INTRODUCTION
Despite its simple two-carbon structure, the olefin ethylene is a potent modulator of plant growth and development (Ecker, 1995). The plant hormone ethylene is involved in many aspects of the plant life cycle, including seed germination, root hair development, root nodulation, flower senescence, abscission, and fruit ripening (reviewed in Johnson and Ecker, 1998). The production of ethylene is tightly regulated by internal signals during development and in response to environmental stimuli from biotic (e.g., pathogen attack) and abiotic stresses, such as wounding, hypoxia, ozone, chilling, or freezing. To understand the roles of ethylene in plant functions, it is important to know how this gaseous hormone is synthesized, how its production is regulated, and how the signal is transduced. Morphological changes in dark-grown (etiolated) seedlings treated with ethylene or its metabolic precursor, 1-aminocyclopropane-1-carboxylic acid (ACC), have been termed the triple response. The exaggerated curvature of the apical hook, radial swelling of the hypocotyl, and shortening of the hypocotyl and root are the unmistakable hallmarks of this ethylene response. Over the past decade, the triple response phenotype has been used to screen for mutants that are defective in ethylene responses (Bleecker et al., 1988; Guzman and Ecker, 1990). Etiolated Arabidopsis seedlings with minor or no phenotypic response upon ethylene application are termed ethylene-insensitive (ein) or ethylene-resistant (etr) mutants. Mutants have also been identified that display a constitutive triple response in the absence of ethylene (Kieber et al., 1993; Roman and Ecker, 1995). This class can be divided into subgroups based on whether or not the constitutive triple response can be suppressed by inhibitors of ethylene perception and biosynthesis, such as silver thiosulfate and aminoethoxyvinyl glycine (AVG). Mutants that are unaffected by these inhibitors are termed constitutive triple-response (ctr) mutants, whereas mutants whose phenotype reverts to normal morphology are termed ethylene-overproducer (eto) mutants, which are defective in the regulation of hormone biosynthesis. The genetic hierarchy among ethylene biosynthesis and signaling pathway components in Arabidopsis has been established by epistasis analysis using these mutants (Solano and Ecker, 1998; Stepanova and Ecker, 2000).
The intent of this review is not to cover all aspects of ethylene biology but to focus on recent findings. In particular, we examine interaction of ethylene and two other plant growth regulators, jasmonic acid (JA) and salicyclic acid (SA), and their roles in mediating responses to biotic and abiotic stresses. We begin by summarizing what is currently known about the mechanism and regulation of ethylene biosynthesis and by providing an update of our current understanding of the ethylene signaling pathway.
BIOSYNTHESIS OF ETHYLENE: MECHANISMS AND REGULATION
The biochemistry of ethylene biosynthesis has been a subject of intensive study in plant hormone physiology (reviewed in Kende, 1993). Major breakthroughs in the ethylene synthesis pathway were the establishment of S-adenosylmethionine (S-AdoMet) and ACC as the precursors of ethylene (Figure 1) (reviewed in Yang and Hoffman, 1984). On the basis of this knowledge, the enzymes that catalyze these reactions were characterized and purified using biochemistry approaches. The first successes in molecular cloning of the ACC synthase (ACS) (Sato and Theologis, 1989) and ACC oxidase (ACO) (Hamilton et al., 1991; Spanu et al., 1991) genes led to the demonstration that these enzymes belong to a multigene family and are regulated by a complex network of developmental and environmental signals responding to both internal and external stimuli (reviewed in Johnson and Ecker, 1998).
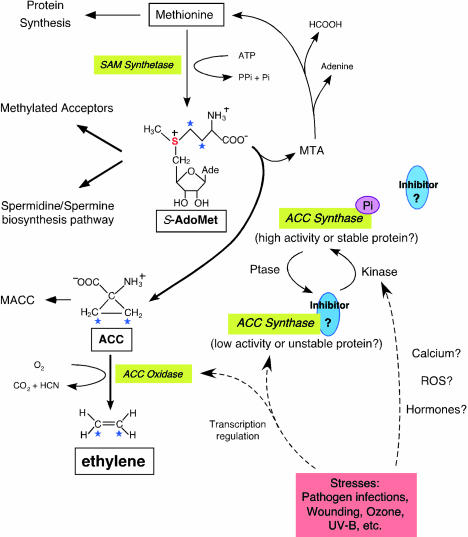
Figure 1.
Biosynthetic Pathway and Regulation of Ethylene.
The formation of S-AdoMet is catalyzed by SAM synthetase from the methionine at the expense of one molecule of ATP per molecule of S-AdoMet synthesized. S-AdoMet is the methyl group donor for many cellular molecules (Methylated Acceptors), including nucleic acids, proteins, and lipids. In addition, S-AdoMet is the precursor of the polyamine synthesis pathway (Spermidine/Spermine biosynthesis pathway). ACC is the immediate precursor of ethylene. The rate-limiting step of ethylene synthesis is the conversion of S-AdoMet to ACC by ACC synthase under most conditions. MTA is the by-product generated along with ACC production by ACC synthase. Recycling of MTA back to methionine conserves the methylthio group and is able to maintain a constant concentration of cellular methionine even when ethylene is rapidly synthesized. Malonylation of ACC to malonyl-ACC (MACC) deprives the ACC pool and reduces the ethylene production. ACC oxidase catalyses the final step of ethylene synthesis using ACC as substrate and generates carbon dioxide and cyanide. Transcriptional regulation of both ACC synthase and ACC oxidase is indicated by dashed arrows. Reversible phosphorylation of ACC synthase is hypothesized and may be induced by unknown phosphatases (Ptase) and kinases, the latter presumably activated by stresses. Both native and phosphorylated form (ACC synthase-Pi) of ACC synthase are functional, although the native ACC synthase may be less stable or active in vivo. A hypothetical inhibitor is associated with ACC synthase at the carboxyl end and may be dissociated from the enzyme if it is modified by phosphorylation at the vicinity.
Mechanistic View of Ethylene Synthesis
S-AdoMet is the precursor for ethylene biosynthesis (reviewed in Yang and Hoffman, 1984; Kende, 1993). In addition to being an essential building block of protein synthesis, nearly 80% of cellular methionine is converted to S-AdoMet by S-AdoMet synthetase (SAM synthetase, EC 2.5.1.6) at the expense of ATP utilization (Ravanel et al., 1998). S-AdoMet is the major methyl donor in plants and is used as a substrate for many biochemical pathways, including polyamines and ethylene biosynthesis (Ravanel et al., 1998). In addition, S-AdoMet is involved in methylation reactions that modify lipids, proteins, and nucleic acids. On the basis of the Yang cycle, the first committed step of ethylene biosynthesis is the conversion of S-AdoMet to ACC by ACC synthase (S-adenosyl-l-methionine methylthioadenosine-lyase, EC4.4.14) (reviewed in Yang and Hoffman, 1984; Kende, 1993). In addition to ACC, ACC synthase (ACS) also produces 5′-methylthioadenosine (MTA) in this reaction, which is then converted to methionine by using a modified methionine cycle (reviewed in Bleecker and Kende, 2000). This salvage pathway preserves the methyl group for another round of ethylene production. Therefore, ethylene can be synthesized continuously without demanding an increasing pool of methionine. At the same time, the sulfur group of the methionine is also conserved. Finally, ACC is oxidized by ACC oxidase to form ethylene, CO2, and cyanide, which is detoxified to β-cyanoalanine by β-cyanoalanine synthase (β-CAS, EC 4.4.1.9) to prevent toxicity of accumulated cyanide during high rates of ethylene synthesis (Figure 1).
The rate-limiting step of ethylene synthesis is the conversion of S-AdoMet to ACC by ACC synthase (reviewed in Kende, 1993). The observations that expression of the ACS genes is highly regulated by a variety of signals and that active ACC synthase is labile and present at low levels suggest that ethylene biosynthesis is tightly controlled. Both positive and negative feedback regulation of ethylene biosynthesis have been reported in different plant species (reviewed in Kende, 1993; Nakatsuka et al., 1998; Barry et al., 2000). Different isoforms of ACS appear to be the principle targets. For example, in tomato, Le-ACS2 and Le-ACS4 are positively regulated, and Le-ACS6 is negatively regulated by ethylene synthesized during fruit ripening (Nakatsuka et al., 1998). Most studies addressing ACS regulation have focused on ACS gene expression in response to various endogenous cues and environmental stimuli. The only feature found in common is that the ACS enzymes are spatially and temporally regulated and are controlled by various internal and external signals.
ACC Synthase: A Multigene Family in Plants
Early attempts to purify plant ACC synthases were hampered by their low abundance and labile nature (reviewed in Kende, 1993). ACC synthase is encoded by a multigene family whose structure resembles the subgroup-I family of pyridoxal 5′-phosphate (PLP)–dependent aminotransferases (Mehta et al., 1993). PLP is an essential co-factor for ACS activity that is pre-bound in the active site of unliganded enzymes. The crystal structure of ACS from apple has been determined and reveals that the enzyme forms a homodimer (Capitani et al., 1999). Not only are 11 invariant residues conserved between aminotransferases and ACC synthases, but also the tertiary arrangement of the conserved residues near the active site can be superimposed on that of aminotransferase enzymes. Most of the conserved residues among the isoforms of the ACS family are located on the dimer surface and are clustered near the active site of the enzyme. The substrate specificity of these enzymes may arise from the relative distances between conserved residues in the active site, which is supported by the different spatial position of Y85 of apple ACC synthase and Y70 of aminotransferase in the active site. The ACS dimer is aligned by a local twofold axis, with the most variable carboxylic region protruding away from the center. There are two distinct domains of each monomer, a large and a small domain that are defined by the tertiary structure. The large domain spans the central region of the enzyme and contains the strictly conserved secondary structures found among families of PLP-dependent enzymes. The small domain, which shows greater variability in structure between ACC synthases and aminotransferases, consists of the most amino and carboxyl regions of the protein. The active site with a bound PLP cofactor is predicted to lie between the cleft formed by the two domains. Interestingly, some of the residues in the active site that interact with PLP (Y85 in apple ACC synthase; Y70 and R292 in aminotransferase) are provided from the neighboring subunit, supporting the hypothesis that active ACC synthase functions as a dimer (Tarun and Theologis, 1998). Random and site-directed mutations introduced in LeACS2 have been used to study the relationship between ACS structure and function (Tarun et al., 1998). Mutations introduced at these conserved residues render the enzyme inactive (White et al., 1994; Tarun et al., 1998). Coexpression of two mutated ACC synthases with compensatory mutations partially rescues the activity in a bacterial system (Tarun et al., 1998).
Regulation of ACC Synthase: Gene Expression
Because of their central role in ethylene biosynthesis, the regulation of ACC synthases has been an intensively studied. Since the cloning of ACS from zucchini (Cucurbita) (Sato and Theologis, 1989), many ACS genes have been identified and cloned from different plant species, including tomato, winter squash, apple, carnation, mung bean, and Arabidopsis (reviewed in Johnson and Ecker, 1998; Ge et al., 2000). An emerging paradigm is that different isoforms of ACC synthase are differentially regulated (Oetiker et al., 1997; Peck and Kende, 1998; Barry et al., 2000). Although studies of ACS genes from other species, particularly those of the tomato ACS family, have been informative, the ACS genes identified from Arabidopsis can be used to exemplify this point. In Arabidopsis, seven ACS genes have been characterized (Liang et al., 1992; Van der Straeten et al., 1992; Arteca and Arteca, 1999; Samach et al., 2000). ACS2 is induced by cycloheximide, wounding, and 2 h of ethylene treatment. The ethylene-induced expression gradually decreases with prolonged ethylene exposure, suggesting negative feedback regulation of ACS2 (Van der Straeten et al., 1992; Liang et al., 1996). ACS4 is induced in seedlings by cycloheximide, indoleacetic acid, and wounding (Liang et al., 1992; Abel et al., 1995). ACS5 is induced by lithium chloride and a low concentration of cytokinin only in etiolated seedlings (Liang et al., 1996; Vogel et al., 1998b). ACS6 can be induced specifically by cyanide treatment, exposure to ozone in light-grown leaves, and mechanical strain by touching; it can also be induced by cycloheximide, indoleacetic acid, and ethylene (Vahala et al., 1998; Arteca and Arteca, 1999; Overmyer et al., 2000; Smith and Arteca, 2000). ACS10 was identified as one of the early targets of CONSTANS, which promotes flowering of Arabidopsis in response to light (Samach et al., 2000). Because cycloheximide treatment induces most of the ACS isoforms, the implication is that ACS transcripts are short-lived and negatively regulated by some unknown labile repressor(s) (Liang et al., 1992). An alternate explanation is that cycloheximide treatment results in retention of mRNA on the ribosomes; therefore, the steady state of ACS mRNA is relatively increased.
In Arabidopsis, ACS1 and ACS3 do not show ACS activity in either bacterial or yeast expression systems (Liang et al., 1995). ACS1 is missing a highly conserved tripeptide, TNP (Thr-Asn-Pro), which is located near the active site and may be essential for ACS activity (Liang et al., 1995). Deletion of this tripeptide from ACS2 inactivates it. On the other hand, ACS3 is believed to be a pseudogene resulting from a partial duplication of ACS1. It is intriguing to speculate about the role of ACS1 in planta, given that it is expressed and induced by several signals that activate other ACS genes. It is possible that ACS1 may function as a regulator of ACS activity through dimerization with other ACS enzymes.
Regulation of ACS: Post-Translational Regulation
Pharmacological evidence supports the possibility of post-translational regulation of ACS activity (Chappell et al., 1984; Felix et al., 1991; Spanu et al., 1994). Addition of fungal elicitors to tomato suspension cell culture induces a rapid increase in ACS activity, which is rapidly inactivated by addition of the protein kinase inhibitors K-252a or staurosporine (Spanu et al., 1994). Moreover, treatment with the protein phosphatase inhibitor calyculin A not only stimulates ACS activity of the tomato cell culture without elicitors but also greatly enhances the effect of elicitor treatment. These effects on ACS activity by both types of inhibitors require de novo protein synthesis, because cycloheximide blocks the effects. Because the kinetics are very fast, occurring usually within minutes, the phosphorylation event appears to be a primary effect of increased ACS activity. These results suggest that de novo protein synthesis is required to sustain or stabilize the increased ACS activity upon elicitor induction and that phosphorylation may be involved. However, phosphorylation may not play a part in the catalytic activity of ACS per se. It is known that ACS is unstable in vivo and present at low abundance, so it is conceivable that phosphorylation of ACS may increase its stability to sustain the elevated activity. Consistent with this view, purified ACS from bacterial expression system remains active (Li and Mattoo, 1994; White et al., 1994). Alternatively, an unknown repressor of ACS activity may be inactivated by phosphorylation.
Two recent studies further highlight the possibility of post-translational regulation of ACS (Vogel et al., 1998b; Tatsuki and Mori, 2001). Three ethylene overproduction mutants, eto1, eto2 and eto3, have been identified from Arabidopsis (Guzman and Ecker, 1990; Kieber et al., 1993). eto1 is a recessive mutation, whereas eto2 and eto3 are dominant. The constitutive triple-response phenotype of the eto mutants can be suppressed by silver thiosulfate or AVG, suggesting that these mutants are affected in the regulation of ethylene biosynthesis. It has been shown that low doses of cytokinin (0.5 to 10 μM) stimulate ethylene production in etiolated seedlings of Arabidopsis and induce morphological changes resembling the triple response by ethylene treatment (Cary et al., 1995). On the basis of these observations, five complementation groups, termed cytokinin-insensitive mutants (cin1 to cin5), have been found by screening for mutations causing insensitivity to cytokinin treatment (Vogel et al., 1998a). Recessive mutations in one of these complementation groups, cin5, were mapped very close to eto2 and ACS5 (Liang et al., 1992), and CIN5 was subsequently found to correspond to ACS5 (Vogel et al., 1998b). Sequencing of ACS5 from the eto2 mutant background has revealed that a single base insertion leads to an alteration of the last 12 residues of ACS5. The dominant mutation of eto2-1 changes the last 12 residues from RVSYTDRVPDER to PGFMDRSCT. The other mutant alleles of eto2/cin5 are recessive, loss-of-function mutations. Although the steady state eto2-1 mRNA shows little change, ethylene production in eto2-1 etiolated seedlings is nearly 20-fold that of the wild type, suggesting that the increased activity is not the result of gene expression (Vogel et al., 1998b). In addition, cytokinin-mediated ethylene production does not correlate with an induction of ACS5 mRNA, with more ethylene produced than the level of induced ACS5 expression by cytokinin. The implication of these results is that cytokinin regulates ethylene synthesis by modifying ACS post-translationally. The altered residues in eto2-1 may be the targets for such modification. The identification of such mutations in ACS5 further supports the notion that ACS isoforms can play essential roles in regulating tissue-specific and hormone-inducible ethylene biosynthesis.
Interestingly, tomato LeACS2 protein has been found to be phosphorylated in response to wounding (Tatsuki and Mori, 2001). The phosphorylation site of LeACS2 was identified as Ser-460 by phosphoamino acid analysis, site-directed mutagenesis of recombinant LeACS2, and use of synthetic ACS peptides. The phosphorylation of LeACS2 in vivo has also been demonstrated by immunoprecipitation with anti-LeACS2 antibody from the wounding fruit extract and in vitro by using a wounding-induced kinase activity to phosphorylate recombinant protein. In addition, the kinase activity for LeACS2 is calcium-dependent. Alignment of ACS from different species reveals that a tripeptide (R/K)(L/V)(S) is conserved within the divergent carboxyl region of most ACS enzymes. The phosphorylated Ser-460 residue (RLS460) in LeACS2 is located at an equivalent position as the Ser-461 (RVS461) residue in ACS5 of Arabidopsis, which is mutated in the eto2-1 mutant. Finally, phosphatase treatment of the native wound-induced LeACS2 does not show a significant change in catalytic activity. These results strongly suggest that the C-terminal peptide in the ACS5/ETO2 contains a target for negative regulation that may be modified post-translationally by phosphorylation at Ser-461. Because phosphorylation of LeACS2 does not affect its activity, the decreased turnover of ACS may result in an increase in ethylene production. Considering that eto2-1 mutant has significantly increased ACS activity, it is possible that the de-repression effect seen in eto2-1 is the consequence by the phosphorylation of Ser-461 of ACS5 or by the deletion of this putative negative regulatory domain.
On the basis of these findings, a hypothetical model for the regulation of ETO2/ACS5 is postulated (Figure 1). In this model, the presence of a protein inhibitor is suggested that physically interacts with ACS5 via the C terminus of ACS5 and negatively regulates its activity by any of several possible mechanisms. This interaction may block the active site of ACS5 or change its conformation, resulting in inaccessibility of ACS5 to its substrate. Alternatively, binding of the inhibitor may cause dissociation of the ACS5 dimer, or it may destabilize ACS5 and lead to protein degradation. Finally, the inhibitor protein may simply act as a scaffold that provides a platform for other negative regulators for ACS5. Phosphorylation of ACS5 may play a role in dissociation of the proposed inhibitor. On the basis of the presence of the highly conserved serine residue in ACS enzymes from many different species, negative regulation may represent a general mechanism to modulate very rapid (seconds) changes in ACS activity without a requirement for ACS gene transcription. The recessive nature of the Arabidopsis ethylene overproduction mutant eto1 (Guzman and Ecker, 1990) suggests that ETO1 is a strong candidate for the proposed regulator of ACS5/ETO2 activity.
For the past decade, studies of ethylene biosynthesis have focused on the isolation and characterization of ACS and ACO genes from a wide variety of plant species, with an eye to understanding the signals that govern the differential expression of these genes/enzymes. Important questions remain about the regulation of these genes. For example, how do hormones, such as cytokinin, and a host of biotic and abiotic stresses, such as wounding or pathogen attack, evoke the rapid ethylene evolution from plant cells? With the completion of the Arabidopsis genome sequence, a number of new ACS and ACO homologs have been identified that provide new fodder for future studies. This raises the question of why plants require multigene families for ACS and ACO and whether these proteins have equivalent biochemical activities and regulation. Such regulatory molecules will likely include transcription factors that activate or modulate ACS gene expression as well as enzymes such as kinases and phosphatases that may post-translationally modulate ACS activity. Some of the next major challenges in ethylene biosynthesis research are to understand the biochemical mechanisms of ACS/ACO regulation and to identify the components involved in this regulation.
ETHYLENE SIGNALING
After its synthesis, ethylene is perceived and its signal transduced through transduction machinery to trigger specific biological responses. On the basis of the highly reproducible triple response in dark-grown Arabidopsis seedlings, a number of mutants impaired in their response to ethylene have been identified. Cloning and characterization of the genes disrupted in these mutants are leading to a complete picture of the ethylene signal transduction pathway (see Figure 2).
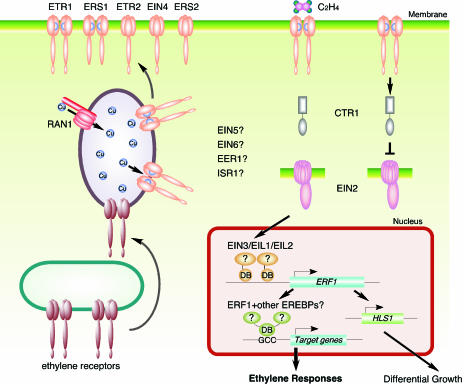
Figure 2.
Model of the Ethylene Signal Transduction Pathway.
There are five ethylene receptors in Arabidopsis, ETR1, ETR2, EIN4, ERS1, and ERS2. ETR1 and ERS1 contain three transmembrane domains and a conserved histidine kinase domain, and have been shown to function as homodimers. ETR2, EIN4, and ERS2 have four membrane-spanning regions and a degenerate histidine kinase domain. Only ETR1, ETR2, and EIN4 have receiver domains at their C termini. Ethylene binding occurs at the N-terminal transmembrane domain of the receptors, and a copper co-factor is required for the binding. RAN1, a copper transporter, is involved in delivery of copper to the ethylene receptor. In the absence of an ethylene signal, ethylene receptors activate a Raf-like kinase, CTR1, and CTR1 in turn negatively regulates the downstream ethylene response pathway, possibly through a MAP-kinase cascade. Binding of ethylene inactivates the receptors, resulting in deactivation of CTR1, which allows EIN2 to function as a positive regulator of the ethylene pathway. EIN2 contains the N-terminal hydrophobic domain similar to the Nramp metal transporter proteins and the novel hydrophilic C terminus. EIN2 positively signals downstream to the EIN3 family of transcription factors located in the nucleus. EIN3 binds to the promoter of ERF1 gene and activates its transcription in an ethylene-dependent manner. Transcription factors ERF1 and other EREBPs can interact with the GCC box in the promoter of target genes and activate downstream ethylene responses.
Perception
Ethylene is perceived by a family of five membrane-localized receptors that are homologous to bacterial two-component histidine kinases involved in sensing environmental changes. The system typically consists of two proteins: a histidine kinase as the sensor that autophosphorylates an internal histidine residue in response to environmental signals, and a response regulator that activates the downstream components upon receiving a phosphate from the histidine residue of the sensor on its aspartate residue (Wurgler-Murphy and Saito, 1997; Pirrung, 1999). Five ethylene receptors exist in Arabiodpsis: ETR1, ETR2, ERS1, ERS2, and EIN4 (Chang et al., 1993; Hua et al., 1995; Hua and Meyerowitz, 1998; Sakai et al., 1998). Among these receptors, only ETR1, ETR2, and EIN4 contain a receiver domain that shows similarity to bacterial response regulators at the C-terminal part of the protein. Since homodimerization of ETR1 and ERS1 has been observed in plants (Schaller et al., 1995; Hall et al., 2000), receptors that do not have receiver domain, ERS1 and ERS2, have been postulated to use the receiver domains of other proteins by forming heterodimers with them (Hua et al., 1998). On the basis of the structural similarities of the sensor domain, regardless of the presence of the receiver domain, the receptor family can be further divided into two subfamilies. The ETR1-like subfamily, consisting of ETR1 and ERS1, features three membrane-spanning regions at the N-terminal region, where ethylene binding occurs (Schaller and Bleecker, 1995; Hall et al., 2000), and a well-conserved histidine kinase domain at the C-terminal part of the protein. The ETR2-like subfamily, which includes ETR2, EIN4, and ERS2, is predicted to have four hydrophobic extensions at the N terminus and a degenerate histidine kinase domain that lacks one or more elements considered necessary for catalytic activity, implying that these receptors may function differently. The fact that members of a family of photoreceptors, the phytochromes, have a histidine kinase domain related to two-component systems but exhibit serine/threonine kinase activity (Fankhauser et al., 1999) supports the notion that the ETR2 class of receptors may function not as histidine kinases but possibly as serine/threonine kinases.
Genetic and biochemical analyses of the ethylene receptors have lent insight into the mechanism of regulation in planta. etr1, etr2, and ein4 were initially identified as dominant ethylene-insensitive plants (Bleecker et al., 1988; Roman et al., 1995; Hua et al., 1998; Sakai et al., 1998). Similar missense mutations introduced into the N-terminal transmembrane domain of ERS1 and ERS2 cause the same ethylene insensitive phenotype, suggesting their role in ethylene perception (Hua et al., 1995). Isolation of the loss-of-function alleles of ETR1, ETR2, EIN4, and ERS2 by screening for intragenic suppressors of the dominant receptor mutants provides genetic evidence of how the ethylene receptors actually work (Hua and Meyerowitz, 1998). The absence of phenotypes in single-receptor mutants suggests that in spite of the structural differences, there is functional redundancy (or compensation of function) among the receptors. The constitutive triple response observed in a quadruple-receptor mutant indicates that the receptors negatively regulate this ethylene response. Consistent with these elegant genetic studies is the observation that the dominant ethylene-insensitive mutant etr1 binds less ethylene (Schaller and Bleecker, 1995; Rodriguez et al., 1999). The synthesis of the results from both genetic and biochemical studies leads one to conclude that ethylene receptors are inactivated by ethylene binding. Interestingly, members of both ETR1 and ETR2 subfamilies have also been identified in other plant species. In tomato, the Never Ripe (NR) gene encodes a receptor similar to the ETR1 class with no receiver domain, whereas LeETR4 is an ETR2 class member with a receiver domain. Reduction in the expression level of LeETR4 leads to enhanced ethylene responses in tomato plants, and overexpression of NR can compensate for the loss of LeETR4 and eliminates the ethylene sensitivity. These results reveal that mechanisms of ethylene perception are likely conserved among flowering plants (Tieman et al., 2000).
Further characterization of ethylene binding to ETR1 has revealed that it occurs in a hydrophobic pocket located at the N terminus of the receptors and requires a transition metal, copper, as a cofactor (Schaller and Bleecker, 1995; Rodriguez et al., 1999). Addition of copper ions was required for the recovery of ETR1 ethylene binding activity in yeast extracts. Subsequently, it was shown that copper copurifies in stoichiometric amounts with the ethylene binding domain extracted from membranes of yeast overexpressing the ETR1 binding domain. Both ethylene binding activity and copurification of copper were eliminated when the etr1-1 mutation, a conversion of Cys-65 to a Tyr, was introduced into the protein. Further evidence for a role of copper in ethylene signaling comes from the characterization of the Arabidopsis RESPONSIVE-TO-ANTAGONIST (RAN1) gene (Hirayama et al., 1999). Two weak mutant alleles, ran1-1 and ran1-2, were identified in a screen for mutants that displayed an ethylene-like triple response in response to treatment with the potent ethylene antagonist transcyclooctene. More importantly, the mutant allele ran1-3/ctr2 or co-suppression of the RAN1 gene led to a constitutive ethylene response phenotype. This is consistent with a loss-of-receptor function (Hirayama et al., 1999; Woeste and Kieber, 2000). This phenotype can be partially rescued by exogenous copper application. Cloning and subsequent functional analysis of RAN1 revealed that it encodes a copper transporter that shares similarity with copper-transporting P-type ATPases such as the yeast Ccc2p and human Menkes/Wilson disease proteins (Hirayama et al., 1999). Taken together, these findings indicate that RAN1 is involved in delivery of copper to the ethylene receptor and that this copper-delivery pathway is required to create functional ethylene receptors in plants.
Signaling
In a screen for Arabidopsis mutants that display the constitutive triple-response phenotype, only one complementation group, ctr1, proved to be unaffected by ethylene synthesis inhibitors or ethylene antagonists. Genetic epistatic analysis has placed CTR1 downstream of the ethylene receptors in the ethylene signaling pathway. The recessive nature and constitutive phenotype of the ctr1 mutant indicate that CTR1 is a negative regulator of downstream signaling events (Kieber et al., 1993). Cloning of the CTR1 gene revealed that it belongs to the Raf family of Ser/Thr protein kinases that initiate mitogen-activated protein (MAP)-kinase signaling cascades in mammals (Kyriakis et al., 1992; Pelech and Sanghera, 1992). The similarity of CTR1 to known MAPKKKs implies that ethylene signaling may operate through a MAP-kinase cascade. Although many genes with homology to MAPKKs and MAPKs have been identified in the Arabidopsis genome sequence, to date none has been associated with ethylene signaling. Thus far, no intermediate components have been identified genetically or biochemically to act between the receptors and the CTR1 kinase. In fact, yeast two-hybrid and in vitro binding have shown that the kinase domain of ETR1 and ERS1 can directly interact with CTR1 (Clark et al., 1998). Because the response regulator domain of ETR1 can also interact with several Arabidopsis histidine-containing phosphotransfer proteins (Urao et al., 2000), the in vivo relevance of these in vitro interactions needs to be confirmed.
Genetic epistasis analysis of ethylene response mutants has shown that EIN2 acts downstream of CTR1 and upstream of EIN3. Null mutations in EIN2 result in the complete loss of ethylene responsiveness throughout plant development, suggesting that EIN2 is an essential positive regulator in the ethylene signaling pathway. EIN2 encodes a novel integral membrane protein (Alonso et al., 1999). The N-terminal hydrophobic domain of EIN2 shows similarity to members of the NRAMP family, which includes metal–ion transporters such as the yeast Smf1p, Drosophila Malvolio, and mammalian DCT1. The C-terminal hydrophilic region has no homology to any known protein, although it does have motifs typically involved in protein–protein interactions. Overexpression of the C-terminal portion of the protein (EIN2 CEND) in an ein2 null background results in constitutive activation of some but not all ethylene responses and restores the ability of the mutant to respond to paraquat and JA but not ethylene. These results suggest that the N-terminal portion of EIN2 is necessary for sensing the ethylene signal from upstream components in the pathway, whereas EIN2 CEND is required for transducing the signal to the downstream components. Interestingly, ein2 mutants have been independently isolated in several different genetic screens designed to identify components of other signaling pathways. For example, ein2 mutants have been found in screens for defects in auxin transport inhibitor resistance (Fujita and Syono, 1996), cytokinin response (Su and Howell, 1992), ABA hypersensitivity (Beaudoin et al., 2000; Ghassemian et al., 2000), and delayed senescence (Oh et al., 1997). In addition, ein2 mutants also show altered sensitivity to several bacterial and fungal pathogens (see discussion below). At least in some cases, such as cytokinin resistance and delayed senescence, the abnormalities observed in ein2 are simply the result of its ethylene insensitivity.
Nuclear Events
Many ethylene responses involve changes in gene expression. The cloning of EIN3 provided direct evidence for nuclear regulation in the early ethylene signal transduction pathway (Chao et al., 1997). EIN3 encodes a novel nuclear-localized protein that belongs to a multigene family in Arabidopsis. Among six members of this family, three of them, EIN3, EIN3-like 1 (EIL1), and EIL2, can rescue the ein3 mutant phenotypes. This indicates that not only EIN3 but also EIL1 and EIL2 are involved in ethylene signal transduction, explaining why null mutations in ein3 cause only partial ethylene insensitivity. Overexpression of EIN3 in an ein2 null mutant background causes constitutive activation of the ethylene response, similar to overexpression of the EIN2 CEND, confirming that EIN3 acts downstream of EIN2. EIN3 gene expression is not induced by ethylene. This result indicates that EIN3 may be regulated by ethylene at the protein level. EIN3-like transcription factors have also been identified in other plant species. The tobacco EIN3-like gene, TEIL, has been cloned. Plants that overexpress the TEIL cDNA exhibit constitutive triple-response phenotypes (Kosugi and Ohashi, 2000). Tomato orthologs of EIN3-like genes, LeEIL1, LeEIL2, and LeEIL3, have also been cloned (Tieman et al., 2001). Each complements the ein3-1 mutation in transgenic Arabidopsis, indicating that all are likely involved in ethylene responses. Antisense tomato plants with reduced expression of a single LeEIL gene did not exhibit significant changes in ethylene response. However, reduced expression of multiple tomato LeEIL genes reduced significantly the sensitivity to ethylene, providing evidence of functional redundancy.
A search for target promoters for the EIN3 family of proteins led to the identification of the primary ethylene response element in the promoter of the ERF1 gene (Solano et al., 1998). In vitro DNA binding studies revealed that homodimers of either EIN3 or EIL1 proteins were able to bind primary ethylene response elements in the promoters of ERF1 (Solano et al., 1998) and other unrelated transcription factors (EDF1) (A.N. Stepanova and J.R. Ecker, unpublished data). ERF1 belongs to a large family of plant-specific transcription factors referred to as ethylene-response-element binding proteins (EREBPs). EREBPs were originally identified on the basis of their ability to bind to the GCC box, a DNA motif associated with ethylene- and pathogen-induced gene expression. EIN3 is both necessary and sufficient to stimulate ERF1 expression. Moreover, overexpression of ERF1 in an ein3 background leads to constitutive activation of a subset of ethylene phenotypes. These results indicate that ERF1 may regulate one branch of the ethylene response pathway downstream of EIN3. Interestingly, although a large number of EREBPs have been found in the Arabidopsis genomes and other plant species (Riechmann and Meyerowitz, 1998), only a very few of them have been shown to be regulated by ethylene (Thara et al., 1999; Yamamoto et al., 1999). SA, JA, salt, drought, and other stress are among the growing number of stimuli known to regulate the expression of these genes (Ohme-Takagi and Shinshi, 1995; Buttner and Singh, 1997; Suzuki et al., 1998; Thara et al., 1999; Fujimoto et al., 2000; Gu et al., 2000; Park et al., 2001). Thus, in keeping with the more general role for the EREBPs, the GCC box is likely to be a more general transcriptional regulatory element that is not specific to the ethylene response. Definitive proof that any of these EREBPs (also called ERFs) other than ERF1 function in the ethylene response awaits further experimental evidence.
New Ethylene Mutants
Several novel ethylene-related mutants have recently been identified. The Arabidopsis mutant enhanced ethylene response (eer) was identified by novel genetic screen using subthreshold levels of ethylene (Larsen and Chang, 2001). The eer1 mutant displays increased ethylene sensitivity in the hypocotyl and stem but reduced sensitivity in root. Like the eto class of ethylene overproducer mutants, the eer1 mutant phenotype is suppressed by treatment with the ethylene biosynthesis inhibitor AVG. Similarly the eer1 phenotype is completely suppressed by the ethylene-insensitive mutations etr1-1 and ein2-1. However, eer1 displays a highly exaggerated (atypical) triple-response phenotype and shows an additive effect when combined with the constitutive ethylene response mutant ctr1-3, suggesting that the eer1 phenotype is not simply the result of ethylene overproduction. eer1 seedlings have significantly elevated levels of basic-chitinase expression, suggesting that eer1 may be highly sensitive to low levels of endogenous ethylene. Interestingly, like ran1 mutant, eer1 shows ethylene-like responses to ethylene receptor antagonists. Although the specific step at which EER1 acts has not been established, these results suggest that EER1 may act in addition to CTR1 to oppose ethylene responses in the hypocotyl and stem. It is possible that EER1 can regulate ethylene receptor function or is involved in an alternate ethylene signaling pathway that bypasses the requirement for functional CTR1. Cloning and characterization of the EER1 should help to elucidate its role in the ethylene response.
There is also new information about an old tomato ethylene-related mutant called epinastic (epi). Dark-grown epi seedlings display a phenotype similar to the triple response in the absence of ethylene (Barry et al., 2001). Double mutant analysis between epi and dominant ethylene-insensitive receptor mutant NR revealed that epi likely acts downstream of ethylene receptor NR. Interestingly, unlike ctr1, epi does not demonstrate a global constitutive ethylene response, suggesting a role for EPI either in the regulation of a subset of ethylene responses regulating the cell expansion or in an independent pathway required for normal growth. In addition, epi does not show linkage to either of the two previously reported tomato CTR1 homologs, LeCTR1 and LeCTR2 (Giovannoni et al., 1999). Cloning and characterization of the genes corresponding to these new mutants, eer1 and epi, as well as the existing ethylene-insensitive mutants ein5 and ein6, will certainly expand our knowledge of the ethylene signal transduction pathway.
ETHYLENE IN PLANT DISEASE RESISTANCE
Plants have evolved sophisticated detection and defense systems to protect themselves from pathogen invasion. Ethylene seems to play an important role in various plant disease resistance pathways. However, depending on the type of pathogen and plant species, the role of ethylene can be dramatically different. Plants deficient in ethylene signaling may show either increased susceptibility or increased resistance. For example, in soybean, mutants with reduced ethylene sensitivity produce less severe chlorotic symptoms when challenged with the virulent strains Pseudomonas syringae pv glycinea and Phytophthora sojae, whereas virulent strains of the fungi Septoria glycines and Rhizoctonia solani cause more severe symptoms (Hoffman et al., 1999). Similarly, in Arabidopsis, the ethylene-insensitive mutant ein2 develops only minimal disease symptoms as the result of enhanced disease tolerance when infected by virulent P. syringae pv tomato or Xanthomonas campestris pv campestris (Bent et al., 1992). However, the ein2 mutant also displays enhanced susceptibility to the necrotrophic fungus Botrytis cinerea (Thomma et al., 1999). On the basis of these observations, ethylene seems to inhibit symptom development in necrotrophic pathogen infection but enhances the cell death caused by other type of pathogen infection. In fact, Arabidopsis protoplasts isolated from the etr1-1 mutant display reduced cell death from the fungal toxin fumonisin B1 (Asai et al., 2000), and presence of the ein2 mutation reduces cell death in the accelerated cell death 5 (acd5) mutant (Greenberg et al., 2000), supporting a role for ethylene in the regulation of programmed cell death.
Ethylene in Gene-for-Gene Resistance
Upon pathogen infection, the avirulence signal (avr) carried by pathogens is recognized by a specific plant resistance (R) gene product (Hammond-Kosack and Jones, 1997). This avr/R interaction is called gene-for-gene resistance and often triggers a strong defense mechanism that includes the programmed cell death of plant cells at the site of infection (known as the hypersensitive response), resulting in efficient containment of the pathogen. In tomato, it has been demonstrated that a direct interaction between the R gene Pto and the avirulence gene avrPto in the P. s. tomato strain determines gene-for-gene specificity in this plant–pathogen interaction (Scofield et al., 1996; Tang et al., 1996; Frederick et al., 1998). Recently, a transcription factor, Pti4, has been identified on the basis of its specific interaction with Pto (Gu et al., 2000). Interestingly, this Pti4 protein shares extensive similarity with the amino acid sequences of EREBPs and can specifically bind the GCC-box cis element present in the promoter of many ethylene-regulated pathogen-related (PR) genes. Expression of Pti4 in tomato leaves is rapidly induced by ethylene, and this induction precedes expression of GCC-box-containing PR genes. Moreover, phosphorylation of Pti4 by the Pto kinase enhances its binding to the GCC box. These results provide evidence that the ethylene response is linked to gene-for-gene resistance in tomato.
Interactions among the SA and JA and Ethylene Responses
Activation of the hypersensitive response triggers a long-lasting response known as systemic acquired resistance, which provides immunity against subsequent infections caused by a broad spectrum of pathogens (Ryals et al., 1994). In many cases, systemic acquired resistance is characterized by an increase in endogenous salicylic acid (SA) levels and expression of a subset of PR genes, as well as enhanced resistance to a broad spectrum of virulent pathogens. However, some pathogens can induce plant defense responses via activation of the ethylene and JA signal transduction pathways. Arabidopsis plants with defects in ethylene perception (ein2) or JA signaling (coi1) fail to induce a subset of PR gene expression, including the plant defensin gene PDF1.2, a basic chitinase (PR-3), and an acidic hevein-like protein (PR-4), resulting in enhanced susceptibility toward certain pathogens (Penninckx et al., 1998). Interestingly, the induction of PDF1.2 requires both intact JA and ethylene signaling, whereas the majority of other responses mediated by these hormones are specific to only one of the signals. This suggests that the ethylene and JA pathways interact with each other, co-regulating expression of some genes involved in plant defense. Because only a small subset of genes is affected by both signals, the interaction between these two pathways is likely to be downstream, possibly at the level of the specific defense gene promoters. Nevertheless, ethylene and JA signaling may also function independently to regulate distinct processes in defense response. A recent study has shown that pathogen- or elicitor-induced accumulation of the defense compound 3-indolylmethylglucosinolate is mediated by JA but not by ethylene or SA (Brader et al., 2001), indicating that ethylene and JA pathways may have different roles in disease resistance.
Although SA-dependent and JA/ethylene-dependent pathways induce expression of different sets of PR genes and result in plant resistance to different pathogens, there appear to be considerable interactions between these two pathways in systemic acquired resistance (see Figure 3). Here, use of the word “cross-talk” is reserved for communications between two separate, linear signal transduction pathways that are simultaneously activated in the same cell. Therefore, the components of the two signaling pathways have to be (1) shown to be expressed in the same cell and (2) demonstrated to physically interact under normal physiological conditions (Noselli and Perrimon, 2000). A recent survey of changes in the expression levels of 2375 selected genes upon pathogen infection or SA, JA, and ethylene treatment had revealed that although some genes are affected by one signal or another, many respond to two or more defense signals (Schenk et al., 2000). These results indicate the existence of a substantial network of regulatory interaction and coordination among different plant defense pathways. For example, two Arabidopsis mutants that constitutively express PR genes, cpr5 and cpr6, express both PR-1 and PDF1.2 genes in the absence of pathogen infection. Although the constitutive expression of PR-1 is dependent on SA, it is only partially suppressed by the npr1 (for non-expressor of PR-1) mutation, a gene that is required downstream of SA to activate PR-1 gene expression, indicating the existence of a SA-mediated, NPR1-independent response (Clarke et al., 2000). Only when ethylene signaling is also blocked by ein2 in addition to npr1 mutation in cpr5 and cpr6 mutants is PR-1 gene expression abolished completely. Furthermore, ein2 potentiates SA accumulation in cpr5 and dampens SA accumulation in cpr6. These results suggest the existence of interactions between ethylene- and SA-dependent signaling through an NPR1-independent pathway. Interestingly, a suppressor of npr1, ssi1, which completely bypasses NPR1 function, constitutively expresses the JA/ethylene-dependent marker PDF1.2 gene in an SA-dependent manner (Shah et al., 1999), suggesting that SSI1, together with CPR5 and CPR6, may participate in the interactions between the SA- and JA/ethylene-dependent pathways.
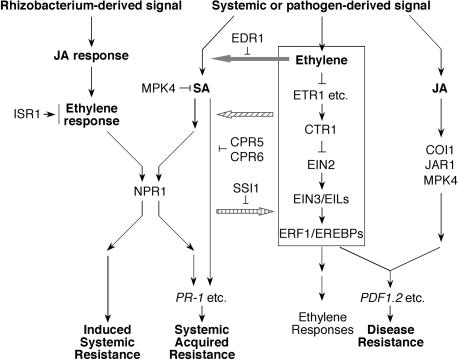
Figure 3.
Interactions between the Ethylene Signal Transduction Pathway and Plant Disease Resistance.
The ethylene signal transduction pathway can interact with the JA pathway to co-regulate expression of a subset of defense-related PR genes, for example, PDF1.2, involved in plant disease resistance. Meanwhile, there are considerable interactions between JA/ethylene- and SA-dependent pathways in systemic acquired resistance. In edr1 mutant, ethylene potentiates SA-mediated PR-1 gene expression. In the absence of CPR5 and CPR6, the ethylene pathway can also activate SA-dependent PR-1 gene expression independent of NPR1 to promote systemic acquired resistance. In the ssi1 mutant, the JA/ethylene-dependent PDF1.2 gene is constitutively expressed. Moreover, the ethylene pathway is also required for the rhizobacteria-mediated induced systemic resistance, which is independent of SA and pathogenesis-related gene activation. Ethylene signaling acts downstream of the JA pathway but upstream of NPR1 in ISR activation. Plants that lack ISR1 fail to develop ISR and display ethylene insensitivity. Arrows indicate positive regulation, and open blocks indicate negative regulation.
Recently, a null mutation in the EDR1 gene has been shown to enhance resistance to P. syringae and Erisyphe cichoracearum, and causes rapid activation of defense-related genes such as PR-1 (Frye et al., 2001). This enhanced disease resistance depends on the SA-induced defense response pathway and is independent of the JA/ethylene pathway. However, PR-1 gene expression, which is SA-dependent, is highly induced by ethylene treatment in edr1 mutant plants, whereas it is almost undetectable in wild-type plants. This again suggests that there is significant interaction between the ethylene and SA-dependent pathways. In this case, ethylene potentiates SA-mediated PR-1 gene expression, and EDR1 negatively regulates this process. Removal of EDR1 produces a dramatic effect of ethylene on SA-dependent responses, resulting in enhanced disease resistance in edr1 mutant plants. EDR1 encodes a putative MAPKKK similar to CTR1, but unlike the ctr1 mutant, edr1 does not display ethylene response phenotypes.
There are many other examples of similar interaction between the SA and JA/ethylene pathways. Perturbations in SA-dependent signaling have been reported to affect JA/ethylene-dependent signaling represented by PDF1.2 expression (Penninckx et al., 1996, 1998; Clarke et al., 1998, 2000; Dewdney et al., 2000; Gupta et al., 2000; Rao et al., 2000). It has been noticed that there is a correlation between a decrease in SA levels and increased PDF1.2 expression, indicating that SA may have an inhibitory effect on JA/ethylene biosynthesis or signaling (Jirage et al., 2001). Consistent with this observation, PDF1.2 mRNA accumulates at higher levels in mutants defective in SA signaling compared with levels in the wild type after B. cinerea infections (Clarke et al., 2001). This may also explain why mutants that disrupt SA-mediated responses become sensitized for activation of the JA/ethylene pathway (Clarke et al., 1998, 2000; Gupta et al., 2000). On the other hand, JA/ethylene can also repress the expression of SA-induced genes by inhibiting SA accumulation. For example, the mpk4 (for MAP kinase 4) mutant, which has elevated SA levels and constitutive activation of SA-dependent signaling, failed to induce the expression of PDF1.2 gene upon JA treatment (Petersen et al., 2000). This could result from high SA levels antagonizing JA/ethylene signaling as described above. However, when the mpk4 mutant is crossed to plants carrying the nahG transgene, which encodes an enzyme that degrades SA, activation of PDF1.2 expression is still blocked in the nahG mpk4 double mutant. These results suggest that block in JA/ethylene signaling relieves the suppression of SA signaling. Nevertheless, it should be pointed out that because both JA- and ethylene-dependent pathways are involved in regulating PDF1.2, changes in this gene expression may not always reflect an alteration in the ethylene-dependent pathway. In fact, although mpk4 dwarfism was similar to that of the ethylene constitutive triple-response mutant ctr1, MPK4 does not act in the ethylene response pathway between CTR1 and EIN2 (Petersen et al., 2000). Recent studies of an ethylene pathway gene ERF1 have shown that activation of ethylene responses by ERF1 overexpression in Arabidopsis plants is sufficient to confer resistance to B. cinerea but reduces SA-mediated tolerance against P. s. tomato DC3000 (Berrocal-Lobo et al., 2002), suggesting a negative regulation between ethylene and SA responses.
Despite the above-mentioned antagonistic interactions, there are examples in which both ethylene- and SA-dependent pathways cooperate on defense-related responses. In Arabidopsis, both ethylene and SA signal transduction pathways are necessary to mount an effective defense response against Plectosphaerella cucumerina (Berrocal-Lobo et al., 2002). In tomato, the transgenic ethylene-underproducing ACC deaminase line (ACD) and the ethylene-insensitive mutant Nr show reduced accumulation of SA upon X. campestris pv vesicatoria infection, resulting in less severe disease symptoms (O'Donnell et al., 2001). Taken together, these results demonstrate that both positive and negative interactions between ethylene and SA pathways can be established, depending on the type of pathogen or specific defense responses. This is consistent with the results that ein2 mutation increases SA accumulation in the cpr5 mutant but decreases SA levels in the cpr6 mutant, which represents a distinct resistance pathway regulated by CPR5 (Clarke et al., 2000).
Ethylene-Dependent Induced Systemic Resistance
Besides systemic acquired resistance, colonization of roots by certain rhizosphere bacteria confers another form of systemic disease resistance called induced systemic resistance (ISR) (Pieterse et al., 1998). Because SA-nonaccumulating nahG plants can mount ISR, ISR is independent of SA accumulation and pathogenesis-related gene activation (Pieterse et al., 1998). Although ISR requires responsiveness to both ethylene and JA, it is not accompanied by an increase in their production (Pieterse et al., 2000). However, NPR1, a component in the SA pathway, is required to mount the ISR. Because ethylene can induce ISR in jar1 mutants, it is thought that components of the ethylene response act downstream of JA in ISR signaling. Recently, it has been shown that ISR and systemic acquired resistance can be activated simultaneously, resulting in an additive level of protection against P. syringae (van Wees et al., 2000). Some Arabidopsis ecotypes, for example, RLD, fail to develop ISR, a trait that is associated with a relatively high level of susceptibility to P. syringae. This trait has been mapped to a single recessive locus (isr1) (Ton et al., 1999). Interestingly, this locus also cosegregates with significant root ethylene insensitivity in the seedling triple-response assay (Ton et al., 2001). Therefore, the susceptibility of the RLD ecotype to P. syringae may be directly linked to ethylene-insensitive phenotype, suggesting that the ISR1 locus is involved in the response to ethylene. This study also reports that the ecotype Wassilewskija (Ws) is similarly reduced in sensitivity to ethylene and that the ethylene-insensitive gene is an allele of ISR1. This result, however, is contradictory to the observation that Ws is known to be more sensitive to ethylene than most ecotypes, including Columbia, the reference strain for most of the ethylene response mutants (Roman et al., 1995).
ETHYLENE AND ABIOTIC STRESSES
Besides its physiological roles in different developmental stages, ethylene was originally regarded as a stress hormone because its synthesis is induced by a variety of stress signals, such as mechanical wounding, chemicals and metals, drought, extreme temperatures, and pathogen infection (Kende, 1993; Johnson and Ecker, 1998). Stress-induced ethylene production is typically controlled by accelerating the conversion of S-AdoMet to ACC, suggesting that the expression of ACC synthase is the major target of regulation. Among the environmental stresses, such as ozone, UV irradiation, and wounding, stimulation for ethylene synthesis has been reported to involve the generation of reactive oxygen species (Surplus et al., 1998; Orozco-Cardenas and Ryan, 1999; Pellinen et al., 1999). Reactive oxygen species (ROS or AOS for active oxygen species), including superoxide anions, hydroxyl radicals, and hydrogen peroxide, cause damage to cellular organelles by lipid peroxidation. In addition, ROS, in particular hydrogen peroxide, have been shown to function as signaling molecules (Levine et al., 1994). Recent results using mutants defective in ethylene, JA, and SA pathways to investigate the mechanisms underlying the wound- and ozone-induced responses have suggested that abiotic stress–induced responses share characteristics with pathogen defense pathways (Figure 4). As discussed below, interactions among SA, JA, and ethylene have been found to modulate responses to ROS.
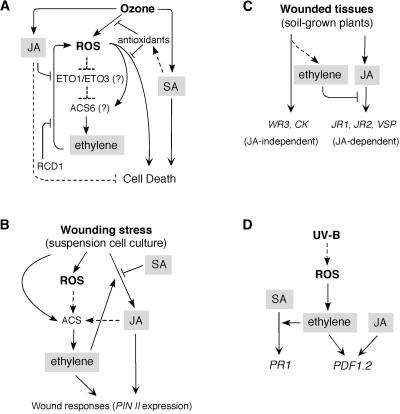
Figure 4.
Interactions among Ethylene, JA, and SA in Abiotic Stresses.
(A) Ozone stress.
(B) Wound response in tomato suspension cell culture.
(C) Wounded tissues (local responses).
(D) UV-B stress.
ACS, ACC synthase; ETO1 (ETO3), ethylene overproducer; RCD1, radical-induced cell death. Arrows indicate positive regulation, and open blocks indicate negative regulation. Dashed lines indicate possible or indirect interactions.
Ethylene and Ozone
Ozone (O3) has been observed as an air pollutant since the early 1950s and now is recognized as an abiotic elicitor that induces plant defense responses like those brought on by pathogen infection (Sandermann et al., 1998). Acute exposure to ozone leads to an oxidative burst, which evokes a local cell death response similar to that caused by the hypersensitive response upon pathogen infection (Pell et al., 1997). Cell death resulting from the oxidative burst is probably a consequence of direct deleterious effects on cellular membranes and organelles by ROS (Wojtaszek, 1997). Alternatively, it may represent programmed cell death activated by signals derived from ROS, as suggested for the hypersensitive response (Lamb and Dixon, 1997). Generation of ROS has been shown to be the casual effector for hypersensitive response and regulates the accumulation of SA, which is required for both the hypersensitive response and systemic acquired resistance (Sticher et al., 1997).
ROS generated by ozone result from its reactions with water and other cellular components when it enters from the stomata. ROS-induced cell death by ozone is supported by the observed hypersensitivity to ozone of an ascorbic acid (a potent antioxidant)–deficient mutant in Arabidopsis (Conklin et al., 1996). Ozone has been shown to induce the accumulation of SA, which subsequently induces systemic acquired resistance and activates the transcription of some of the ozone-induced genes (Sharma et al., 1996). The observation that a highly ozone-sensitive ecotype of Arabidopsis, Cvi-0, accumulates higher levels of SA prompted Rao and Davis (1999) to investigate the casual relationship between ozone sensitivity and the level of ozone-induced SA. Surprisingly, their results suggest that an optimal threshold of SA induced by ozone is required for protective antioxidant defense against the oxidative burst induced by the same signal. If the SA level is excessive, such as that found in Cvi-0, programmed cell death as a part of the hypersensitive response is activated rapidly and appears like ozone sensitivity.
Ethylene synthesis is one of the earliest responses to ozone stress (Vahala et al., 1998; Overmyer et al., 2000). The expression of ACS6 in Arabidopsis is rapidly activated within 30 min after the onset of ozone exposure. Accordingly, the production of ethylene reaches the maximal rate at one hour after ozone treatment and then declines gradually (Vahala et al., 1998). Compared with the synthesis kinetics of the other two molecules involved in ozone response, JA and SA, ethylene precedes both of them. The production of JA reaches the maximal rate within 5 h, whereas the synthesis of SA is not maximal until 5 h after ozone exposure (H. Tuominen and J. Kangasjärvi, personal communication). Two ethylene overproduction mutants, eto1 and eto3, show greater susceptibility to ozone than does the wild type (M.V. Rao and K.R. Davis, personal communication; H. Tuominen and J. Kangasjärvi, personal communication). This is intriguing because ethylene production in adult plants is comparable among eto1, eto3, and Col-0 wild type (Woeste et al., 1999). However, exposure to ozone dramatically increases ethylene production in both eto1 and eto3 compared with that of wild-type plants. Furthermore, ozone-induced ethylene accumulation is compromised in NahG and npr1 plants. The expression of two ozone pathway–specific molecular markers, PR1 (SA-dependent) and PR4 (ethylene-dependent), is also induced to a greater magnitude in eto1 and eto3 than in the wild type after exposure to ozone, whereas no significant differences in the induction of PDF1.2 (JA-dependent) were observed. Additionally, the induction of PR1 and PR4 by ozone is strongly suppressed in plants bearing both alleles of eto3 and nahG (M.V. Rao and K.R. Davis, personal communication). These results imply that the SA signaling pathway is required for the optimal induction of ethylene synthesis in response to ozone treatment, and that ethylene and SA act synergistically to effect cell death induced by ozone (Figure 4A).
An ozone-sensitive mutant, rcd1 (for radical-induced cell death), has been shown to have a higher susceptibility to the oxidative burst (Overmyer et al., 2000). Compared with wild-type plants, rcd1 is more susceptible to superoxide than hydrogen peroxide and shows prolonged lesions on leaves even after ozone is removed, suggesting a defect in restraining the toxicity of ROS. Interestingly, avirulent pathogens also trigger more severe hypersensitive response–like cell death in rcd1. As described above, ACS6 is induced by ozone and may result in the concomitant production of ACC and ethylene in rcd1. Ethylene production in rcd1 is higher than that of the wild type and continues even after ozone is removed. In contrast, ethylene synthesis returns to the basal level in the wild type when ozone treatment is ended. The prolonged cell death response observed even after ozone treatment is removed in rcd1 can be suppressed by norbornacliene (an ethylene receptor antagonist), application of methyl jasmonate, or by mutations in EIN2, suggesting that ethylene signaling is required for cell death and is antagonized by the JA pathway. The other implication is that RCD1 may function upstream of the ethylene receptor and acts to confine ethylene production once it is initiated. Therefore, it is possible that the hypersensitivity of rcd1 to ozone stress may be a consequence of a defective feedback regulation of ethylene synthesis or elevated ethylene sensitivity.
Ethylene and Wounding
Unlike animals, plants are sessile organisms that cannot defend themselves by avoiding injury. Therefore, wound responses in plants serve mostly to repair the damaged tissues, fend off herbivore attack, and defend against further pathogen infection (Leon et al., 2001). Elicitors used to invoke wound responses in tomato cell culture include oligogalacturonide fragments (OGAs) of cell wall, an 18-mer peptide systemin, and jasmonates. On the other hand, aspirin (acetyl salicylic acid [ASA]) and SA are negative regulators that block the expression of proteinase inhibitor genes (PIN), which are specific molecular markers for wound response (O'Donnell et al., 1996).
Ethylene has been shown to potentiate JA action in the wound response (O'Donnell et al., 1996) (Figure 4B). The biosynthesis of ethylene has been well known to be stimulated by wounding, most likely by the induction of ACS activity (Kende, 1993; Watanabe et al., 2001). It has been confirmed that wounding and elicitors, such as systemin, OGA, and JA, are able to induce ethylene generation in tomato cell culture (O'Donnell et al., 1996). This induction can be repressed by ASA, an inhibitor of JA synthesis and JA signaling pathways. The expression of PIN II (for proteinase inhibitor II), a specific marker for the JA wounding pathway, is linked to the induction of ethylene production by either elicitors or ethylene inhibitor treatment. Using inhibitors of ethylene biosynthesis and perception, as well as ACO antisense transgenic lines, it has been demonstrated that ethylene signaling is required for PIN II induction upon wounding in tomato plants. It has been postulated that both ethylene and JA are required for PIN II induction because direct application of ethylene does not induce PIN II expression. Consistent with this idea, the suppressed expression of PIN II in ASA-treated wounded plants only can be partially rescued by exogenous application of both ethylene and JA. The synergistic effects of JA and ethylene in the wound response is further supported by the finding that ethylene is required for the maximal production of JA. The wound induction of JA accumulation is reduced to <30% by the addition of inhibitors of ethylene biosynthesis. Also, ethylene has been shown to positively regulate the induction of allene oxide synthase, which catalyzes the first step in the biosynthesis of JA, in tomato and Arabidopsis (O'Donnell et al., 1996; Laudert and Weiler, 1998; Sivasankar et al., 2000). On the other hand, ROS and JA stimulate ethylene production by activating ACC synthase gene expression in winter squash (Watanabe and Sakai, 1998). Diphenylene iodonium, an inhibitor of ROS generation, blocks ethylene production but not JA accumulation. These results are consistent with the model that the synthesis of JA and ethylene is positively regulated by each other under wounding stress, whereas aspirin and SA suppress JA synthesis and the subsequent ethylene production in tomato suspension cell culture.
Ethylene seems to play an opposite role in the damaged tissues of soil-grown plants undergoing wound stress (Figure 4C). Two wound response pathways have been proposed, with only one of them being JA-dependent (Titarenko et al., 1997; Leon et al., 1998; Rojo et al., 1998, 1999). In Arabidopsis, evidence for a JA-dependent pathway is based on the expression of a subset of wound-induced genes, including JR1, JR2, and vegetative storage protein (VSP). The expression of these genes upon wounding is suppressed in the JA signaling mutant coi1. The expression of a second class of wound-inducible genes, including choline kinase (CK) and WR3, can be induced by elicitors, such as chitosan and OGA; these compounds do not activate JR1, JR2, and VSP. Furthermore, wounding induces expression of CK and WR3 in coi1 plants. The mRNAs of CK and WR3 accumulate in damaged leaves (local), whereas the maximal level expression of JR1 and VSP is present in unwounded leaves (systemic), suggesting that the JA-dependent pathway is suppressed in wounded tissues. Surprisingly, ethylene accumulation by wounding is via a JA-independent pathway, which seems to play a negative role in the expression of JA-dependent genes in the wounded tissue (Rojo et al., 1999). Inhibitors of ethylene biosynthesis or receptor binding, or mutations that confer reduced ethylene insensitivity, such as ein2, ein3, and etr1, are able to alleviate the suppression of JA-dependent gene expression activated by exogenous JA, treatment of elicitor chitosan, and wounding stress. However, the generation of JA in wounded tissues is not affected by ethylene. The negative role of ethylene in the JA-mediated wound response is opposite what has been suggested by tomato cell culture studies (O'Donnell et al., 1996), in which ethylene augments the JA-dependent induction of PIN II. The difference may lie in the experimental conditions, tissue samples, and different species of plants (Arabidopsis vs. tomato) used for wounding or elicitor treatment. Considering the local and systemic responses to wounding, ethylene may only block the JA-dependent pathway in the local damaged tissues because of its low mobility, although it does not effect JA-dependent functions in the systemic tissues.
Ethylene and UV-B
To a lesser extent than ozone, UV-B also causes an oxidative burst (Surplus et al., 1998). Treatment of Arabidopsis plants with UV-B (280 to 320 nm) light results in decreased expression of the photosynthetic genes Lhcb and psbA, and increased expression of PR-1 and PDF1.2. ROS are required for this altered gene expression because pretreatment of plants with ascorbic acid blocks the induction of PDF1.2 by UV-B. Induction of PDF1.2 is also inhibited in etr1-1 and jar1 mutants, suggesting that ROS lie upstream of the ethylene and JA pathways. Both ethylene and JA are required for the maximal induction of PDF1.2, as evidenced by application of these two growth regulators separately or together, and by examining the signaling defective mutants jar1 and etr1-1. Interestingly, induction of PR-1 is dependent on ethylene, but not on JA, and shows faster kinetics than that of PDF1.2, suggesting that ethylene is an early signal required to activate the SA pathway upon UV-B treatment. These results suggest that ethylene potentiates the response to both SA (PR-1 induction) and JA (PDF1.2 induction) (Figure 4D).
The synergistic and antagonistic interactions among JA, ethylene, and SA in response to abiotic stresses have been reported by several groups, and the following schemes are emerging. In ozone-induced stress, SA and ethylene stimulate cell death, whereas JA protects the stressed plants from deleterious damages by the oxidative burst (Rao and Davis, 1999; Overmyer et al., 2000; Rao et al., 2000). Therefore, it is expected that a similar phenotype will be shared by eto1, jar1, and rcd1 in ozone sensitivity. If ethylene plays a role in potentiating the hypersensitive response induced by SA, the transient induction of ethylene synthesis by stress followed by a controlled quenching would be a way for plants to limit the propagation of cell death. On the other hand, SA plays a central role in the regulation of both survival and death signals, depending on which side the balance is tipped (Rao and Davis, 1999). The role of ethylene in wounding responses is dependent on cell types. In the suspension cell culture of tomato treated with elicitors, ethylene potentiates, while SA represses, the defensive functions of the JA pathway (O'Donnell et al., 1996). On the other hand, ethylene represses the JA signaling pathway in the injured or elicitor-treated foliar tissues locally but has no effect in the systemic tissues (Rojo et al., 1999). In all cases, ethylene is involved in the early responses to different stresses, and its synthesis is required to be extinguished after the stress is removed. Unlike ethylene, JA and SA may have more long-term effects by their antagonistic interaction at the later stage of stress to prevent autocatalytic amplification of cellular damages.
ETHYLENE AND DEVELOPMENT: NODULATION
The legume–rhizobium interaction is a host-specific symbiosis (Stougaard, 2000). Legumes secrete compounds, usually flavonoids, into the rhizosphere that induce the synthesis of lipo-chitin-oligosaccharides (LCO, general term for Nod factors) from rhizobia, which in turn function as morphogens to initiate the development of nitrogen-fixation nodules in the infected root cells. The specificity of this interaction depends on the presentation of host-secreted flavonoids to rhizobia and the recognition of Nod factors by the host plants. The first morphological change after rhizobial infection is the deformation of root hair. A host-derived passage called the infection thread is developed and is used by the rhizobia to invade the root cortex, where it subsequently becomes the nodule primordium. A specialized organ, the nodule, will eventually develop to incorporate the rhizobial symbionts in this compartment. Although it is a beneficial process between plants and their symbionts, feedback inhibition of nodulation is evidenced by restricting the infection to root hair cells. Therefore, not every infection will lead to successful nodulation. Cellular responses to Nod factors, including membrane depolarization and influx of ions, take place within minutes and are followed by a periodic oscillation of cytosolic calcium concentration, or calcium spiking. Calcium spiking induced by Nod factors is a legume-specific activity and is known to be essential for nodule development, because legume mutants that fail to show calcium spiking are defective in forming nodules (Ehrhardt et al., 1996; Wais et al., 2000).
It has been suggested that ethylene is involved in the nodule development of Rhizobium–legume symbiosis (Penmetsa and Cook, 1997; Fernandez-Lopez et al., 1998; Oldroyd et al., 2001). A hyper-nodulation mutant, sickle (skl), identified in Medicago truncatula, shows increases of more than an order of magnitude in the number of persistent infections and mature nodules (Penmetsa and Cook, 1997). Because not all infection events result in nodule formation, SKL is thought to be involved in the regulation of rhizobial infection arrest. skl plants also show ethylene-insensitive phenotypes, such as delayed senescence of petals and leaves, decreased abscission of seedpods and leaves, as well as insensitivity of seedlings to ACC treatment. Experiments applying ACC and AVG at different times after the initiation of infection have demonstrated that ethylene inhibits a step before or at the onset of infection initiation and does not have an inhibitory effect after the formation of nodule primordia, suggesting that the role of ethylene is to regulate the persistence of the initial infection. To have an infection successfully leading to nodule formation, either ethylene production in the infected root hair cell must not occur or the cells must be insensitive to endogenous ethylene. Interestingly, a bacterial toxin, rhizobitoxine (an analog of the ACS inhibitor AVG), is produced by the legume symbiont Bradyrhizobium elkanii and has been shown to enhance the nodulation on its host, Macroptilium atropurpureum (Yasuta et al., 1999; Yuhashi et al., 2000).
If ethylene functions to regulate the threshold concentration of Nod factors and subsequent calcium spiking in the root cells, then mutation in skl apparently decreases the required threshold; thus, SKL can be regarded as an enhancer for Nod factor sensitivity. Because calcium spiking induced by Nod factor continues for 2 h after initiation, it is possible that application of ethylene affects the stability of Nod factor or other effectors that regulate calcium spiking, whereas the skl mutation decreases this effect. Alternatively, as suggested by Oldroyd et al. (2001), skl may have pleiotropic effects on calcium spiking independent of ethylene, or ethyl-ene could modulate the frequency of calcium spiking. The results suggest that the inhibitory effect of ethylene is to perturb the calcium spiking induced by Nod factors, which may subsequently be required for nodule development. Increasing the concentration of Nod factors can counter the ethylene inhibition. Two model legumes, Lotus japonicus and M. truncatula, have been used to generate new mutants whose functions can be assigned to different stages of the nodulation process (Wais et al., 2000; Stougaard, 2001). Genetic analysis using the available nodulation mutants in combination with ethylene mutant skl (Penmetsa and Cook, 1997) may provide further insight into the role of ethylene in nodule formation.
ETHYLENE SIGNALING IN METAZOANS?
Recent studies of the marine sponge (invertebrate) and mammalian cell cultures (vertebrate) have raised an interesting question: does ethylene play any role in metazoans? Although ethylene has long been known as a plant hormone, it is not produced in species outside the plant kingdom. However, ethylene is one of the major alkene in seawater produced by photochemical reactions of the dissolved organic carbon and is present in the growth environment of sponges. Two ethylene-inducible genes have been identified from sponges (Suberites domuncula) (Krasko et al., 1999). The first one shares >80% similarity with an ethylene-responsive gene, HEVER (for Heven ethylene responsive), in Hevea brasiliensis (rubber tree) (Sivasubramaniam et al., 1995). The expression of HEVER is induced by stress treatment with SA and ethephon, an ethylene releasing agent. Another ethylene-induced gene encodes a putative Ca2+/calmodulin-dependent protein kinase. The latter seems to be consistent with the observation that ethylene treatment of sponge primmorph cell culture (“organ-like” aggregates of single sponge cells) (Custodio et al., 1998) induces a burst of cytosolic calcium concentration. More recently, mammalian cell cultures treated with ethephon show a dramatic increase of cytosolic calcium influx (Perovic et al., 2001). It is known that acidic gas accompanies ethylene release from ethephon in solution. Because it is unclear whether the calcium influx could result from the damage caused to cell membranes by an acidic gas, the roles of ethylene remain to be determined in the mammalian system. Interestingly, a putative human ACS gene has been recently identified, although the corresponding product does not have ACS activity, most likely due to the absence of two conserved residues (Tyr-85 and Gln-83 in apple ACS) in the active site (Koch et al., 2001).
CONCLUSION
A wealth of ethylene mutants in a variety of plant species and the results derived from the epistasis studies have provided us with the knowledge to draw a more complete picture of the ethylene pathway and its interaction with other hormones. Although many of the genes corresponding to these mutants have been cloned, their functional characterization is still at an early stage. Future studies to characterize the interaction among pathway components will reveal more detailed information about how ethylene synthesis and signaling are regulated and how they may interact with components of other pathways. One particularly useful approach using the whole genome-based DNA chip technology will be obviously an effective means to examine the regulation of expression of ethylene and other hormone/stress signaling genes. The interaction of ethylene with SA and JA signaling pathways in biotic and abiotic stresses demonstrates the complex nature of the plant's decisions and the different outcomes. The challenge will be the characterization of what makes either positive or negative interaction under different conditions for the ethylene and other signaling pathways, especially JA and SA, in pathogen infection and environmental stresses.
Acknowledgments
The authors thank Drs. Sarah Liljegren, Linda Walling, and Jaakko Kangasjärvi for critical reading of the manuscript, and colleagues in the Plant Biology Laboratory for helpful discussions and comments. They are also grateful for the unpublished results from the laboratories of Drs. Keith Davis and Jaakko Kangasjärvi and for insightful communications with Dr. Hannele Tuominen. This work was supported by grants from the U.S. Department of Energy and the National Science Foundation to J.R.E., and by a National Institutes of Health Postdoctoral Fellowship to H.L.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001768.
References
- Abel, S., Nguyen, M.D., Chow, W., and Theologis, A. (1995). ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. Structural characterization, expression in Escherichia coli, and expression characteristics in response to auxin. J. Biol. Chem. 270 19093–19099. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 2148–2152. [DOI] [PubMed] [Google Scholar]
- Arteca, J.M., and Arteca, R.N. (1999). A multi-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase (ACS6) in mature Arabidopsis leaves. Plant Mol. Biol. 39 209–219. [DOI] [PubMed] [Google Scholar]
- Asai, T., Stone, J.M., Heard, J.E., Kovtun, Y., Yorgey, P., Sheen, J., and Ausubel, F.M. (2000). Fumonisin B1–induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12 1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, C.S., Fox, E.A., Yen, H., Lee, S., Ying, T., Grierson, D., and Giovannoni, J.J. (2001). Analysis of the ethylene response in the epinastic mutant of tomato. Plant Physiol. 127 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, C.S., Llop-Tous, M.I., and Grierson, D. (2000). The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 123 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin, N., Serizet, C., Gosti, F., and Giraudat, J. (2000). Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F., Innes, R.W., Ecker, J.R., and Staskawicz, B.J. (1992). Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol. Plant-Microbe Interact. 5 372–378. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo, M., Molina, A., and Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29 23–32. [DOI] [PubMed] [Google Scholar]
- Bleecker, A.B., Estelle, M.A., Somerville, C., and Kende, H. (1988). Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241 1086–1089. [DOI] [PubMed] [Google Scholar]
- Bleecker, A.B., and Kende, H. (2000). Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16 1–18. [DOI] [PubMed] [Google Scholar]
- Brader, G., Tas, E., and Palva, E.T. (2001). Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiol. 126 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner, M., and Singh, K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein, interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitani, G., Hohenester, E., Feng, L., Storici, P., Kirsch, J.F., and Jansonius, J.N. (1999). Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene. J. Mol. Biol. 294 745–756. [DOI] [PubMed] [Google Scholar]
- Cary, A.J., Liu, W., and Howell, S.H. (1995). Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol. 107 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C., Kwok, S.F., Bleecker, A.B., and Meyerowitz, E.M. (1993). Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science 262 539–544. [DOI] [PubMed] [Google Scholar]
- Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., and Ecker, J.R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89 1133–1144. [DOI] [PubMed] [Google Scholar]
- Chappell, J., Hahlbrock, K., and Boller, T. (1984). Rapid induction of ethylene biosynthesis in cultured parsley cells by fungal elicitor and its relationship to the induction of phenylalanine ammonia-lyase. Planta 161 475–480. [DOI] [PubMed] [Google Scholar]
- Clark, K.L., Larsen, P.B., Wang, X., and Chang, C. (1998). Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc. Natl. Acad. Sci. USA 95 5401–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.D., Aarts, N., Feys, B.J., Dong, X., and Parker, J.E. (2001). Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J. 26 409–420. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.D., Volko, S.M., Ledford, H., Ausubel, F.M., and Dong, X. (2000). Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin, P.L., Williams, E.H., and Last, R.L. (1996). Environmental stress sensitivity of an ascorbic acid–deficient Arabidopsis mutant. Proc. Natl. Acad. Sci. USA 93 9970–9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio, M.R., Prokic, I., Steffen, R., Koziol, C., Borojevic, R., Brummer, F., Nickel, M., and Muller, W.E. (1998). Primmorphs generated from dissociated cells of the sponge Suberites domuncula: A model system for studies of cell proliferation and cell death. Mech. Ageing Dev. 105 45–59. [DOI] [PubMed] [Google Scholar]
- Dewdney, J., Reuber, T.L., Wildermuth, M.C., Devoto, A., Cui, J., Stutius, L.M., Drummond, E.P., and Ausubel, F.M. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24 205–218. [DOI] [PubMed] [Google Scholar]
- Ecker, J.R. (1995). The ethylene signal transduction pathway in plants. Science 268 667–675. [DOI] [PubMed] [Google Scholar]
- Ehrhardt, D.W., Wais, R., and Long, S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85 673–681. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284 1539–1541. [DOI] [PubMed] [Google Scholar]
- Felix, G., Grosskopf, D.G., Regenass, M., and Boller, T. (1991). Rapid changes of protein-phosphorylation are involved in transduction of the elicitor signal in plant cells. Proc. Natl. Acad. Sci. USA 88 8831–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Lopez, M., Goormachtig, S., Gao, M., D'Haeze, W., Van Montagu, M., and Holsters, M. (1998). Ethylene-mediated phenotypic plasticity in root nodule development on Sesbania rostrata. Proc. Natl. Acad. Sci. USA 95 12724–12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick, R.D., Thilmony, R.L., Sessa, G., and Martin, G.B. (1998). Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol. Cell 2 241–245. [DOI] [PubMed] [Google Scholar]
- Frye, C.A., Tang, D., and Innes, R.W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 98 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell 12 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, H., and Syono, K. (1996). Genetic analysis of the effects of polar auxin transport inhibitors on root growth in Arabidopsis thaliana. Plant Cell Physiol. 37 1094–1101. [DOI] [PubMed] [Google Scholar]
- Ge, L., Liu, J.Z., Wong, W.S., Hsiao, W.L.W., Chong, K., Xu, Z.K., Yang, S.F., Kung, S.D., and Li, N. (2000). Identification of a novel multiple environmental factor-responsive 1-aminocyclopropane-1-carboxylate synthase gene, NT-ACS2, from tobacco. Plant Cell Environ. 23 1169–1182. [Google Scholar]
- Ghassemian, M., Nambara, E., Cutler, S., Kawaide, H., Kamiya, Y., and McCourt, P. (2000). Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni, J., Yen, H., Shelton, B., Miller, S., Vrebalov, J., Kannan, P., Tieman, D., Hackett, R., Grierson, D., and Klee, H. (1999). Genetic mapping of ripening and ethylene-related loci in tomato. Theor. Appl. Genet. 98 1005–1013. [Google Scholar]
- Greenberg, J.T., Silverman, F.P., and Liang, H. (2000). Uncoupling salicylic acid–dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y.Q., Yang, C., Thara, V.K., Zhou, J., and Martin, G.B. (2000). Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, V., Willits, M.G., and Glazebrook, J. (2000). Arabidopsis thaliana EDS4 contributes to salicylic acid (SA)–dependent expression of defense responses: Evidence for inhibition of jasmonic acid signaling by SA. Mol. Plant-Microbe Interact. 13 503–511. [DOI] [PubMed] [Google Scholar]
- Guzman, P., and Ecker, J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A.E., Findell, J.L., Schaller, G.E., Sisler, E.C., and Bleecker, A.B. (2000). Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol. 123 1449–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, A.J., Bouzayen, M., and Grierson, D. (1991). Identification of a tomato gene for the ethylene-forming enzyme by expression in yeast. Proc. Natl. Acad. Sci. USA 88 7434–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1997). Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 575–607. [DOI] [PubMed] [Google Scholar]
- Hirayama, T., Kieber, J.J., Hirayama, N., Kogan, M., Guzman, P., Nourizadeh, S., Alonso, J.M., Dailey, W.P., Dancis, A., and Ecker, J.R. (1999). RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease–related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97 383–393. [DOI] [PubMed] [Google Scholar]
- Hoffman, T., Schmidt, J.S., Zheng, X., and Bent, A.F. (1999). Isolation of ethylene-insensitive soybean mutants that are altered in pathogen susceptibility and gene-for-gene disease resistance. Plant Physiol. 119 935–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, J., Chang, C., Sun, Q., and Meyerowitz, E.M. (1995). Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269 1712–1714. [DOI] [PubMed] [Google Scholar]
- Hua, J., and Meyerowitz, E.M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94 261–271. [DOI] [PubMed] [Google Scholar]
- Hua, J., Sakai, H., Nourizadeh, S., Chen, Q.G., Bleecker, A.B., Ecker, J.R., and Meyerowitz, E.M. (1998). EIN4 and ERS2 are members of the putative ethylene-receptor gene family in Arabidopsis. Plant Cell 10 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D., Zhou, N., Cooper, B., Clarke, J.D., Dong, X., and Glazebrook, J. (2001). Constitutive salicylic acid–dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J. 26 395–407. [DOI] [PubMed] [Google Scholar]
- Johnson, P.R., and Ecker, J.R. (1998). The ethylene gas signal transduction pathway: A molecular perspective. Annu. Rev. Genet. 32 227–254. [DOI] [PubMed] [Google Scholar]
- Kende, H. (1993). Ethylene biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44 283–307. [Google Scholar]
- Kieber, J.J., Rothenberg, M., Roman, G., Feldmann, K.A., and Ecker, J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72 427–441. [DOI] [PubMed] [Google Scholar]
- Koch, K.A., Capitani, G., Gruetter, M.G., and Kirsch, J.F. (2001). The human cDNA for a homologue of the plant enzyme 1-aminocyclopropane-1-carboxylate synthase encodes a protein lacking that activity. Gene 272 75–84. [DOI] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2000). Cloning and DNA-binding properties of a tobacco Ethylene-insensitive3 (EIN3) homolog. Nucleic Acids Res. 28 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasko, A., Schroder, H.C., Perovic, S., Steffen, R., Kruse, M., Reichert, W., Muller, I.M., and Muller, W.E. (1999). Ethylene modulates gene expression in cells of the marine sponge Suberites domuncula and reduces the degree of apoptosis. J. Biol. Chem. 274 31524–31530. [DOI] [PubMed] [Google Scholar]
- Kyriakis, J.M., App, H., Zhang, X.F., Banerjee, P., Brautigan, D.L., Rapp, U.R., and Avruch, J. (1992). Raf-1 activates MAP kinase-kinase. Nature 358 417–421. [DOI] [PubMed] [Google Scholar]
- Lamb, C., and Dixon, R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 251–275. [DOI] [PubMed] [Google Scholar]
- Larsen, P.B., and Chang, C. (2001). The Arabidopsis eer1 mutant has enhanced ethylene responses in the hypocotyl and stem. Plant Physiol. 125 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudert, D., and Weiler, E.W. (1998). Allene oxide synthase: A major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 15 675–684. [DOI] [PubMed] [Google Scholar]
- Leon, J., Rojo, E., and Sanchez-Serrano, J.J. (2001). Wound signalling in plants. J. Exp. Bot. 52 1–9. [DOI] [PubMed] [Google Scholar]
- Leon, J., Rojo, E., Titarenko, E., and Sanchez-Serrano, J.J. (1998). Jasmonic acid–dependent and –independent wound signal transduction pathways are differentially regulated by Ca2+/calmodulin in Arabidopsis thaliana. Mol. Gen. Genet. 258 412–419. [DOI] [PubMed] [Google Scholar]
- Levine, A., Tenhaken, R., Dixon, R., and Lamb, C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79 583–593. [DOI] [PubMed] [Google Scholar]
- Li, N., and Mattoo, A.K. (1994). Deletion of the carboxyl-terminal region of 1-aminocyclopropane-1-carboxylic acid synthase, a key protein in the biosynthesis of ethylene, results in catalytically hyperactive, monomeric enzyme. J. Biol. Chem. 269 6908–6917. [PubMed] [Google Scholar]
- Liang, X., Abel, S., Keller, J.A., Shen, N.F., and Theologis, A. (1992). The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 89 11046–11050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X., Oono, Y., Shen, N.F., Kohler, C., Li, K., Scolnik, P.A., and Theologis, A. (1995). Characterization of two members (ACS1 and ACS3) of the 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Gene 167 17–24. [DOI] [PubMed] [Google Scholar]
- Liang, X., Shen, N.F., and Theologis, A. (1996). Li(+)-regulated 1-aminocyclopropane-1-carboxylate synthase gene expression in Arabidopsis thaliana. Plant J. 10 1027–1036. [DOI] [PubMed] [Google Scholar]
- Mehta, P.K., Hale, T.I., and Christen, P. (1993). Aminotransferases: Demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214 549–561. [DOI] [PubMed] [Google Scholar]
- Nakatsuka, A., Murachi, S., Okunishi, H., Shiomi, S., Nakano, R., Kubo, Y., and Inaba, A. (1998). Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol. 118 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noselli, S., and Perrimon, N. (2000). Signal transduction. Are there close encounters between signaling pathways? Science 290 68–69. [DOI] [PubMed] [Google Scholar]
- O'Donnell, P.J., Calvert, C., Atzorn, R., Wasternack, C., Leyser, H.M.O., and Bowles, D.J. (1996). Ethylene as a signal mediating the wound response of tomato plants. Science 274 1914–1917. [DOI] [PubMed] [Google Scholar]
- O'Donnell, P.J., Jones, J.B., Antoine, F.R., Ciardi, J., and Klee, H.J. (2001). Ethylene-dependent salicylic acid regulates an expanded cell death response to a plant pathogen. Plant J. 25 315–323. [DOI] [PubMed] [Google Scholar]
- Oetiker, J.H., Olson, D.C., Shiu, O.Y., and Yang, S.F. (1997). Differential induction of seven 1-aminocyclopropane-1-carboxylate synthase genes by elicitor in suspension cultures of tomato (Lycopersicon esculentum). Plant Mol. Biol. 34 275–286. [DOI] [PubMed] [Google Scholar]
- Oh, S.A., Park, J.H., Lee, G.I., Paek, K.H., Park, S.K., and Nam, H.G. (1997). Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. 12 527–535. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd, G.E., Engstrom, E.M., and Long, S.R. (2001). Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13 1835–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cardenas, M., and Ryan, C.A. (1999). Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 96 6553–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer, K., Tuominen, H., Kettunen, R., Betz, C., Langebartels, C., Sandermann, H., Jr., and Kangasjärvi, J. (2000). Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12 1849–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.J., Shin, R., Park, J.M., Lee, G.J., Yoo, T.H., and Paek, K.H. (2001). A hot pepper cDNA encoding a pathogenesis-related protein 4 is induced during the resistance response to tobacco mosaic virus. Mol. Cell 11 122–127. [PubMed] [Google Scholar]
- Peck, S.C., and Kende, H. (1998). Differential regulation of genes encoding 1-aminocyclopropane-1-carboxylate (ACC) synthase in etiolated pea seedlings: Effects of indole-3-acetic acid, wounding, and ethylene. Plant Mol. Biol. 38 977–982. [DOI] [PubMed] [Google Scholar]
- Pelech, S.L., and Sanghera, J.S. (1992). Mitogen-activated protein kinases: Versatile transducers for cell signaling. Trends Biochem. Sci. 17 233–238. [DOI] [PubMed] [Google Scholar]
- Pell, E.J., Schlagnhaufer, C.D., and Arteca, R.N. (1997). Ozone-induced oxidative stress: Mechanisms of action and reaction. Physiol. Plant. 100 264–273. [Google Scholar]
- Pellinen, R., Palva, T., and Kangasjärvi, J. (1999). Short communication: Subcellular localization of ozone-induced hydrogen peroxide production in birch (Betula pendula) leaf cells. Plant J. 20 349–356. [DOI] [PubMed] [Google Scholar]
- Penmetsa, R.V., and Cook, D.R. (1997). A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275 527–530. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A., Eggermont, K., Terras, F.R., Thomma, B.P., De Samblanx, G.W., Buchala, A., Metraux, J.P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid–independent pathway. Plant Cell 8 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A., Thomma, B.P., Buchala, A., Metraux, J.P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perovic, S., Seack, J., Gamulin, V., Muller, W.E., and Schroder, H.C. (2001). Modulation of intracellular calcium and proliferative activity of invertebrate and vertebrate cells by ethylene. BMC Cell. Biol. 2 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M., van Wees, S.C., van Pelt, J.A., Knoester, M., Laan, R., Gerrits, H., Weisbeek, P.J., and van Loon, L.C. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J., van Pelt, J.T.J., Parchmann, S., Mueller, M.J., Buchala, A.J., Metraux, J.-P., and van Loon, L.C. (2000). Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol. Mol. Plant Pathol. 57 123–134. [Google Scholar]
- Pirrung, M.C. (1999). Histidine kinases and two-component signal transduction systems. Chem. Biol. 6 R167–R175. [DOI] [PubMed] [Google Scholar]
- Rao, M.V., and Davis, K.R. (1999). Ozone-induced cell death occurs via two distinct mechanisms in Arabidopsis: The role of salicylic acid. Plant J. 17 603–614. [DOI] [PubMed] [Google Scholar]
- Rao, M.V., Lee, H., Creelman, R.A., Mullet, J.E., and Davis, K.R. (2000). Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel, S., Gakiere, B., Job, D., and Douce, R. (1998). The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. USA 95 7805–7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379 633–646. [DOI] [PubMed] [Google Scholar]
- Rodriguez, F.I., Esch, J.J., Hall, A.E., Binder, B.M., Schaller, G.E., and Bleecker, A.B. (1999). A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283 996–998. [DOI] [PubMed] [Google Scholar]
- Rojo, E., Leon, J., and Sanchez-Serrano, J.J. (1999). Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J. 20 135–142. [DOI] [PubMed] [Google Scholar]
- Rojo, E., Titarenko, E., Leon, J., Berger, S., Vancanneyt, G., and Sanchez-Serrano, J.J. (1998). Reversible protein phosphorylation regulates jasmonic acid–dependent and –independent wound signal transduction pathways in Arabidopsis thaliana. Plant J. 13 153–165. [DOI] [PubMed] [Google Scholar]
- Roman, G., and Ecker, J.R. (1995). Genetic analysis of a seedling stress response to ethylene in Arabidopsis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 350 75–81. [DOI] [PubMed] [Google Scholar]
- Roman, G., Lubarsky, B., Kieber, J.J., Rothenberg, M., and Ecker, J.R. (1995). Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics 139 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J., Uknes, S., and Ward, E. (1994). Systemic acquired-resistance. Plant Physiol. 104 1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, H., Hua, J., Chen, Q.G., Chang, C., Medrano, L.J., Bleecker, A.B., and Meyerowitz, E.M. (1998). ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 95 5812–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sandermann, H., Ernst, D., Heller, W., and Langebartels, C. (1998). Ozone: An abiotic elicitor of plant defence reactions. Trends Plant Sci. 3 47–50. [Google Scholar]
- Sato, T., and Theologis, A. (1989). Cloning the mRNA encoding 1-aminocyclopropane-1-carboxylate synthase, the key enzyme for ethylene biosynthesis in plants. Proc. Natl. Acad. Sci. USA 86 6621–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, G.E., and Bleecker, A.B. (1995). Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270 1809–1811. [DOI] [PubMed] [Google Scholar]
- Schaller, G.E., Ladd, A.N., Lanahan, M.B., Spanbauer, J.M., and Bleecker, A.B. (1995). The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J. Biol. Chem. 270 12526–12530. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.W., and Staskawicz, B.J. (1996). Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274 2063–2065. [DOI] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., and Klessig, D.F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, Y.K., Leon, J., Raskin, I., and Davis, K.R. (1996). Ozone-induced responses in Arabidopsis thaliana: The role of salicylic acid in the accumulation of defense-related transcripts and induced resistance. Proc. Natl. Acad. Sci. USA 93 5099–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankar, S., Sheldrick, B., and Rothstein, S.J. (2000). Expression of allene oxide synthase determines defense gene activation in tomato. Plant Physiol. 122 1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramaniam, S., Vanniasingham, V.M., Tan, C.T., and Chua, N.H. (1995). Characterisation of HEVER, a novel stress-induced gene from Hevea brasiliensis. Plant Mol. Biol. 29 173–178. [DOI] [PubMed] [Google Scholar]
- Smith, J.M., and Arteca, R.N. (2000). Molecular control of ethylene production by cyanide in Arabidopsis thaliana. Physiol. Plant. 109 180–187. [Google Scholar]
- Solano, R., and Ecker, J.R. (1998). Ethylene gas: Perception, signaling and response. Curr. Opin. Plant Biol. 1 393–398. [DOI] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu, P., Grosskopf, D.G., Felix, G., and Boller, T. (1994). The apparent turnover of 1-aminocyclopropane-1-carboxylate synthase in tomato cells is regulated by protein-phosphorylation and dephosphorylation. Plant Physiol. 106 529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu, P., Reinhardt, D., and Boller, T. (1991). Analysis and cloning of the ethylene-forming enzyme from tomato by functional expression of its mRNA in Xenopus laevis oocytes. EMBO J. 10 2007–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova, A.N., and Ecker, J.R. (2000). Ethylene signaling: From mutants to molecules. Curr. Opin. Plant Biol. 3 353–360. [DOI] [PubMed] [Google Scholar]
- Sticher, L., Mauch-Mani, B., and Metraux, J.P. (1997). Systemic acquired resistance. Annu. Rev. Phytopathol. 35 235–270. [DOI] [PubMed] [Google Scholar]
- Stougaard, J. (2000). Regulators and regulation of legume root nodule development. Plant Physiol. 124 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard, J. (2001). Genetics and genomics of root symbiosis. Curr. Opin. Plant Biol. 4 328–335. [DOI] [PubMed] [Google Scholar]
- Su, W.P., and Howell, S.H. (1992). A single genetic-locus, ckr1, defines Arabidopsis mutants in which root-growth is resistant to low concentrations of cytokinin. Plant Physiol. 99 1569–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surplus, S.L., Jordan, B.R., Murphy, A.M., Carr, J.P., Thomas, B., and Mackerness, S.A.H. (1998). Ultraviolet-B-induced responses in Arabidopsis thaliana: Role of salicylic acid and reactive oxygen species in the regulation of transcripts encoding photosynthetic and acidic pathogenesis-related proteins. Plant Cell Environ. 21 685–694. [Google Scholar]
- Suzuki, K., Suzuki, N., Ohme-Takagi, M., and Shinshi, H. (1998). Immediate early induction of mRNAs for ethylene-responsive transcription factors in tobacco leaf strips after cutting. Plant J. 15 657–665. [DOI] [PubMed] [Google Scholar]
- Tang, X., Frederick, R.D., Zhou, J., Halterman, D.A., Jia, Y., and Martin, G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274 2060–2063. [DOI] [PubMed] [Google Scholar]
- Tarun, A.S., Lee, J.S., and Theologis, A. (1998). Random mutagenesis of 1-aminocyclopropane-1-carboxylate synthase: A key enzyme in ethylene biosynthesis. Proc. Natl. Acad. Sci. USA 95 9796–9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun, A.S., and Theologis, A. (1998). Complementation analysis of mutants of 1-aminocyclopropane-1-carboxylate synthase reveals the enzyme is a dimer with shared active sites. J. Biol. Chem. 273 12509–12514. [DOI] [PubMed] [Google Scholar]
- Tatsuki, M., and Mori, H. (2001). Phosphorylation of tomato 1-aminocyclopropane-1-carboxylic acid synthase, Le-ACS2, at the C-terminal region. J. Biol. Chem. 276 28051–28057. [DOI] [PubMed] [Google Scholar]
- Thara, V.K., Tang, X., Gu, Y.Q., Martin, G.B., and Zhou, J.M. (1999). Pseudomonas syringae pv tomato induces the expression of tomato EREBP-like genes Pti4 and Pti5 independent of ethylene, salicylate and jasmonate. Plant J. 20 475–483. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P., Eggermont, K., Tierens, K.F., and Broekaert, W.F. (1999). Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman, D.M., Ciardi, J.A., Taylor, M.G., and Klee, H.J. (2001). Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J. 26 47–58. [DOI] [PubMed] [Google Scholar]
- Tieman, D.M., Taylor, M.G., Ciardi, J.A., and Klee, H.J. (2000). The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc. Natl. Acad. Sci. USA 97 5663–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko, E., Rojo, E., Leon, J., and Sanchez-Serrano, J.J. (1997). Jasmonic acid–dependent and –independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol. 115 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, J., Davison, S., Van Wees, S.C., Van Loon, L., and Pieterse, C.M. (2001). The Arabidopsis isr1 locus controlling rhizobacteria-mediated induced systemic resistance is involved in ethylene signaling. Plant Physiol. 125 652–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, J., Pieterse, C.M., and Van Loon, L.C. (1999). Identification of a locus in Arabidopsis controlling both the expression of rhizobacteria-mediated induced systemic resistance (ISR) and basal resistance against Pseudomonas syringae pv. tomato. Mol. Plant-Microbe Interact. 12 911–918. [DOI] [PubMed] [Google Scholar]
- Urao, T., Miyata, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2000). Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett. 478 227–232. [DOI] [PubMed] [Google Scholar]
- Vahala, J., Schlagnhaufer, C.D., and Pell, E.J. (1998). Induction of an ACC synthase cDNA by ozone in light-grown Arabidopsis thaliana leaves. Physiol. Plant. 103 45–50. [Google Scholar]
- Van der Straeten, D., Rodrigues-Pousada, R.A., Villarroel, R., Hanley, S., Goodman, H.M., and Van Montagu, M. (1992). Cloning, genetic mapping, and expression analysis of an Arabidopsis thaliana gene that encodes 1-aminocyclopropane-1-carboxylate synthase. Proc. Natl. Acad. Sci. USA 89 9969–9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees, S.C., de Swart, E.A., van Pelt, J.A., van Loon, L.C., and Pieterse, C.M. (2000). Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97 8711–8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J.P., Schuerman, P., Woeste, K., Brandstatter, I., and Kieber, J.J. (1998. a). Isolation and characterization of Arabidopsis mutants defective in the induction of ethylene biosynthesis by cytokinin. Genetics 149 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J.P., Woeste, K.E., Theologis, A., and Kieber, J.J. (1998. b). Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA 95 4766–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wais, R.J., Galera, C., Oldroyd, G., Catoira, R., Penmetsa, R.V., Cook, D., Gough, C., Denarie, J., and Long, S.R. (2000). Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. USA 97 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T., and Sakai, S. (1998). Effects of active oxygen species and methyl jasmonate on expression of the gene for a wound-inducible 1-aminocyclopropane-1-carboxylate synthase in winter squash (Cucurbita maxima). Planta 206 570–576. [Google Scholar]
- Watanabe, T., Seo, S., and Sakai, S. (2001). Wound-induced expression of a gene for 1-aminocyclopropane-1-carboxylate synthase and ethylene production is regulated by both reactive oxygen species and jasmonic acid in Cucurbita maxima. Plant Physiol. Biochem. 39 121–127. [Google Scholar]
- White, M.F., Vasquez, J., Yang, S.F., and Kirsch, J.F. (1994). Expression of apple 1-aminocyclopropane-1-carboxylate synthase in Escherichia coli: Kinetic characterization of wild-type and active-site mutant forms. Proc. Natl. Acad. Sci. USA 91 12428–12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste, K.E., and Kieber, J.J. (2000). A strong loss-of-function mutation in RAN1 results in constitutive activation of the ethylene response pathway as well as a rosette-lethal phenotype. Plant Cell 12 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste, K.E., Ye, C., and Kieber, J.J. (1999). Two Arabidopsis mutants that overproduce ethylene are affected in the posttranscriptional regulation of 1-aminocyclopropane-1-carboxylic acid synthase. Plant Physiol. 119 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek, P. (1997). Oxidative burst: An early plant response to pathogen infection. Biochem. J. 322 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurgler-Murphy, S.M., and Saito, H. (1997). Two-component signal transducers and MAPK cascades. Trends Biochem. Sci. 22 172–176. [DOI] [PubMed] [Google Scholar]
- Yamamoto, S., Suzuki, K., and Shinshi, H. (1999). Elicitor-responsive, ethylene-independent activation of GCC box–mediated transcription that is regulated by both protein phosphorylation and dephosphorylation in cultured tobacco cells. Plant J. 20 571–579. [DOI] [PubMed] [Google Scholar]
- Yang, S.F., and Hoffman, N.E. (1984). Ethylene biosynthesis and its regulation in higher-plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 35 155–189. [Google Scholar]
- Yasuta, T., Satoh, S., and Minamisawa, K. (1999). New assay for rhizobitoxine based on inhibition of 1-aminocyclopropane-1-carboxylate synthase. Appl. Environ. Microbiol. 65 849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuhashi, K., Ichikawa, N., Ezura, H., Akao, S., Minakawa, Y., Nukui, N., Yasuta, T., and Minamisawa, K. (2000). Rhizobitoxine production by Bradyrhizobium elkanii enhances nodulation and competitiveness on Macroptilium atropurpureum. Appl. Environ. Microbiol. 66 2658–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]