developmental pathways controlled by homologs of the eukaryotic translation initiation factor eIF3 in plants. On the basis of recent advances in transcription profiling, it is expected that many more BL-induced genes soon will be identified, facilitating a comparative analysis of promoter sequences and subsequent identification of putative BR response elements, for example, by linker scanning mutagenesis and promoter deletions.
bin2, a New Component of BR Signaling
As mentioned above, a second brassinosteroid insensitive 2 (bin2) mutant has been identified recently in Arabidopsis (Li et al., 2001b). Screening 150,000 ethyl methane sulphonate-mutagenized seeds led to the isolation of only two bin2 mutant alleles, bin2-1 and bin2-2. The bin2 mutants are semidominant dwarfs that exhibit insensitivity to BL, but hypersensitivity to ABA. Homozygous bin2 plants show a bri1-like extreme dwarf phenotype and lack transcriptional downregulation of the CPD gene in the presence of BL, whereas the heterozygous bin2 plants are semidwarfs and display ∼50% reduction in CPD transcript levels in comparison to the wild type (Li et al., 2001b). It will be interesting to determine whether the bin2 mutation also leads to accumulation of BR intermediates, as seen in the bri1 mutants.
BIN2 has been cloned by a map-based approach and found to encode an Arabidopsis ortholog of human glycogen synthase kinase β and Drosophila SHAGGY protein kinase (Li and Nam, 2002). Concurrent with this, the ultracurvata1 (ucu1) Arabidopsis mutant displaying aberrant leaf morphology was also found to carry a mutation in the same kinase gene (Perez-Perez et al., 2002). In both bin2 and ucu1, missense mutations resulted in a semidominant phenotype. Therefore, Li et al. (2001b) proposed that bin2 represents either a gain-of-function neomorphic mutation or a hypermorphic mutation defining a negative regulator function in BR signaling. The hypermorphic nature of the bin2 mutation is indicated by the activity of the mutant bin2 kinase, which is increased by ∼33% of that of its wild-type form (Li and Nam, 2002). An allelic series of BIN2 overexpression lines showed that plants with higher expression of BIN2 had more-pronounced dwarfism. Overexpression of BIN2 in a weak bri1 mutant background also generated a more-pronounced dwarf phenotype (Li and Nam, 2002). In contrast, co-suppression of BIN2 transcription resulted in partial suppression of the weak bri1 phenotype (Li and Nam, 2002). Taken together, these data indicate that BIN2 is a negative regulator of BL signaling. Li and Nam (2002) speculate that BRI1 may interact with and phosphorylate BIN2 to inactivate it. Nonetheless, BIN2 appears to lack close homology to the consensus sequence surrounding the preferred serines that BRI1 is likely to phosphorylate (Oh et al., 2000), and BRI1 is thus unlikely to directly interact with the BIN2 kinase. Li and Nam (2002) suggest that BIN2 is constitutively active in the absence of BRs and phosphorylates positive BR signaling proteins to inactivate them.
BR Signaling and Mutant Phenotypes
Considering the high degree of genetic redundancy detected in the Arabidopsis genome, it is remarkable that mutations affecting BR signaling and biosynthesis were so easily obtained and have such a dramatic phenotype. However, it is worth acknowledging that phenotypes of loss-of-function mutations might represent only that proportion of gene products that is not affected by functional redundancy. If this assumption is correct, other functions that are controlled by steroid hormone signaling might be revealed, for example, by overexpression studies.
Overexpression of BL biosynthesis genes and BRI1-GFP generate similar phenotypes, suggesting that the production of steroid hormones and their BRI1 receptor may represent limiting factors of BL signaling. This would indicate a dose-dependent regulation, which contradicts the fact that all known mutations affecting BR perception and biosynthesis are recessive, except for bin2, which shows a dose-dependent dominant phenotype. Thus, the phenotypes resulting from overexpression of these functions could either reflect an artificial excess (i.e., over the threshold of normal responses that are not physiologically relevant for normal growth and development) or suggest the existence of yet-uncharacterized regulatory functions. The phenotypes of transgenic lines that overproduce BR intermediates may reflect, for example, a consequence of the induction of programmed cell death during xylem formation (xylogenesis), which is characterized by a dramatic increase in BR concentration stimulating the production of tracheary elements (Yamamoto et al., 1997). A great reduction of xylem differentiation observed in the BR-deficient cpd mutant suggests a potential inhibition of the cell death pathway required for normal xylogenesis (Szekeres et al., 1996). Would deregulation of induced cell death explain the phenotype of overexpressor lines that show etiolated petioles and leaves? The fact that BR-deficient mutants exhibit reduced senescence and xylogenesis supports this assumption, providing an intriguing model for further exploration of the developmental role of BR signaling.
Drosophila Toll and BL Signaling
The Drosophila Toll gene and Toll signaling pathway represent one of the well-characterized regulatory pathways involving a LRR protein. The Toll gene encodes an LRR receptor (Figure 4A) that controls both embryo development (Morisato and Anderson, 1995) and innate immunity to bacteria and fungi (Imler and Hoffmann, 2001). Activation of Toll leads to nuclear translocation of a Rel-related transcription factor, Dorsal. The levels of Toll and Dorsal are uniform throughout the syncytial embryo. However, the generation of the Toll ligand is asymmetric, and thus Toll signaling provides a ventral-to-dorsal gradient of Dorsal nuclear localization leading to differential regulation of downstream genes defining embryonic polarity.
The Toll gene product shows structural similarity to BRI1, including an interrupted feature of the LRR domain (see Figure 4A). In addition, the Pelle kinase, which is closely related to BRI1 evolutionarily, interacts with Toll. In both embryo development and immunity response, the presumed ligand for the Toll LRR is Spätzle that is secreted and proteolytically processed by several proteases. This indicates that Toll is not the bona fide receptor in either establishing the dorsoventral axis or in the immune response, but acts as an important downstream signaling component in both pathways.
The immunity response mediated by Toll is activated by a circulating peptidoglycan-recognition protein (PGRP-SA) that binds peptidoglycans of Gram-positive bacteria (Michel et al., 2001). How this leads to Toll activation through the proteolysis of Spätzle is still a mystery, but the available data suggest that PGRP-SA is the receptor for this signaling pathway. In the ventralization pathway, Toll activation leads to phosphorylation and localized recruitment of the adaptor protein Tube and activation of the Pelle kinase that controls the degradation of the IκB-like inhibitor Cactus and nuclear translocation of the Rel transcription factor Dorsal. In immunity responses, Dorsal-related immune factor is analogously activated and translocated to the nucleus to promote the expression of antimicrobial peptides, such as Drosomycin.
It is tantalizing to speculate that BRI1 might perform a signaling function similar to that of Toll, namely not as a BR receptor but as a critical signaling component that is downstream of the primary BL perception event. It is conceivable that BL is recognized by an SBP “receptor,” which is either itself activated by proteolysis or acts as the recognition protein that activates proteolysis of a potential BRI1 ligand. The carboxypeptidase BRS could therefore play a role in either processing the SBP into a BL binding form (see Figure 5) or act in a yet-uncharacterized proteolytic cascade. It is also exciting to speculate that similarities to the Toll pathway in the activation of a transcription factor may be found downstream of BRI1 in BR signaling.
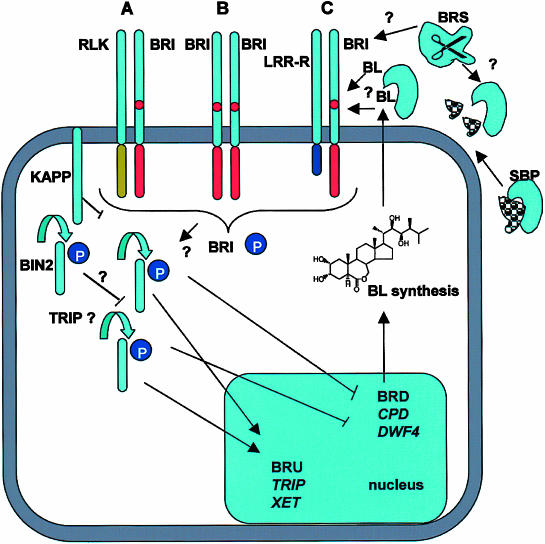
Figure 5.
Schematic Models Indicate a Central Role for BRI1 in BL Signaling.
Three scenarios for formation of a possible BRI1 receptor complex are shown:
(A) BRI1 interacts with another LRR-RLK.
(B) BRI1 forms a homodimer.
(C) BRI1 interacts with an LRR-receptor-like protein.
In each case, the amino-acid island of BRI1 is implicated in BL sensing, either by directly binding BL or by binding an SBP. KAPP, a kinase-associated protein phosphatase, is likely to be associated with the receptor complex to inactivate it. BL activation of BRI1 leads to autophosphorylation and subsequent phosphorylation of other substrates, e.g., TRIP1 (transforming growth factor β receptor interacting protein homolog). BL-mediated activation of BRI1 leads to suppression of genes controlling BR biosynthesis, such as CPD and DWF4 (BRD, BR-downregulated genes), but promotes the expression of BR-upregulated genes (BRU). A putative role for a sterol binding protein and potential processing of this protein by BRS facilitating BL binding and BRI1 activation is indicated. Potentially, BRS may also modify BRI. (See main text for further details).
CLAVATA and BR Signaling Pathways
As with the Drosophila Toll signaling pathway, there are some similarities between elements of the BR and CLAVATA signaling pathways in Arabidopsis. Mutations in three CLAVATA loci, CLV1, CLV2, and CLV3, result in similar phenotypes of enlarged shoot meristems. CLV1 is a LRR-RLK (Clark et al., 1997; Figure 4B), and CLV2 is a LRR receptor–like protein (Jeong et al., 1999; Figure 4A), whereas CLV3 is a small secreted peptide (Fletcher et al., 1999). Both genetic (Fletcher et al., 1999) and biochemical (Trotochaud et al., 1999) studies indicate that CLV1 forms a heterodimeric receptor complex with CLV2. Further biochemical (Trotochaud et al., 2000) and controlled expression analysis of CLV3 (Brand et al., 2000) has shown that CLV3 interacts as a multimeric ligand with a CLV1 complex, and that CLV1 kinase activity is required for CLV3 binding (Trotochaud et al., 2000). The CLV1 complex exists in two distinct states, a smaller disulphide-linked presumed heterodimer of CLV1 and CLV2, and a larger complex that includes the heterodimer, a kinase-associated protein phosphatase (KAPP) and a rho GTPase–related protein (Trotochaud et al., 1999). It is postulated that the ρ-related protein may provide the signaling mechanism to downstream targets, whereas KAPP acts as a negative regulator (Williams et al., 1997; Stone et al., 1998). The fact that the CLV1/CLV2 heterodimer is linked via disulfide bonds suggests a role for cysteines at either end of the LRRs (Figure 4). CLV signaling directly or indirectly downregulates the expression of Wuschel (WUS), which negatively regulates the differentiation of meristematic stem cells. WUS is required for expression of CLV3. Thus, downregulation of WUS leads to a decrease of CLV3 levels, which in turn reduces the CLV1/CLV2 activation that leads to upregulation of WUS, ensuring a delicate balance in the control of meristem differentiation (Clark, 2001).
Relevant to BR signaling, KAPP has been observed to interact with a number of LRR-RLKs, including BRI1 (Schumacher and Chory, 2000). It will be intriguing, therefore, to learn whether overexpression of KAPP or a KAPP homolog leads to a bri1-like phenotype and upregulation of genes in BR biosynthesis. Initial results from such a study indicated that only a clavata phenotype was observed when overexpressing KAPP (Williams et al., 1997). Furthermore, it remains to be determined whether BRI1 is capable of forming heterodimeric complexes with other LRR receptor–like proteins or LRR-RLKs in analogy with CLV1/CLV2. In respect to global control of steroid metabolism, it will be also interesting to learn whether BRI1 can interact with SBPs or some steroid-conjugated signaling factors showing analogy to the SONIC/HEDGEHOG proteins that play crucial roles in the regulation of cell differentiation pathways in mammals (Porter et al., 1996; Lewis et al., 2001).
BL SIGNALING MECHANISM, CONCLUSIONS AND PROSPECTS
According to the speculative models discussed above, BR perception may imply the formation of BRI1-associated receptor complexes (Figure 5). The BRI1 complex might interact either directly with BL or with a secreted and processed sterol binding/carrier protein that could undergo BRS-mediated proteolytic processing. Nonetheless, it is currently unknown whether BRI1 is capable of forming a heterodimeric receptor complex with other LRR-RLKs or LRR receptor–like proteins. A recent meeting report on the existence of a BRI1-interacting kinase, which is a LRR-RLK and forms a RLK heterodimer with BRI1, supports model A in Figure 5 (Eckartdt et al., 2001). BL is known to promote autophosphorylation and activation of BRI1, triggering downstream responses, which may be modulated by BRI1 binding an inhibitory KAPP-like protein phosphatase. A phosphorylation cascade, including, for example, MAPK(K/K) kinases, may ensure signal amplification leading to the activation of transcription factors that control activation (e.g., XET) or repression (e.g., BR biosynthesis) of BR-responsive genes. Alternatively, BRI1 may directly phosphorylate transcription factors and other signaling components (e.g., TRIP) modulating diverse cellular responses. BIN2 appears to play a role as negative regulator acting downstream of BRI1 in this signaling pathway.
Further genetic screens for mutations that suppress or enhance the above-described mutations and/or confer resistance to the BR biosynthesis inhibitor brassinozole are expected to unravel new elements of BR signaling. In addition, biochemical and protein interaction studies will help in the identification of signaling partners of BRI1 and BIN2 kinases. Also expected to be derived from further studies of BL signaling is a major input toward gaining detailed insight into the regulation of nongenomic steroid signaling, which may facilitate the understanding of similar pathways in other organisms. In particular, it is hoped that the study of plant steroid hormone signaling pathways, based on the use of comparative genome analysis, will uncover the conservation of some signaling functions that also play a pivotal role in nongenomic control of sterol homeostasis and steroid hormone metabolism in mammals. As a practical goal, further exploration of BR signaling and its interaction with other hormonal and developmental pathways is predicted to provide new strategies for the regulation of growth and improvement in yield of important crops.
Acknowledgments
We thank T. Montoya, J. Castle, T. Nomura, and T. Yokota for unpublished communications and for helping in the preparation of figures. We also thank S. Clouse and the Rick Seed Stock Centre (U.C. Davis) for providing seed of Arabidopsis bri1-1 and tomato cu-3 mutants. We thank the editors and reviewers for their helpful comments on the text. GB's lab has obtained financial support from Biotechnology and Biological Sciences Research Council, The Royal Society, and The British Council. This review also contributes to the Human Frontier Science Program Organization Project Grant RG00162/2000M.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001461.
References
- Adam, G., and Marquardt, V. (1986). Brassinosteroids. Phytochemistry 25 1787–1799. [Google Scholar]
- Altmann, T. (1999). Molecular physiology of brassinosteroids revealed by the analysis of mutants. Planta 208 1–11. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Arteca, R.N., and Bachman, J.M. (1987). Light inhibition of brassinosteroid-induced ethylene production. J. Plant Physiol. 129 13–18. [Google Scholar]
- Arteca, R.N., Tsai, D.S., Schlagnhaufer, C., and Mandava, N.B. (1983). The effect of brassinosteroid on auxin-induced ethylene production by etiolated mung bean segments. Physiol. Plant. 59 539–544. [Google Scholar]
- Asami, T., Min, Y.K., Nagata, N., Yamagishi, K., Takatsuto, S., Fujioka, S., Murofushi, N., Yamaguchi, I., and Yoshida, S. (2000). Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato, M., Herrlich, P., and Schutz, G. (1995). Steroid hormone receptors—Many actors in search of a plot. Cell 83 851–857. [DOI] [PubMed] [Google Scholar]
- Bishop, G.J., Harrison, K., and Jones, J.D.G. (1996). The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell 8 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., Nomura, T., Yokota, T., Harrison, K., Noguchi, T., Fujioka, S., Takatsuto, S., Jones, J.D.G., and Kamiya, Y. (1999). The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc. Natl. Acad. Sci. USA 96 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., and Yokota, T. (2001). Plants steroid hormones, brassinosteroids: Current highlights of molecular aspects on their synthesis/metabolism, transport, perception and response. Plant Cell Physiol. 42 114–120. [DOI] [PubMed] [Google Scholar]
- Bouquin, T., Meier, C., Foster, R., Nielsen, M.E., and Mundy, J. (2001). Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 127 450–458. [PMC free article] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289 617–619. [DOI] [PubMed] [Google Scholar]
- Braun, P., and Wild, A. (1984). The influence of brassinosteroid on growth and parameters of photosynthesis of wheat and mustard plants. J. Plant Physiol. 116 189–196. [DOI] [PubMed] [Google Scholar]
- Cerana, R., Bonetti, A., Marre, M.T., Romani, G., Lado, P., and Marre, E. (1983). Effects of a brassinosteroid on growth and electrogenic proton extrusion in Azuki bean epicotyls. Physiol. Plant. 59 23–27. [Google Scholar]
- Choe, S.W., Dilkes, B.P., Fujioka, S., Takatsuto, S., Sakurai, A., and Feldmann, K.A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22 alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S.W., Fujioka, S., Noguchi, T., Takatsuto, S., Yoshida, S., and Feldmann, K.A. (2001). Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26 573–582. [DOI] [PubMed] [Google Scholar]
- Clark, S.E. (2001). Cell signaling at the shoot meristem. Nature Rev. Mol. Cell Biol. 2 276–284. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 575–585. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D. (1997). Molecular genetic analysis of brassinosteroid action. Physiol. Plant. 100 702–709. [Google Scholar]
- Clouse, S.D., Langford, M., and McMorris, T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse, S.D., and Sasse, J.M. (1998). Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 427–451. [DOI] [PubMed] [Google Scholar]
- Crozier, A., Yokota, T., Bishop, G.J., and Kamiya, Y. (2000). Biosynthesis of hormones and elicitor molecules. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 850–929.
- Dhaubhadel, S., Chaudhary, S., Dobinson, K.F., and Krishna, P. (1999). Treatment with 24-epibrassinolide, a brassinosteroid, increases the basic thermotolerance of Brassica napus and tomato seedlings. Plant Mol. Biol. 40 333–342. [DOI] [PubMed] [Google Scholar]
- Eckartdt, N.A., Takashi, A., Benning, C., Cubas, P., Goodrich, J., Jacobsen, S.E., Masson, P., Nambara, E., Simon, R., Somerville, S., and Wasteneys, G. (2001). Arabidopsis Research 2001. Meeting Report. Plant Cell 13 1973–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephritikhine, G., Fellner, M., Vannini, C., Lapous, D., and BarbierBrygoo, H. (1999). The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. 18 303–314. [DOI] [PubMed] [Google Scholar]
- Falkenstein, E., Tillmann, H.C., Christ, M., Feuring, M., and Wehling, M. (2000). Multiple actions of steroid hormones—A focus on rapid, nongenomic effects. Pharmacol. Rev. 52 513–555. [PubMed] [Google Scholar]
- Fletcher, L.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 1911–1914. [DOI] [PubMed] [Google Scholar]
- Friedrichsen, D., and Chory, J. (2001). Steroid signaling in plants: from the cell surface to the nucleus. Bioessays 23 1028–1036. [DOI] [PubMed] [Google Scholar]
- Friedrichsen, D.M., Joazeiro, C.A.P., Li, J.M., Hunter, T., and Chory, J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuns, J.M.C. (1978). Steroid hormones and plant growth and development. Phytochemistry 17 1–14. [Google Scholar]
- Goetz, M., Godt, D.E., and Roitsch, T. (2000). Tissue-specific induction of the mRNA for an extracellular invertase isoenzyme of tomato by brassinosteroids suggests a role for steroid hormones in assimilate partitioning. Plant J. 22 515–522. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5 1003–1011. [DOI] [PubMed] [Google Scholar]
- Grove, M.D., Spencer, G.F., Rohwedder, W.K., Mandava, N., Worley, J.F., Jr., J.D.W., Steffens, G.L., Flippen-Anderson, J.L., and Carter Cook, J. (1979). Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281 216–217. [Google Scholar]
- Guan, M., and Roddick, J.G. (1988. a). Epibrassinolide-inhibition of development of excised, adventitious and intact roots of tomato (Lycopersicon esculentum): Comparison with the effects of steroidal estrogens. Physiol. Plant. 74 720–726. [Google Scholar]
- Guan, M., and Roddick, J.G. (1988. b). Comparison of the effects of epibrassinolide and steroidal estrogens on adventitious root growth and early shoot development in mung bean cuttings. Physiol. Plant. 73 426–431. [Google Scholar]
- Hashimoto, C., Hudson, K.L., and Anderson, K.V. (1988). The Toll gene of Drosophila, required for dorsal-ventral embryonic polarity, appears to encode a transmembrane protein. Cell 52 269–279. [DOI] [PubMed] [Google Scholar]
- He, Z.H., Wang, Z.Y., Li, J.M., Zhu, Q., Lamb, C., Ronald, P., and Chory, J. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288 2360–2363. [DOI] [PubMed] [Google Scholar]
- Hu, Y.X., Bao, F., and Li, J.Y. (2000). Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 24 693–701. [DOI] [PubMed] [Google Scholar]
- Imler, J.L., and Hoffmann, J.A. (2001). Toll receptors in innate immunity. Trends Cell Biol. 11 304–311. [DOI] [PubMed] [Google Scholar]
- Iwasaki, T., and Shibaoka, H. (1991). Brassinosteroids act as regulators of tracheary-element differentiation in isolated Zinnia mesophyll cells. Plant Cell Physiol. 32 1007–1014. [Google Scholar]
- Jeong, S., Trotochaud, A.E., and Clark, S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11 1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J.R., and Clouse, S.D. (2001). Expression of a plant gene with sequence similarity to animal TGF-beta receptor interacting protein is regulated by brassinosteroids and required for normal plant development. Plant J. 26 35–45. [DOI] [PubMed] [Google Scholar]
- Jinn, T.L., Stone, J.M., and Walker, J.C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14 108–117. [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., Thomas, C.M., Hammondkosack, K.E., Balintkurti, P.J., and Jones, J.D.G. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266 789–793. [DOI] [PubMed] [Google Scholar]
- Jones, J.L., and Roddick, J.G. (1988). Steroidal oestrogens and androgens in relation to reproductive development in higher plants. J. Plant Physiol. 133 510–518. [Google Scholar]
- Kang, J.G., Yun, J., Kim, D.H., Chung, K.S., Fujioka, S., Kim, J.I., Dae, H.W., Yoshida, S., Takatsuto, S., Song, P.S., and Park, C.M. (2001). Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell 105 625–636. [DOI] [PubMed] [Google Scholar]
- Kauschmann, A., Jessop, A., Koncz, C., Szekeres, M., Willmitzer, L., and Altmann, T. (1996). Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 9 701–713. [Google Scholar]
- Klahre, U., Noguchi, T., Fujioka, S., Takatsuto, S., Yokota, T., Nomura, T., Yoshida, S., and Chua, N.H. (1998). The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10 1677–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1994). The leucine-rich repeat—A versatile binding motif. Trends Biochem. Sci. 19 415–421. [DOI] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1995). A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374 183–186. [DOI] [PubMed] [Google Scholar]
- Koka, C.V., Cerny, R.E., Gardner, R.G., Noguchi, T., Fujioka, S., Takatsuto, S., Yoshida, S., and Clouse, S.D. (2000). A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 122 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek, D.T., and Mandava, N.B. (1983. a). Influence of spectral quality on the growth-response of intact bean plants to brassinosteroid, a growth promoting steroidal lactone. 1. Stem elongation and morphogenesis. Physiol. Plant. 57 317–323. [Google Scholar]
- Krizek, D.T., and Mandava, N.B. (1983. b). Influence of spectral quality on the growth-response of intact bean plants to brassinosteroid, a growth promoting steroidal lactone. 2. Chlorophyll content and partitioning of assimilate. Physiol. Plant. 57 324–329. [Google Scholar]
- Lease, K.A., Lau, N.Y., Schuster, R.A., Torii, K.U., and Walker, J.C. (2001). Receptor serine/threonine protein kinases in signaling: analysis of the ERECTA receptor-like kinase of Arabidopsis thaliana. New Phytol. 151 133–143. [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger, G. (2001). Brassinosteroids and gibberellins promote tobacco seed germination by distinct pathways. Planta 213 758–763. [DOI] [PubMed] [Google Scholar]
- Lewis, P.M., Dunn, M.P., McMahon, J.A., Logan, M., Martin, J.F., St.-Jacques, B., and McMahon, A.P. (2001). Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell 105 599–612. [DOI] [PubMed] [Google Scholar]
- Li, J., Lease, K.A., Tax, F.E., and Walker, J.C. (2001. a). BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98 5916–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.M., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J.M., and Chory, J. (1999). Brassinosteroid actions in plants. J. Exp. Bot. 50 275–282. [Google Scholar]
- Li, J.M., Nagpal, P., Vitart, V., McMorris, T.C., and Chory, J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272 398–401. [DOI] [PubMed] [Google Scholar]
- Li, J.M., and Nam, K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li, J.M., Nam, K.H., Vafeados, D., and Chory, J. (2001. b). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandava, B.N. (1988). Plant growth promoting brassinosteroids. Annu. Rev. Plant Physiol. Plant Mol. Biol 39 23–52. [Google Scholar]
- Mandava, B.N., Sasse, J.M., and Yopp, J.H. (1981). Brassinolide, a growth promoting steroidal lactone.2. Activity in selected gibberellin and cytokinin bioassays. Physiol. Plant. 53 453–461. [Google Scholar]
- Marumo, S., Hattori, H., Nanoyama, Y., and Munakata, K. (1968). The presence of novel plant growth regulators in leaves of Distylium racemosum Sieb et Zucc. Agric. Biol. Chem. 32 528–529. [Google Scholar]
- Mathur, J., et al. (1998). Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 14 593–602. [DOI] [PubMed] [Google Scholar]
- Mayumi, K., and Shibaoka, H. (1995). A possible double role for brassinolide in the reorientation of cortical microtubules in the epidermal cells of Azuki bean epicotyls. Plant Cell Physiol. 36 173–181. [Google Scholar]
- McCarty, D.R., and Chory, J. (2000). Conservation and innovation in plant signaling pathways. Cell 103 201–209. [DOI] [PubMed] [Google Scholar]
- Michel, T., Reichhart, J.M., Hoffmann, J.A., and Royet, J. (2001). Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414 756–759. [DOI] [PubMed] [Google Scholar]
- Mitchell, J.W., Mandava, N., Worley, J.F., Plimmer, J.R., and Smith, M.V. (1970). Brassins: A new family of plant hormones from rape pollen. Nature 225 1065–1066. [DOI] [PubMed] [Google Scholar]
- Morisato, D., and Anderson, K.V. (1995). Signaling pathways that establish the dorsal-ventral pattern of the Drosophila embryo. Annu. Rev. Genet. 29 371–399. [DOI] [PubMed] [Google Scholar]
- Mussig, C., and Altmann, T. (2001). Brassinosteroid signaling in plants. Trends Endocrinol. Metab. 12 398–402. [DOI] [PubMed] [Google Scholar]
- Nagata, N., Asami, T., and Yoshida, S. (2001). Brassinazole, an inhibitor of brassinosteroid biosynthesis, inhibits development of secondary xylem in cress plants (Lepidium sativum). Plant Cell Physiol. 42 1006–1011. [DOI] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Tax, F.E., Yoshida, S., and Feldmann, K.A. (2000). Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 124 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H., Feldmann, K.A., and Tax, F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T., Kitasaka, Y., Takatsuto, S., Reid, J.B., Fukami, M., and Yokota, T. (1999). Brassinosteroid/sterol synthesis and plant growth as affected by lka and lkb mutations of pea. Plant Physiol. 119 1517–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T., Nakayama, M., Reid, J.B., Takeuchi, Y., and Yokota, T. (1997). Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Physiol. 113 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, M.H., Ray, W.K., Huber, S.C., Asara, J.M., Gage, D.A., and Clouse, S.D. (2000). Recombinant brassinosteroid insensitive 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol. 124 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez, J.M., Ponce, M.R., and Micol, J.L. (2002). The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev. Biol. 242 161–173. [DOI] [PubMed] [Google Scholar]
- Porter, J.A., Young, K.E., and Beachy, P.A. (1996). Cholesterol modification of hedgehog signaling proteins in animal development. Science 274 255–259. [DOI] [PubMed] [Google Scholar]
- Romani, G., Marre, M.T., Bonetti, A., Cerana, R., Lado, P., and Marre, E. (1983). Effects of a brassinosteroid on growth and electrogenic proton extrusion in maize root segments. Physiol. Plant. 59 528–532. [Google Scholar]
- Sakurai, A., and Fujioka, S. (1993). The current status of physiology and biochemistry of brassinosteroids—A review. Plant Growth Regul. 13 147–159. [Google Scholar]
- Sakurai, A., Yokota, T., and Clouse, S.D., eds (1999). Brassinosteroids Steroidal Plant Hormones. (Tokyo: Springer).
- Sasse, J.M. (1997). Recent progress in brassinosteroid research. Physiol. Plant. 100 696–701. [Google Scholar]
- Schumacher, K., and Chory, J. (2000). Brassinosteroid signal transduction: Still casting the actors. Curr. Opin. Plant Biol. 3 79–84. [DOI] [PubMed] [Google Scholar]
- Shimada, Y., Fujioka, S., Miyauchi, N., Kushiro, M., Takatsuto, S., Nomura, T., Yokota, T., Kamiya, Y., Bishop, G.J., and Yoshida, S. (2001). Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 126 770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.Y., Wang, G.L., Chen, L.L., Kim, H.S., Pi, L.Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.X., Zhu, L.H., Fauquet, C., and Ronald, P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene Xa21. Science 270 1804–1806. [DOI] [PubMed] [Google Scholar]
- Steber, C.M., and McCourt, P. (2001). A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 125 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, J.M., Trotochaud, A.E., Walker, J.C., and Clark, S.E. (1998). Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol. 117 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres, M., and Koncz, C. (1998). Biochemical and genetic analysis of brassinosteroid metabolism and function in Arabidopsis. Plant Physiol. Biochem. 36 145–155. [Google Scholar]
- Szekeres, M., Németh, K., Koncz-Kálmán, Z., Mathur, J., Kauschmann, A., Altmann, T., Rédei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85 171–182. [DOI] [PubMed] [Google Scholar]
- Torii, K.U. (2000). Receptor kinase activation and signal transduction in plants: An emerging picture. Curr. Opin. Plant Biol. 3 361–367. [DOI] [PubMed] [Google Scholar]
- Torii, K.U., and Clark, S.E. (2000). Receptor-like kinases in plant development. Adv. Bot. Res. Incorporating Adv. Plant Pathol. 32 225–267. [Google Scholar]
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., and Komeda, Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud, A.E., Hao, T., Wu, G., Yang, Z.B., and Clark, S.E. (1999). The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud, A.E., Jeong, S., and Clark, S.E. (2000). CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science 289 613–617. [DOI] [PubMed] [Google Scholar]
- Uozu, S., Tanaka-Ueguchi, M., Kitano, H., Hattori, K., and Matsuoka, M. (2000). Characterization of XET-related genes of rice. Plant Physiol. 122 853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410 380–383. [DOI] [PubMed] [Google Scholar]
- Wehling, M. (1997). Specific nongenomic actions of steroid hormones. Annu. Rev. Physiol. 59 365–393. [DOI] [PubMed] [Google Scholar]
- Wilen, R.W., Sacco, M., Gusta, L.V., and Krishna, P. (1995). Effects of 24-epibrassinolide on freezing and thermotolerance of bromegrass (Bromus inermis) cell cultures. Physiol. Plant. 95 195–202. [Google Scholar]
- Williams, R.W., Wilson, J.M., and Meyerowitz, E.M. (1997). A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc. Natl. Acad. Sci. USA 94 10467–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., Campbell, P., Vargheese, A.K., and Braam, J. (1996). The Arabidopsis XET-related gene family: Environmental and hormonal regulation of expression. Plant J. 9 879–889. [DOI] [PubMed] [Google Scholar]
- Yamamoto, R., Demura, T., and Fukuda, H. (1997). Brassinosteroids induce entry into the final stage of tracheary element differentiation in cultured Zinnia cells. Plant Cell Physiol. 38 980–983. [DOI] [PubMed] [Google Scholar]
- Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2000). Loss of function of a rice brassinosteroid insensitive 1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12 1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota, T. (1997). The structure, biosynthesis and function of brassinosteroids. Trends Plant Sci. 2 137–143. [Google Scholar]
- Yopp, J.H., Mandava, N.B., and Sasse, J.M. (1981). Brassinolide, a growth promoting steroidal lactone.1. Activity in selected auxin bioassays. Physiol. Plant. 53 445–452. [Google Scholar]
- Yoshizumi, T., Nagata, N., Shimada, H., and Matsui, M. (1999). An Arabidopsis cell cycle-dependent kinase-related gene, CDC2b, plays a role in regulating seedling growth in darkness. Plant Cell 11 1883–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek, D.M., and Clouse, S.D. (1994). Molecular cloning and characterization of a brassinosteroid regulated gene from elongating soybean (Glycine max L.) epicotyls. Plant Physiol. 104 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]