INTRODUCTION
To build a plant requires strict control of stem cell populations as well as specification of appropriate cell fates once cells enter a differentiation program. It is now accepted that both intracellular and intercellular signaling play important roles in controlling cell division patterns and cell specification (Westhoff et al., 1998; Scheres, 2001). The molecular nature of this signaling, however, has long been elusive. Recent molecular and genetic studies have begun to reveal the signaling mechanisms that regulate cell differentiation in both shoot and root. In this review, we discuss recent advances in the areas of stem cell control in both vegetative and floral meristems as well as in pattern formation in roots. Our knowledge of the molecular mechanisms underlying these processes has been obtained primarily from Arabidopsis. Whether common mechanisms operate in other species awaits experimental proof by the isolation and characterization of orthologous genes.
SIGNALING AT THE TOP END: MAINTENANCE OF THE SHOOT APICAL MERISTEM
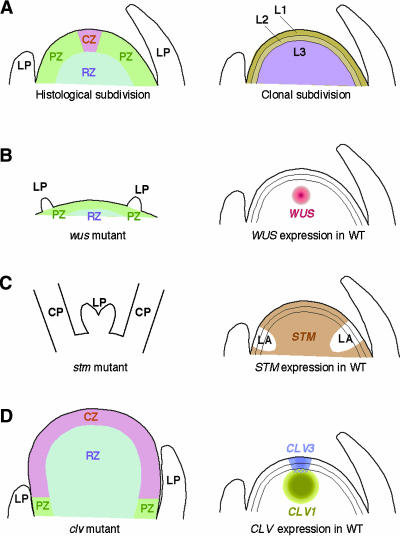
The shoot apical meristem (SAM) is the source of cells for all aerial organs produced after germination. The SAM is first formed at the globular stage of embryogenesis and develops into a dome shape in the mature embryo. Upon germination, the SAM starts a highly coordinated cell division program that continues throughout vegetative growth. Based on histological studies, the SAM of mature plants has been subdivided into three domains: the central zone, the peripheral zone, and the rib zone (Figure 1A, left) (for reviews, see Steeves and Sussex, 1989; Howell, 1998). As its name indicates, the central zone is a small, centrally located region toward the top of the SAM. The central zone contains a population of stem cells that divide relatively slowly. As these cells divide, their peripheral daughters are displaced gradually from the central zone and enter the peripheral and rib zones. Cells in the peripheral zone and the rib zone are rich in cytoplasm and divide rapidly. Later, these cells are recruited into the cell division programs of lateral organs and the stem, respectively.
Figure 1.
Schemes Depicting Wild-Type and Mutant Arabidopsis SAM Structures and Expression Patterns of Key Regulatory Genes.
(A) Subdivision of SAM domains based on histological observations (left) and clonal relationships of cell layers (right).
(B) SAM structure of loss-of-function wus mutants (left) and wild-type WUS gene expression (right) (Laux et al., 1996; Mayer et al., 1998).
(C) SAM structure of strong loss-of-function stm mutants (left) and wild-type STM gene expression (right) (Barton and Poethig, 1993; Endrizzi et al., 1996; Long et al., 1996). In strong stm mutants, the region corresponding to the wild-type SAM is reduced to a small number of cells between the cotyledon petioles. Leaf primordia arise in the center of this region.
(D) SAM structure caused by a loss-of-function mutation in CLV1, CLV2, or CLV3 (left) and gene expression patterns of CLV1 and CLV3 (right) (Clark et al., 1993, 1995, 1997; Kayes and Clark, 1998; Fletcher et al., 1999).
CP, cotyledon petiole; CZ, central zone; LA, leaf anlagen; LP, leaf primordium; L1, L2, and L3, epidermal, subepidermal, and underlying layers, respectively; PZ, peripheral zone; RZ, rib zone; WT, wild type.
Cells in the SAM also can be grouped according to their clonal relationships: the epidermal, subepidermal, and underlying layers (Figure 1A, right) (Satina et al., 1940; reviewed by Steeves and Sussex, 1989). Cells in the epidermal and subepidermal layers divide in a plane perpendicular to the layers (anticlinal division). These cells are ultimately incorporated into the epidermal and subepidermal layers of lateral organs (i.e., leaves and floral organs). Cells in the underlying layer divide in a more complex manner, and their daughter cells differentiate into the inner tissues of lateral organs as well as into the pith in the stem. Despite clear clonal distinctions between the three layers, their cell divisions are highly coordinated with each other, indicating an intimate intercellular communication that allows programmed development of organs with fixed shape and size.
Key Signaling Components
Maintaining a strict balance between the number of stem cells and the programmed differentiation of their progeny is critical to SAM function. In Arabidopsis, a stem cell population persists even after the transition from vegetative to reproductive growth, allowing the inflorescence SAM to produce an indeterminate number of flowers on its flanks. The maintenance of stem cells requires the WUSCHEL (WUS) homeodomain transcription factor. Loss-of-function wus mutants have defects in the SAM at all developmental stages (Laux et al., 1996; Mayer et al., 1998). The wus vegetative shoot apex is flat and lacks intensely stained cells typically found in the wild-type SAM (Figure 1B, left). Leaf primordia are formed slowly and fail to develop into mature leaves. At later stages, inflorescence stems emerge from the flank of the defective shoot apex as well as from leaf axils, giving a “wuschel” (tousled) appearance to the mutants. From these observations, it has been proposed that the primary role of WUS is to confer stem cell fate (Laux et al., 1996; Mayer et al., 1998). Expression analyses revealed that WUS mRNA is localized to a few cells underlying the stem cells (Figure 1B, right). Therefore, WUS controls stem cell fate in a non-cell-autonomous manner (Mayer et al., 1998).
shoot-meristemless (stm) mutants exhibit defects in SAM formation/maintenance similar to those of wus (Barton and Poethig, 1993; Clark et al., 1996; Endrizzi et al., 1996). Plants with strong stm alleles fail to establish a SAM during embryogenesis (Figure 1C, left). Leaf primordia are formed occasionally between the fused cotyledon petioles, but they do not develop into typical rosette leaves. The STM gene encodes a putative transcription factor with a homeobox DNA binding domain (Long et al., 1996). STM is transcribed throughout the SAM except in the regions corresponding to incipient leaf primordia (Figure 1C, right). All of these features indicate that the primary function of STM is either to inhibit cells from entering differentiation programs or to promote cell proliferation in the center of the SAM (Endrizzi et al., 1996; Long et al., 1996).
Mutations in three CLAVATA loci (CLV1, CLV2, and CLV3) result in a phenotype opposite to those of wus and stm (Clark et al., 1993, 1995; Kayes and Clark, 1998). The SAM in the mutant embryo is slightly larger than that of wild-type embryos. During postembryonic growth, organ initiation is retarded, whereas the SAM gradually increases in size by accumulating undifferentiated cells (Figure 1D, left). These mutant phenotypes indicate a role for the CLV genes in restricting the size of the stem cell population. All three CLV genes have been identified, and their protein products likely constitute a single receptor–ligand complex (Figure 2), consistent with the three mutants having an almost identical phenotype in the SAM (Clark et al., 1997; Fletcher et al., 1999; Jeong et al., 1999). The CLV1 protein is a receptor-like kinase composed of a Leu-rich repeat–containing extracellular domain with putative receptor function and a cytoplasmic Ser kinase domain linked through a transmembrane domain (Clark et al., 1997). CLV2 is structurally similar to CLV1 but lacks a cytoplasmic kinase domain (Jeong et al., 1999). CLV3 encodes a small polypeptide that contains a putative signal sequence for secretion. Otherwise, CLV3 shows no apparent homology with other proteins with known biochemical functions (Fletcher et al., 1999).
Figure 2.
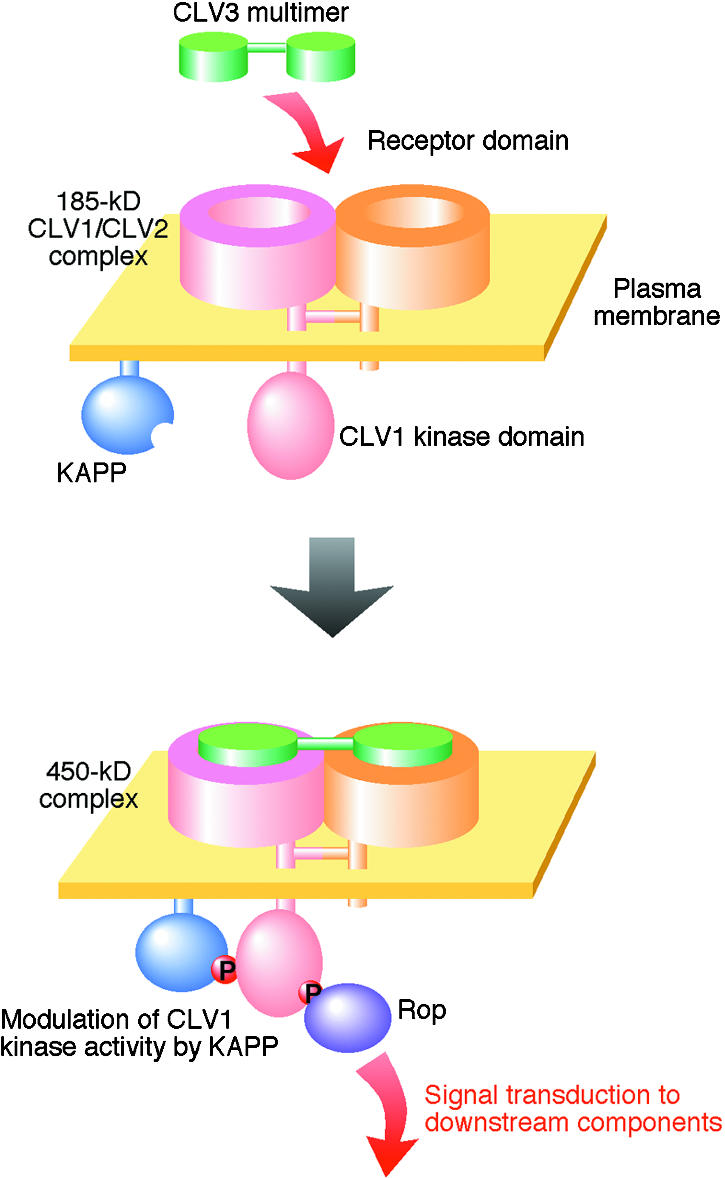
Predicted Signal Transduction Mechanism by the CLV Signaling Complex.
CLV1 and CLV2 form a 185-kD heterodimer via thioester bonds on the plasma membrane. Their N-terminal Leu-rich repeat (LRR) regions are considered to be facing outside the cell to form a receptor domain, whereas the C-terminal kinase domain is located in the cytoplasm. Here, the LRR region of each CLV monomer is assumed to take a cylindrical form, based on three-dimensional modeling of plant-specific LRRs to a known LRR crystal structure (Kajava, 1998). Free CLV3 ligand likely forms a multimer, although its molecular nature is not known (a homodimer-like structure is assumed in this scheme). Upon binding of the CLV3 ligand to the CLV1/CLV2 receptor, the CLV1 kinase domain is phosphorylated (P), probably by other CLV1/CLV2 complexes. The phosphorylated kinase domain then is recognized by several protein molecules, including a Rho GTPase-related protein (Rop) and a kinase associated protein phosphatase (KAPP), forming a 450-kD complex. Rop is presumed to act via a mechanism analogous to the mitogen-activated protein kinase cascade. KAPP is anchored to the inner surface of the plasma membrane through its uncleaved signal peptide. KAPP negatively regulates the Rop-mediated signal transduction pathway, probably through its phosphatase activity. The scheme is drawn based on the publications by Stone et al. (1994)(1998), Williams et al. (1997), and Trotochaud et al. (1999)(2000).
In both cauliflower and Arabidopsis, the CLV1 protein is present in two complexes of 185 and 450 kD (Trotochaud et al., 1999) (Figure 2). Convincing but indirect evidence indicates that the 185-kD complex is a disulfide-linked heterodimer of CLV1 and CLV2 (Trotochaud et al., 1999). The larger 450-kD complex includes the 185-kD complex, the CLV3 peptide, and at least two other noncovalently bound subunits: a kinase-associated protein phosphatase (KAPP) and a Rho GTPase–related protein (Rop) (Trotochaud et al., 1999, 2000). KAPP is likely to be a modulator of CLV1 kinase activity, whereas Rho may participate in the downstream signal transduction pathway analogous to the animal mitogen-activated protein kinase cascade (Hirt, 1997; Williams et al., 1997; Stone et al., 1998; Trotochaud et al., 1999). Assembly of the 450-kD complex requires the CLV3 peptide, because CLV1 was found exclusively in the 185-kD complex in strong clv3 mutants (Trotochaud et al., 1999). Furthermore, a CLV1/CLV2 receptor expressed on the yeast cell surface has been shown to bind CLV3 from a cauliflower extract (Trotochaud et al., 2000). All of these observations are consistent with a model in which the CLV3 peptide binds to and activates the 185-kD CLV1/CLV2 heterodimer through autophosphorylation, which then becomes a 450-kD complex that includes KAPP and Rop (Figure 2) (Trotochaud et al., 1999). In cauliflower, 76% of CLV3 was found in the 450-kD complex, whereas the remaining 24% was found in a 25-kD multimer (Trotochaud et al., 2000). It is not known, however, what CLV3 partner is in the 25-kD multimer or whether CLV3 binds to the CLV1/CLV2 receptor as a multimer or as a monomer.
CLV1 and CLV3 are expressed in distinct regions of the SAM (Figure 1D, right). Although CLV3 mRNA accumulates specifically in the stem cells in the central zone, CLV1 is expressed in the center of the rib zone. The CLV1 expression domain overlaps that of CLV3 only slightly (Clark et al., 1997; Fletcher et al., 1999). This is consistent with the possible extracellular secretion of the CLV3 peptide (Fletcher et al., 1999): CLV3 may be secreted from the stem cells and perceived by the CLV1-expressing cells just below the stem cells. CLV2 is expressed in most organs and may have additional roles in other signaling pathways (Kayes and Clark, 1998; Jeong et al., 1999).
Stem Cell Maintenance
How do the key regulatory genes act in maintaining the SAM? Recent molecular genetic studies have revealed interdependence between the WUS and CLV pathways (Brand et al., 2000; Schoof et al., 2000) (Figure 3A). First, the CLV signal downregulates WUS expression. This is based on the observations that (1) the size of the WUS expression domain is enlarged in clv mutant backgrounds (Schoof et al., 2000); and (2) ectopic expression of CLV3 eliminates WUS-expressing cells, thereby causing a wus-like phenotype (Brand et al., 2000). Second, because CLV3 expression is specific to the stem cells, whose maintenance requires WUS, the CLV pathway may be regulated indirectly by WUS (Laux et al., 1996; Mayer et al., 1998). This has been confirmed by the ectopic expression of WUS. Transgenic plants expressing WUS under either the CLV1 or AINTEGUMENTA (ANT) promoters accumulate undifferentiated cells that express CLV3 (Schoof et al., 2000).
Figure 3.
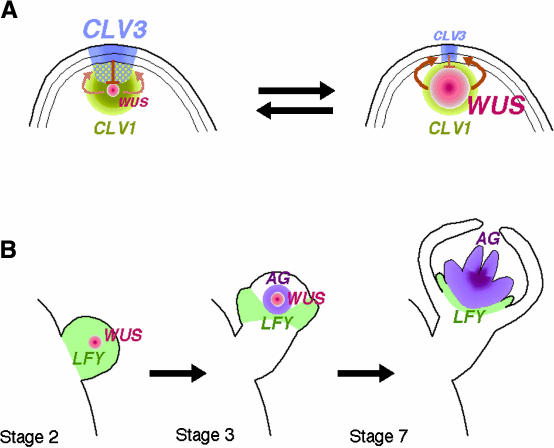
Models of Stem Cell Regulation.
(A) Stem cell maintenance in the vegetative and inflorescence meristems (Brand et al., 2000; Schoof et al., 2000). The size of the stem cell population is controlled by a regulatory loop between WUS and CLV. When the number of stem cells is increased, more CLV3 ligand is released from the stem cells, which is perceived by the CLV1/CLV2 receptor kinase in underlying layers. This results in fewer cells expressing WUS, thereby attenuating stem cell–promoting activity (left). In contrast, when the number of stem cells is decreased, less CLV3 ligand is released. Consequently, more cells start to express WUS, thereby promoting stem cell identity (right).
(B) Termination of the stem cell population in a FM (Lenhard et al., 2001; Lohmann et al., 2001). In early flower primordia (stage 2, left), the meristem identity gene LFY is expressed throughout the primordia (Weigel et al., 1992), whereas WUS is expressed in the center to reserve a stem cell population for later developmental stages (Mayer et al., 1998). Later, in stage 3 (middle), LFY and WUS together activate AG expression in the center of the primordia. As flower development proceeds (stage 7, right), WUS expression becomes downregulated by AG. AG expression persists in the center of the primordia and determines floral organ identities in whorls 3 and 4 (Drews et al., 1991).
Based on this relationship, a mechanism involving a self-regulatory loop has been proposed for stem cell maintenance (Figure 3A). If the number of stem cells is increased in the central zone, more CLV3 peptide is produced, which then signals the CLV1-expressing cells in the rib zone to downregulate WUS expression. Fewer WUS-expressing cells would reduce the number of stem cells. Conversely, if too few stem cells are left in the SAM, signaling by CLV3 would be attenuated, which would lead to more cells expressing WUS and hence more stem cells (Schoof et al., 2000). In ANT promoter::WUS transgenic plants, WUS expression was uncoupled from the self-regulatory loop, resulting in a phenotype similar to that of loss-of-function clv mutants, despite ectopic CLV3 expression.
Both WUS and CLV3 can signal across cell layers. For CLV3 signaling, this probably is accomplished by apoplastic movement of the CLV3 peptide (Fletcher et al., 1999). The molecular mechanism for the non-cell-autonomous action of WUS is not yet known. Likewise, it is not clear how STM acts in relation to the WUS/CLV regulatory pathway. Based on its expression pattern in the mature SAM, STM appears to reserve the region composed of undifferentiated cells upon which WUS and CLV expression patterns are specified. During embryogenesis, however, WUS expression can be detected as early as the 16-cell-embryo stage, when neither STM expression nor a visible SAM structure has been established (Mayer et al., 1998). Furthermore, neither WUS nor STM expression in the embryo requires the function of the other (Mayer et al., 1998). Double mutant analyses have demonstrated that although either the stm or the clv phenotype can be rescued partially by mutation of the other, the wus mutation enhances the defects caused by weak stm alleles (Clark et al., 1996; Endrizzi et al., 1996). These observations indicate that the WUS/CLV signaling pathway and STM act at different levels but are not correlated in a simple epistatic order. Moreover, their interdependence may change depending on the developmental stage.
When Stem Cells Come to an End
The young flower primordium retains stem cells at its apex; therefore, it is called a floral meristem (FM). Both mutant phenotypes and gene expression patterns indicate that the WUS/CLV signaling pathway acts to maintain stem cells in the FM. wus mutant flowers form normal numbers of sepals and petals, but the central two whorls are replaced with a single stamen (Laux et al., 1996). This defect results from the inability of wus to maintain a sufficient quantity of stem cells to form the correct numbers of stamens and carpels. In contrast, the FM of clv mutants accumulates stem cells and gives rise to a flower with increased numbers of floral organs, especially carpels (Clark et al., 1993, 1995; Kayes and Clark, 1998). WUS is expressed in a few cells in the center of the FM, whereas CLV1 and CLV3 are expressed in the center and apex of the FM, respectively, similar to their expression patterns in the SAM (Clark et al., 1997; Mayer et al., 1998; Fletcher et al., 1999).
In the wild-type FM, the ability of the stem cells to maintain a constant population size must end late in flower development, because the central part of the FM is programmed to become a determinate number of carpels. The AGAMOUS (AG) gene has been implicated in this process because, in addition to having defects in floral organ specification, ag mutant flowers produce indeterminate numbers of floral organs (Bowman et al., 1989). This phenotype requires functional WUS, because flowers of ag wus double mutants show defects similar to those of wus single mutants (Laux et al., 1996).
Recent genetic analyses by two groups have demonstrated a role for AG in the regulation of WUS (Lenhard et al., 2001; Lohmann et al., 2001). In the wild-type FM, expression of both WUS and CLV3 diminishes as flower development proceeds and then disappears completely by the time carpel primordia initiate. In the FM of ag mutants, both WUS and CLV expression remain long after floral organ development is completed (Lenhard et al., 2001; Lohmann et al., 2001). The repression of WUS by AG appears to be independent of CLV signaling, because in the FM of ag clv1 double mutants, WUS expression is not only prolonged but expanded spatially (Lohmann et al., 2001). Conversely, the ectopic expression of WUS in various regions of the FM results in indeterminate organ formation in the corresponding floral region (Lenhard et al., 2001; Lohmann et al., 2001). All of these observations indicate a requirement for AG in the downregulation of WUS and hence in the termination of stem cell maintenance.
The involvement of AG in stem cell termination raises the question of how AG expression is induced in the FM at the correct time and place. There is strong evidence that AG expression depends on WUS and on the floral meristem identity gene LEAFY (LFY) (Lenhard et al., 2001; Lohmann et al., 2001). The ectopic expression of WUS results in the transcription of a β-glucuronidase (GUS) reporter controlled by the AG cis regulatory region. This is consistent with the ability of ectopic WUS to form an enlarged stem cell population and an enhanced “C” function mediated by AG, as shown by an indeterminate number of stamens and carpels. Both GUS expression and the indeterminate organ formation are largely absent when the same experiments are performed in a lfy mutant background. The interaction of WUS/LFY and AG was investigated further at the molecular level. Both in vitro and in vivo studies demonstrated direct binding of LFY and WUS to the cis elements in the AG second intron (Busch et al., 1999; Lohmann et al., 2001). A quantitative analysis using a yeast expression system indicated that WUS and LFY act synergistically to activate AG expression, even though cooperative binding of the two proteins was not observed in vitro (Lohmann et al., 2001).
Based on these observations, a signaling pathway for stem cell termination has been proposed (Lenhard et al., 2001; Lohmann et al., 2001) (Figure 3B). In young flower primordia, the WUS/CLV signaling pathway maintains a stem cell population in the apex. WUS then activates AG transcription by binding directly to the AG promoter. The transcription of AG by WUS is enhanced synergistically by LFY, which is expressed in flower primordia but not in the SAM. Because WUS expression is limited to a small number of cells in the FM, strong AG expression occurs only at the center of the flower primordium, where it specifies stamen and carpel identities. Later in flower development, AG downregulates WUS expression, thereby terminating the stem cell population. Thus, WUS and AG constitute a negative feedback loop in which WUS activates the transcription of AG, which in turn represses WUS expression. This signaling loop is similar to the WUS/CLV regulatory loop in the SAM. An important difference is that the WUS/AG pathway does not involve cell-to-cell communication. One should note, however, that the WUS/AG pathway requires an existing prepattern, as does the WUS/CLV pathway. Furthermore, in contrast to the WUS/CLV pathway, which continues in the SAM throughout plant development, the WUS/AG pathway appears to function only once in each FM.
SIGNALING UNDER THE GROUND: CELL DIVISION AND DIFFERENTIATION IN THE ROOT
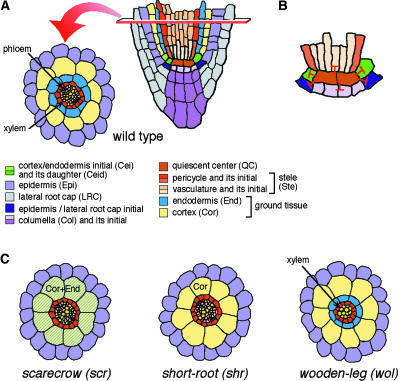
Compared with the SAM, the root meristem (RM) has fewer cells and a simpler structure. In Arabidopsis, the cell division pattern in the RM has been well characterized, and a nearly complete fate map can be drawn for every cell type (Dolan et al., 1993). The mature part of the Arabidopsis root is composed of concentrically organized cell layers, which from inside to outside form vasculature, pericycle, endodermis, cortex, and epidermis (Figure 4A). The vasculature and pericycle together constitute the stele. Cells in each layer have their origin in the “initial cells” located at the RM, which repeat a highly stereotyped sequence of divisions. One of the two daughter cells remains as an initial cell, whereas the other enters an appropriate differentiation pathway. Some of the root cell files share the same clonal origin; for example, epidermis and lateral root cap originate from the same initial cells, and the two ground tissue layers, endodermis and cortex, also share the same initial cells (Figure 4A).
Figure 4.
Schemes of Wild-Type and Mutant Arabidopsis Root Structures.
(A) Wild-type root (Dolan et al., 1993). Cell types are given in the key at bottom. Abbreviations shown in parentheses are used in all subsequent figures.
(B) QC cells function as an organizing center of the RM by inhibiting the differentiation of surrounding initial cells (stem cells) (van den Berg et al., 1997; Umeda et al., 2000).
(C) Defective root radial pattern of three Arabidopsis mutants, scr, shr, and wol (Benfey et al., 1993; Di Laurenzio et al., 1996; Helariutta et al., 2000; Mähönen et al., 2000).
Intercellular Signaling in Root Development
The highly coordinated division of the initial cells suggests the presence of extensive cell-to-cell communication. A laser-ablation study first gave solid evidence for the importance of positional signaling in root pattern formation (van den Berg et al., 1995). When a cortex/endodermis initial cell is ablated, an adjacent pericycle cell invades the ablated position and performs a periclinal division. The outer daughter cell then behaves as a cortex/endodermis initial cell: it undergoes a transverse cell division. The upper daughter cell then divides periclinally, giving rise to the first cells in the cortex and endodermis lineages. These pericycle-derived endodermal cells have differentiated attributes of endodermis, as revealed by the presence of a casparian strip. A similar invasion by neighboring cells, followed by a corresponding cell fate change, was observed when an epidermis/lateral root cap initial cell was ablated (van den Berg et al., 1995).
Respecification of cell fate is not limited to mechanical ablation but seems to occur in nature. Kidner et al. (2000) investigated root cell lineage using genetic mosaics of GUS-positive cells that had been generated by heat shock–induced transposon excision from a CaMV35S::GUS transgene. Analysis of the roots after heat shock treatment revealed that the cells in the cortex/endodermis lineage invaded both the outer epidermis and inner stele layers. Surprisingly, those roots retained a normal root radial pattern with no apparent increase or decrease in the number of cell layers or in the number of cells in each layer. Although heat shock treatment may have enhanced the frequency of cell death, this observation demonstrated that the root radial pattern is not perturbed even when cells in the meristem region are lost accidentally. Clearly, intercellular signaling is key to the self-maintenance capacity of the RM.
Stem Cell Maintenance in the RM
The root initial cells can be thought of as functionally equivalent to the CLV3-expressing stem cells of the SAM. The initial cells are arranged around a few mitotically inactive cells in the center of the RM that constitute a “quiescent center” (QC). The QC appears to play an important role in maintaining RM activity. When a QC cell is laser ablated, the abutting initial cells lose their ability to perform as stem cells and, instead, start to differentiate or divide in a manner characteristic of their daughter cells (van den Berg et al., 1997). Differentiation of initial cells also was observed when laser ablation was performed on a mutant that lacks postembryonic root cell divisions, showing that this function of the QC is independent of cell divisions (van den Berg et al., 1997). These observations indicate that the QC maintains the identity of the surrounding initial cells by inhibiting their differentiation (Figure 4B). A similar conclusion was drawn from a study in which root cell differentiation was promoted genetically through modulation of the expression level of CAK, a cyclin-dependent kinase–activating kinase (Umeda et al., 2000). The ability of the QC to inhibit initial cell divisions is similar to the function of the WUS-expressing cells in the SAM. An important difference, however, is that WUS-expressing cells appear to be replaced continuously, whereas QC cells are maintained for a long period.
Although a few loci have been reported to affect the formation and activity of the RM (Willemsen et al., 1998; Vernoux et al., 2000; Frugier et al., 2001), no putative signaling components have been identified, nor are the root counterparts for WUS and CLV documented. This implies that cell division and the differentiation of shoot and root are controlled by different mechanisms, although both include intercellular signaling to maintain the stem cell population. Alternatively, root stem cells may be maintained by redundant pathways that to date have avoided identification through genetic analyses.
Auxin Signaling and Root Distal Patterning
The plant hormone auxin influences cell division and differentiation as well as cell elongation. Because of its highly pleiotropic effects, however, it has been difficult to determine the role of auxin in each developmental program by genetic approaches. Recently, a physiological study linked the distribution of auxin to patterning of the root apical region (Sabatini et al., 1999). Using a GUS reporter gene fused to a synthetic auxin-responsive promoter (DR5::GUS), a maximum of auxin concentration was localized to the columella initial cells. This auxin distribution was either lost or disturbed in mutants of known auxin signal transducers and transporters. In mutants of the putative auxin efflux carrier PIN-FORMED1, abnormal cell division and elongation were found in the tissue in which the auxin concentration was increased ectopically.
A more dramatic change in root pattern was induced when wild-type plants were grown in the presence of auxin transport inhibitors. The auxin concentration maximum was expanded to include the cortex and apical epidermis together with the original maximum at the columella initials. The change in auxin distribution was accompanied by respecification of cell fates and modification of cell division programs: the cells in the positions of endodermis, cortex, and epidermis were respecified as QC, columella initial, and lateral root cap, respectively. The new columella initials divided, and their daughter cells contained amyloplasts, a functional marker of the columella. Cell fate specification was not dependent on the absolute concentration of auxin but was determined by the position of the auxin concentration maximum relative to the vascular tissue (Sabatini et al., 1999). Similarly, misspecification of epidermis cells as lateral root cap cells has been reported for tornado1 (trn1) and trn2 mutants (Cnops et al., 2000). Although the mutated gene has yet to be identified, TRN1 is likely to be involved in the auxin-dependent cell fate determination pathway, because polar auxin transport was impaired in a trn1 allele (lop1) (Carland and McHale, 1996). These observations strongly suggest a role for auxin distribution in root distal patterning.
Radial Signaling in Ground Tissue Patterning
The radial pattern of cell layers seen in the mature root is determined initially during early embryogenesis. Upon germination, the initial cells in the RM are activated and start a stereotyped cell division sequence that maintains a radial pattern identical to that formed in the embryo. Two lines of experimental data have suggested that the correct radial pattern formation requires a “top-down” flow of positional information from mature cells to the initial cells. First, mutants with defective radial patterns have analogous defects in the embryo and the mature root (Scheres et al., 1995). Second, when intercellular communication between a cortex/endodermis initial cell and the more mature cells above it is blocked by laser ablation, the initial cell ceases to divide correctly (van den Berg et al., 1995). Postembryonic induction experiments with a key regulatory gene, however, indicate that top-down signaling is not essential for ground tissue patterning (our unpublished results).
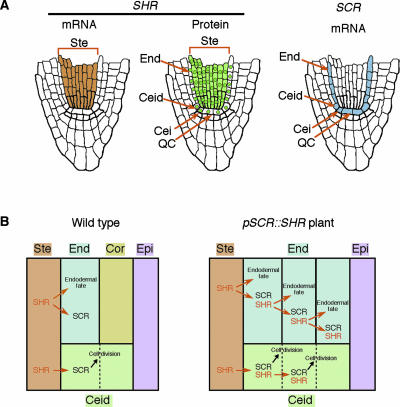
In contrast to the top-down model, analysis of two Arabidopsis mutants, scarecrow (scr) and short-root (shr), has provided compelling evidence for a radial flow of information during root patterning. Both scr and shr loss-of-function mutants lack asymmetric cell division of the cortex/endodermis initial daughter cell, resulting in a single ground tissue layer in place of the normal two layers of cortex and endodermis (Figure 4C) (Benfey et al., 1993; Di Laurenzio et al., 1996; Helariutta et al., 2000). The two mutants differ in that the single layer of scr has differentiated attributes of both cortex and endodermis, whereas that of shr has only cortex characteristics (Figure 4C). Therefore, SCR is necessary for correct cell division of the cortex/endodermis initial daughter cell, whereas SHR is required for both cell division and endodermal cell fate specification. SCR and SHR encode putative transcription factors that belong to the same plant-specific GRAS family (Di Laurenzio et al., 1996; Pysh et al., 1999; Helariutta et al., 2000). Importantly, the SCR and SHR genes are transcribed in mutually exclusive but adjacent layers in the root (Figure 5A).
Figure 5.
Radial Patterning of the Root Ground Tissue.
(A) Localization of SHR mRNA (left), SHR protein (middle), and SCR mRNA (right) in the RM (Di Laurenzio et al., 1996; Helariutta et al., 2000; Wysocka-Diller et al., 2000; Nakajima et al., 2001).
(B) Mechanism of intercellular signaling in root radial pattern formation of wild-type (left) and pSCR::SHR transgenic plants (right) (Nakajima et al., 2001). In wild-type roots (left), the SHR protein is produced in the stele and appears to move to a single adjacent layer, probably through plasmodesmata (red arrows). In the adjacent layer, SHR activates SCR transcription that is essential for the asymmetric cell division of the cortex/endodermis initial daughter (Ceid) cells, resulting in the separation of cortex and endodermis layers. In the mature region, SHR in the adjacent layer confers endodermal cell fate. SHR protein movement is limited to a single cell distance by an as yet unknown mechanism. In pSCR::SHR transgenic plants (right), intercellular SHR signaling is considered to be reinforced repeatedly by the production of SHR protein from the pSCR::SHR transgene. This results in the production of supernumerary layers because of repeated SCR-mediated Ceid divisions and the acquisition of endodermal cell fate in these layers.
Abbreviations for the cell types are given in Figure 4A.
SCR is transcribed in the endodermis as well as in the QC, cortex/endodermis initial, and its daughter (Di Laurenzio et al., 1996; Helariutta et al., 2000). In contrast, SHR is transcribed in the entire stele, including stele initials and pericycle (Helariutta et al., 2000). This expression pattern indicates non-cell-autonomous action of SHR in two processes. First, SHR controls the differentiation of the endodermis, where SHR is not transcribed. Second, SHR is necessary for the correct division of the cortex/endodermis initial daughter cells, which again do not transcribe SHR. Although the downstream targets of SHR for endodermal specification are unknown, there is strong evidence that SHR acts through SCR to effect the division of the cortex/endodermis initial daughter cell (Helariutta et al., 2000; Nakajima et al., 2001).
What is the mechanism of SHR's non-cell-autonomous activity? Most likely, it is dependent on intercellular protein movement (Nakajima et al., 2001). Expression of an SHR::green fluorescent protein (GFP) fusion protein in the shr mutant under the control of the SHR promoter results in complete rescue of the root radial pattern, indicating that the fusion protein is fully functional. In these transgenic roots, GFP fluorescence clearly is localized to the nuclei of the endodermis, the QC, and the cortex/endodermis initial and its daughter, in addition to the nuclei and cytoplasm of the stele cells (Figure 5A). These cells adjacent to the stele are located precisely where mutations in SHR have their effects. The localization of the SHR protein was confirmed by the use of antibodies specific to SHR. The difference in the mRNA and protein localization have led to a model in which SHR transmits positional information from the stele to a single outer layer by its own movement (Figure 5B, left) (Nakajima et al., 2001). Because SCR seems to function in the same cell types in which the gene is transcribed (our unpublished results), intercellular movement does not appear to be a general feature of GRAS proteins. Because SHR is a putative transcription factor and is localized to the nuclei of the adjacent layer, it is likely that SHR is involved directly in the transcriptional control of downstream effector genes.
Although direct transcriptional targets for SHR have yet to be identified, SCR is a good candidate. Because SCR expression is reduced greatly in shr mutants, its transcription must be controlled directly or indirectly by SHR. The single outer layer to which SHR protein appears to move matches exactly the SCR-transcribing cell types (Figure 5A). Consistent with this finding, transgenic plants expressing SHR under the SCR promoter (pSCR::SHR plants) have indeterminate proliferation of the endodermis layers that transcribe SCR (cf. Figures 6A and 6E) (Nakajima et al., 2001). The origin of this phenotype appears to be autocatalytic signal reinforcement, in which SHR activates the SCR promoter in the adjacent layer, which then produces the SHR protein from the pSCR::SHR transgene (Figure 5B, right).
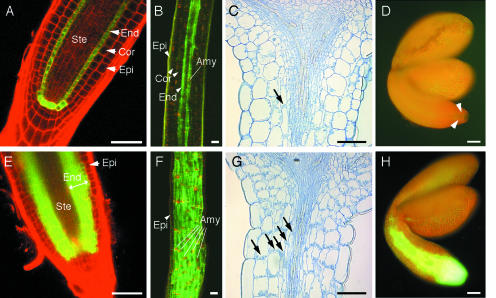
Figure 6.
Comparison of Wild-Type and pSCR::SHR Transgenic Plants.
(A) to (D) Wild type.
(E) to (H) pSCR::SHR transgenic plants.
(A) and (E) Confocal images of roots. Red indicates propidium iodide staining of cell walls. Green indicates GFP fluorescence showing the site of SCR transcription.
(B) and (F) Confocal images of dark-grown hypocotyls. Red indicates autofluorescence of plastids. Green indicates GFP fluorescence showing SCR transcription.
(C) and (G) Longitudinal sections of light-grown hypocotyls. Arrows indicate amyloplasts sedimenting toward the gravity vector. Sedimenting amyloplasts occur specifically in the endodermis of wild-type plants ([C], arrow), whereas they are found in all layers between the stele and the epidermis of transgenic plants ([G], arrows).
(D) and (H) Epifluorescence images of mature embryos. Green represents the site of SCR transcription. In the wild-type embryo, GFP fluorescence is barely detected above the strong red autofluorescence ([D], arrowheads), whereas strong GFP fluorescence is found in the transgenic embryo (H).
Amy, amylose. Abbreviations for the cell types are given in Figure 4A. Bars = 50 μm.
SHR protein movement is not limited to the endodermis but also is directed to the QC (Nakajima et al., 2001). Consistent with this finding, the pSCR::SHR transgenic roots have supernumerary layers of QC, suggesting that, depending on position, SHR specifies not only the endodermis but also QC cell fate (Nakajima et al., 2001).
A Common Mechanism in Root and Shoot Radial Patterning
Radial symmetry is not limited to the root but is found in most plant organs. In Arabidopsis, hypocotyls have a similar radial pattern to that of the root, except that hypocotyls have two layers of cortex instead of the single layer found in the root. Inflorescence stems also have a similar radial pattern, except that the vascular bundles are positioned circumferentially around the central pith tissue. How common are the mechanisms that control root and shoot radial patterning? SCR is transcribed in the cognate layers in roots, stems, and hypocotyls (Wysocka-Diller et al., 2000). Roots and stems of scr mutants show analogous radial pattern defects. Furthermore, scr mutant embryos lack asymmetric cell division in the early heart-stage embryo that first separates the cortex and endodermis cell lineages (Scheres et al., 1995). This results in a mutant hypocotyl with two ground tissue layers instead of the normal three (Scheres et al., 1995; Fukaki et al., 1998).
In the shoot, lack of an endodermis layer results in an inability to respond to gravity, because the shoot endodermis possesses amyloplasts that sediment according to the gravity vector, thereby acting as a gravity-sensing “statolith” (Fukaki et al., 1998). pSCR::SHR plants show very similar transgenic phenotypes in hypocotyls, embryos, and roots. The hypocotyls of the pSCR::SHR plants have supernumerary ground tissue layers similar to those seen in roots. In these supernumerary layers, amyloplasts sediment toward the gravity vector (Figures 6F and 6G). SCR expression also is increased in the transgenic hypocotyl and embryo, in a pattern similar to that seen in the root (Figures 6F and 6H) (Nakajima et al., 2001). All of these observations indicate that a common mechanism operates in ground tissue patterning in both root and shoot.
Root Vascular Patterning
In contrast to the epidermis and ground tissue, the formation of the vascular cylinder requires more complex patterning, including specification of multiple cell types with specialized functions, such as xylem and phloem (Figure 4A). In Arabidopsis, the average number of stele initials is 11, whereas ∼31 cells are seen in cross-sections from mature regions, indicating that more than two formative divisions must take place among the progeny of each initial (Mähönen et al., 2000).
Mutation of the WOODEN-LEG (WOL) gene results in a reduced number of cells in the vascular cylinder of roots and hypocotyls, and all root vascular cells differentiate into protoxylem (Figure 4C, right). Genetic analyses have indicated that WOL controls cell divisions but not cell differentiation. When cell divisions in wol are promoted by the epistatic mutation fass, the root vasculature of the wol fass double mutant produces the full range of cell types (Scheres et al., 1995; Mähönen et al., 2000). WOL encodes a novel two-component His kinase, which recently has been shown to be allelic to the cytokinin receptor CRE1 (Inoue et al., 2001). WOL/CRE1 is expressed in all cells in the root vascular cylinder as well as in the procambium of the embryo (Mähönen et al., 2000). Therefore, WOL/CRE1 has been hypothesized to sense extracellular cytokinin on the vascular cell surface and to transmit a signal to the nucleus (Hwang and Sheen, 2001; Inoue et al., 2001). The signal is thought to act ultimately to promote vascular cell divisions, allowing the differentiation of various cell types.
In root vascular development, the specification of xylem cells precedes that of other vascular cell types, although phloem differentiation becomes visible first (Esau, 1977; Bowman, 1994; Mähönen et al., 2000). Therefore, the wol phenotype has been attributed to a failure to produce a sufficient number of cells that can accommodate cell types other than xylem. To date, little is known about how xylem cell fate is specified in the root vascular cylinder. Numerous studies on leaf vein patterning have suggested that auxin plays a major role in vein pattern formation (for review, see Dengler and Kang, 2001). The specification of root xylem cells just below the existing xylem poles appears to support signaling from the mature xylem tissue. In contrast, classic dissection studies suggested that the root xylem pattern is determined autonomously by the RM (for review, see Raghaven, 2000). These opposing models can be tested by manipulating the number of stele cells along the root axis, possibly through the induction or repression of WOL function.
CONCLUDING REMARKS
Past studies based on physiological and genetic analyses have highlighted the importance of positional information in plant development. The molecular nature of cell-to-cell communication, however, has long been elusive. Components of the predicted signaling networks are beginning to emerge in many developmental studies, and some of these are now understood at the molecular level. In shoot stem cell maintenance, a ligand–receptor interaction appears to constitute an important part of the intercellular signaling pathway, whereas transcription factor movement appears to be responsible for ground tissue patterning. The presence of a large number of CLV3 homologs expressed in a variety of organs suggests that similar ligand–receptor interactions operate in other organs outside the SAM (Cock and McCormick, 2001). On the other hand, a number of transcription factors have been reported to move across cell layers in the SAM and flower organs, although the developmental significance of this movement is obscure (Jackson et al., 1994; Perbal et al., 1996; Sessions et al., 2000). The relative importance of intercellular signaling by ligand–receptor interactions versus protein movement in plant development will be revealed in future studies.
The characterization of various patterning processes has emphasized the importance of plant hormones, especially auxin. In Arabidopsis, cellular auxin levels are thought to be controlled primarily by polar auxin transport, which in turn depends on the distribution of auxin influx and efflux carriers (Estelle, 1998). Not only are efflux carriers expressed in different cell types, but the proteins are targeted to different cell surfaces (Galweiler et al., 1998; Muller et al., 1998). Therefore, cell fate changes caused by modified auxin distribution could lead to redistribution of the efflux carriers, thereby affecting local auxin transport. This may act to maintain auxin homeostasis: a given auxin distribution could stabilize a transporter profile, thus causing the distribution pattern to be perpetuated. Because the Arabidopsis genome contains at least 18 potential auxin carrier genes (Swarup et al., 2000), it is conceivable that auxin distribution in the plant is determined by a complex expression pattern of many transporter genes in combination with their intracellular protein localization. Reverse genetic analysis for each of these genes will contribute to a comprehensive understanding of the role of auxin in plant developmental patterning. It also may lead to an understanding of the genes that control carrier protein expression, which must be responsible for setting up a prepattern of primary importance.
In the past decade, key regulatory genes have been identified based on visible phenotypic alterations. These genes now are undergoing detailed functional studies. Once a major foundation of a signaling pathway is clarified, more elaborate screening procedures can be designed to search specifically for other molecules in the same signaling pathway. Reverse genetic approaches are facilitated greatly by the availability of the entire Arabidopsis genome sequence and recent technical advances in the production of desired knockouts. Microarray analyses also can be used to identify downstream components of signaling pathways. In the next few years, we will obtain a much clearer view of the signaling networks that underlie plant development.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010471.
References
- Barton, M.K., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119 823–831. [Google Scholar]
- Benfey, P.N., Linstead, P.J., Roberts, K., Schiefelbein, J.W., Hauser, M.-T., and Aeschbacher, R.A. (1993). Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development 119 57–70. [DOI] [PubMed] [Google Scholar]
- Bowman, J. (1994). ARABIDOPSIS: An Atlas of Morphology and Development. (New York: Springer-Verlag).
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289 617–619. [DOI] [PubMed] [Google Scholar]
- Busch, M.A., Bomblies, K., and Weigel, D. (1999). Activation of a floral homeotic gene in Arabidopsis. Science 285 585–587. [DOI] [PubMed] [Google Scholar]
- Carland, F.M., and McHale, N.A. (1996). LOP1: A gene involved in auxin transport and vascular patterning in Arabidopsis. Development 122 1811–1819. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121 2057–2067. [Google Scholar]
- Clark, S.E., Jacobsen, S.E., Levin, J.Z., and Meyerowitz, E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122 1567–1575. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 575–585. [DOI] [PubMed] [Google Scholar]
- Cnops, G., Wang, X., Linstead, P., Van Montagu, M., Van Lijsebettens, M., and Dolan, L. (2000). Tornado1 and tornado2 are required for the specification of radial and circumferential pattern in the Arabidopsis root. Development 127 3385–3394. [DOI] [PubMed] [Google Scholar]
- Cock, J.M., and McCormick, S. (2001). A large family of genes that share homology with CLAVATA3. Plant Physiol. 126 939–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler, N., and Kang, J. (2001). Vascular patterning and leaf shape. Curr. Opin. Plant Biol. 4 50–56. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio, L., Wysocka-Diller, J., Malamy, J., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M., Feldmann, K., and Benfey, P. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86 423–433. [DOI] [PubMed] [Google Scholar]
- Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., and Scheres, B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119 71–84. [DOI] [PubMed] [Google Scholar]
- Drews, G.N., Bowman, J.L., and Meyerowitz, E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65 991–1002. [DOI] [PubMed] [Google Scholar]
- Endrizzi, K., Moussian, B., Haecker, A., Levin, J.Z., and Laux, T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10 967–979. [DOI] [PubMed] [Google Scholar]
- Esau, K. (1977). The root: Primary state of growth. In Anatomy of Seed Plants. (New York: John Wiley & Sons), pp. 215–242.
- Estelle, M. (1998). Polar auxin transport: New support for an old model. Plant Cell 10 1775–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, J., Brand, U., Running, M., Simon, R., and Meyerowitz, E. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 1911–1914. [DOI] [PubMed] [Google Scholar]
- Frugier, F., Folmer, S., Blilou, I., Willemsen, V., Wolkenfelt, H., Ferreira, P., and Scheres, B. (2001). HOBBIT, a component of the APC involved in control of cell division and cell fate (abstr. 179). In 12th International Conference on Arabidopsis Research.
- Fukaki, H., Wysocka-Diller, J., Kato, T., Fujisawa, H., Benfey, P.N., and Tasaka, M. (1998). Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 14 425–430. [DOI] [PubMed] [Google Scholar]
- Galweiler, L., Guan, C., Müller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230. [DOI] [PubMed] [Google Scholar]
- Helariutta, Y., Fukaki, H., Wysocka-Diller, J., Nakajima, K., Jung, J., Sena, G., Hauser, M., and Benfey, P. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101 555–567. [DOI] [PubMed] [Google Scholar]
- Hirt, H. (1997). Multiple roles of MAP kinases in signal transduction in plants. Trends Plant Sci. 2 11–15. [Google Scholar]
- Howell, S.H. (1998). Molecular Genetics of Plant Development. (Cambridge, UK: Cambridge University Press).
- Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413 383–389. [DOI] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409 1060–1063. [DOI] [PubMed] [Google Scholar]
- Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 405–413. [Google Scholar]
- Jeong, S., Trotochaud, A.E., and Clark, S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava, A.V. (1998). Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 277 519–527. [DOI] [PubMed] [Google Scholar]
- Kayes, J.M., and Clark, S.E. (1998). CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125 3843–3851. [DOI] [PubMed] [Google Scholar]
- Kidner, C., Sundaresan, V., Roberts, K., and Dolan, L. (2000). Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta 211 191–199. [DOI] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 87–96. [DOI] [PubMed] [Google Scholar]
- Lenhard, M., Bohnert, A., Jurgens, G., and Laux, T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105 805–814. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., Hong, R.L., Hobe, M., Busch, M.A., Parcy, F., Simon, R., and Weigel, D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105 793–803. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Mähönen, A.P., Bonke, M., Kauppinen, L., Riikonen, M., Benfey, P.N., and Helariutta, Y. (2000). A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. [DOI] [PubMed] [Google Scholar]
- Muller, A., Guan, C., Galweiler, L., Tanzler, P., Huijser, P., Marchant, A., Parry, G., Bennett, M., Wisman, E., and Palme, K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, K., Sena, G., Nawy, T., and Benfey, P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413 307–311. [DOI] [PubMed] [Google Scholar]
- Perbal, M., Haughn, G., Saedler, H., and Schwarz-Sommer, Z. (1996). Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development 122 3433–3441. [DOI] [PubMed] [Google Scholar]
- Pysh, L., Wysocka-Diller, J., Camilleri, C., Bouchez, D., and Benfey, P. (1999). The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18 111–119. [DOI] [PubMed] [Google Scholar]
- Raghaven, V. (2000). Developmental Biology of Flowering Plants. (New York: Springer-Verlag).
- Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., and Scheres, B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 463–472. [DOI] [PubMed] [Google Scholar]
- Satina, S., Blakeslee, A.F., and Avery, A. (1940). Demonstration of the three germ layers in the shoot apex of Datura by means of induced polyploidy in periclinal chimeras. Am. J. Bot. 27 895–905. [Google Scholar]
- Scheres, B. (2001). Plant cell identity: The role of position and lineage. Plant Physiol. 125 112–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres, B., Laurenzio, L.D., Willemsen, V., Hauser, M.-T., Janmaat, K., Weisbeek, P., and Benfey, P.N. (1995). Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout embryonic axis. Development 121 53–62. [Google Scholar]
- Schoof, H., Lenhard, M., Haecker, A., Mayer, K.F., Jurgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 635–644. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Yanofsky, M.F., and Weigel, D. (2000). Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 287 419–421. [DOI] [PubMed] [Google Scholar]
- Steeves, T.A., and Sussex, I.M. (1989). Patterns in Plant Development. (Cambridge, UK: Cambridge University Press).
- Stone, J.M., Collinge, M.A., Smith, R.D., Horn, M.A., and Walker, J.C. (1994). Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science 266 793–795. [DOI] [PubMed] [Google Scholar]
- Stone, J.M., Trotochaud, A.E., Walker, J.C., and Clark, S.E. (1998). Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol. 117 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup, R., Marchant, A., and Bennett, M.J. (2000). Auxin transport: Providing a sense of direction during plant development. Biochem. Soc. Trans. 28 481–485. [PubMed] [Google Scholar]
- Trotochaud, A.E., Hao, T., Wu, G., Yang, Z., and Clark, S.E. (1999). The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud, A.E., Jeong, S., and Clark, S.E. (2000). CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science 289 613–617. [DOI] [PubMed] [Google Scholar]
- Umeda, M., Umeda-Hara, C., and Uchimiya, H. (2000). A cyclin-dependent kinase-activating kinase regulates differentiation of root initial cells in Arabidopsis. Proc. Natl. Acad. Sci. USA 97 13396–13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg, C., Willemsen, V., Hage, W., Weisbeek, P., and Scheres, B. (1995). Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378 62–65. [DOI] [PubMed] [Google Scholar]
- van den Berg, C., Willemsen, V., Hendriks, G., Weisbeek, P., and Scheres, B. (1997). Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390 287–289. [DOI] [PubMed] [Google Scholar]
- Vernoux, T., Wilson, R.C., Seeley, K.A., Reichheld, J.P., Muroy, S., Brown, S., Maughan, S.C., Cobbett, C.S., Van Montagu, M., Inze, D., May, M.J., and Sung, A.R. (2000). The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69 843–859. [DOI] [PubMed] [Google Scholar]
- Westhoff, P., Jeske, H., Jürgens, G., Kloppstech, K., and Link, G. (1998). Molecular Plant Development: From Gene to Plant. (Oxford, UK: Oxford University Press).
- Willemsen, V., Wolkenfelt, H., de Vrieze, G., Weisbeek, P., and Scheres, B. (1998). The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125 521–531. [DOI] [PubMed] [Google Scholar]
- Williams, R.W., Wilson, J.M., and Meyerowitz, E.M. (1997). A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc. Natl. Acad. Sci. USA 94 10467–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller, J., Helariutta, Y., Fukaki, H., Malamy, J., and Benfey, P. (2000). Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127 595–603. [DOI] [PubMed] [Google Scholar]