INTRODUCTION
RNA silencing is a sequence-specific RNA degradation mechanism that occurs in a broad range of eukaryotic organisms including fungi (quelling), animals (RNA interference [RNAi]), and plants (post-transcriptional gene silencing). In all these organisms, the process is triggered by double-stranded RNA (dsRNA) and requires a conserved set of gene products (for recent reviews of RNA silencing in plants, see Matzke et al., 2001; Vance and Vaucheret, 2001; Voinnet, 2001; Waterhouse et al., 2001; Baulcombe, 2002; in fungi or animals, see Cogoni and Macino, 2000; Bernstein et al., 2001a; Carthew, 2001; Zamore, 2001). The mechanism for RNA silencing involves an initial processing of the inducing dsRNA into small interfering RNAs (siRNAs) of 21 to 25 nucleotides, corresponding to both sense and antisense strands of the target gene (Hamilton and Baulcombe, 1999). These siRNAs become associated with a protein complex referred to as the RNA-induced silencing complex (RISC), where they serve as guides to select the target RNAs and effect their degradation (Hammond et al., 2000; Zamore et al., 2000). In plants, RNA silencing is typically correlated with methylation within the transcribed regions of the transgene that correspond to target RNA (reviewed in Wassenegger, 2000; Bender, 2001). Methylation of genomic DNA occurs even when the silencing is induced by an RNA virus that replicates exclusively in the cytoplasm (Jones et al., 1998), suggesting communication between the cytoplasm and the nucleus. A good deal of evidence suggests that RNA silencing plays a natural role in defense against foreign nucleic acids, including virus resistance in plants (Covey et al., 1997; Ratcliff et al., 1997, 1999; Mourrain et al., 2000; reviewed in Voinnet, 2001) and in control of transposons in a number of other organisms (Ketting et al., 1999; Tabara et al., 1999; Grishok et al., 2000; Djikeng et al., 2001; Elbashir et al., 2001b; Takeda et al., 2001). Consistent with the antiviral nature of RNA silencing in plants, many plant viruses have evolved proteins that suppress RNA silencing (reviewed in Li and Ding, 2001).
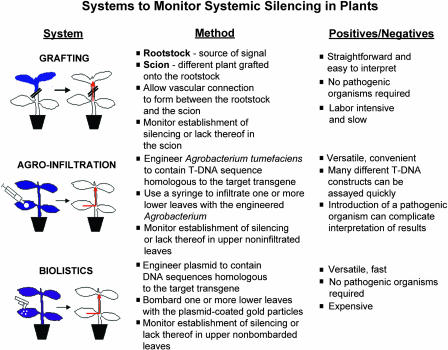
One of the most intriguing aspects of RNA silencing is that it is non-cell-autonomous: in both plants and Caenorhabditis elegans it can be induced locally and then spread to distant sites throughout the organism (Palauqui et al., 1997; Voinnet and Baulcombe, 1997; Fire et al., 1998; Voinnet et al., 1998; Winston et al., 2002). The systemic spread of silencing reflects the existence of an as yet unidentified mobile silencing signal as an integral component of the RNA silencing pathway. In C. elegans, a protein with multiple transmembrane domains has been reported to be required specifically for systemic silencing (Winston et al., 2002). Given the putative membrane localization of this protein, it could act as a receptor or transmembrane channel for the mobile signal. Whereas no mutations specific to systemic silencing have yet been reported in plant systems, certain plant viral proteins interfere with RNA silencing at this step (Voinnet et al., 2000; Guo and Ding, 2002). The mechanisms involved in systemic RNA silencing in plant systems are being actively investigated using grafting and transient expression approaches (Figure 1) in conjunction with a variety of plant viral suppressors of silencing that act at different steps in the silencing pathway. This review focuses on advances in understanding the nature of systemic silencing in plants and the signal(s) that induces silencing at distant sites.
Figure 1.
A Guide to Plant Systems for the Study of Systemic Silencing.
Silencing may spread from silenced rootstocks to scions or from locally silenced regions of a plant to upper parts of the same plant. A general description of the methods to induce systemic silencing in each system as well as the advantages and disadvantages of each system are summarized.
EVIDENCE FOR THE MOBILE SILENCING SIGNAL
The first clue that RNA silencing is non-cell-autonomous came from studies of transgenic plants that silenced developmentally to produce a visible phenotype that was acquired in distinctive but nonclonal spatial patterns (Boerjan et al., 1994; Palauqui et al., 1996). Soon afterward, independent experiments in two different laboratories provided direct evidence for a systemic silencing signal (Palauqui et al., 1997; Voinnet and Baulcombe, 1997). In grafting experiments, systemic silencing was transmitted across a graft junction from spontaneously silenced transgenic tobacco rootstocks to isogenic scions that had not silenced spontaneously (Figure 1; Palauqui et al., 1997). Lines transgenic for one of three different transgenes were examined: the endogenous loci Nia and Nii, encoding nitrate and nitrite reductase respectively, and the exogenous locus uidA, encoding the reporter protein β-glucuronidase (GUS). Similar results were obtained with all, indicating that systemic RNA silencing is not a peculiarity of one particular genetic locus. In all cases, silencing in the scion was specific for the coding sequences that were silenced in the rootstock, demonstrating that the mobile signal is sequence specific. This sequence specificity suggested that the mobile signal is a nucleic acid or includes a nucleic acid.
Independent evidence for the involvement of a systemic signal in RNA silencing has come from the demonstration that systemic silencing can be induced in transgenic tobacco species by using infiltration with Agrobacterium tumefaciens (agro-infiltration) or particle bombardment to deliver exogenous DNA homologous to the transgene (Figure 1; Voinnet and Baulcombe, 1997; Voinnet et al., 1998; Palauqui and Balzergue, 1999). No Agrobacterium or T-DNA could be detected in systemically silenced tissue of agro-infiltrated plants, indicating that the silencing must have been propagated by means of a mobile signal (Voinnet and Baulcombe, 1997). As in the grafting experiments, systemic silencing induced by agro-infiltration and particle bombardment is characterized by sequence specificity and the ability to propagate through a graft junction (Voinnet et al., 1998; Palauqui and Balzergue, 1999; Crete et al., 2001). The current assumption is that the signal produced in these three very different experimental systems is the same, but that might not be the case.
PROPERTIES OF SIGNAL TRANSMISSION
The patterns of systemic silencing suggest that the signal moves both cell-to-cell and through the phloem, mimicking patterns of viral movement through the plant. In 35S green fluorescent protein (GFP) plants, stomatal guard cells that have lost the plasmodesmatal connections to other cells before induction of systemic silencing do not silence, providing evidence that the signal moves cell-to-cell through plasmodesmata (Voinnet et al., 1998). Movement of the signal through the phloem has been most evident from the establishment of systemic silencing along major and minor veins prior to subsequent spread into mesophyll cells (Figures 2 and 3; Palauqui et al., 1997; Voinnet et al., 1998). Phloem movement of the signal is further supported by data showing that systemic silencing initiated from a single leaf is restricted to shoots that emerge from the same side of the stem as the initiating leaf (Voinnet et al., 1998), a pattern reminiscent of phloem transport of dyes and systemic virus (Roberts et al., 1997). The silencing signal can travel relatively long distances in plants: at least several centimeters as shown by propagation of silencing through leafless grafted spacers that cannot silence because homologous sequences are absent (Palauqui et al., 1997; Voinnet et al., 1998). Experiments using agro-infiltration or particle bombardment to induce silencing show that the systemic signal is produced and translocated rapidly, moving out of a leaf within 2 to 3 days (Voinnet et al., 1998; Palauqui and Balzergue, 1999). Although reported to move only upward (Palauqui et al., 1997), the signal moves bidirectionally, but more efficiently upward than downward (Voinnet et al., 1998; Sonoda and Nishiguchi, 2000). Viruses are excluded from meristems after systemic infection of plants (Matthews, 1991), and this is also true for systemic silencing; extreme meristematic zones of shoots, flowers, and roots remain green fluorescent subsequent to extensive and persistent systemic silencing of GFP transgenes (Voinnet et al., 1998). Similarly, silencing is not observed in meristems in Nia or GUS-silenced plants (Beclin et al., 1998). It has not been shown whether the mobile silencing signal fails to get into the meristem or whether meristematic tissue is unable to silence in response to the signal.
Figure 2.
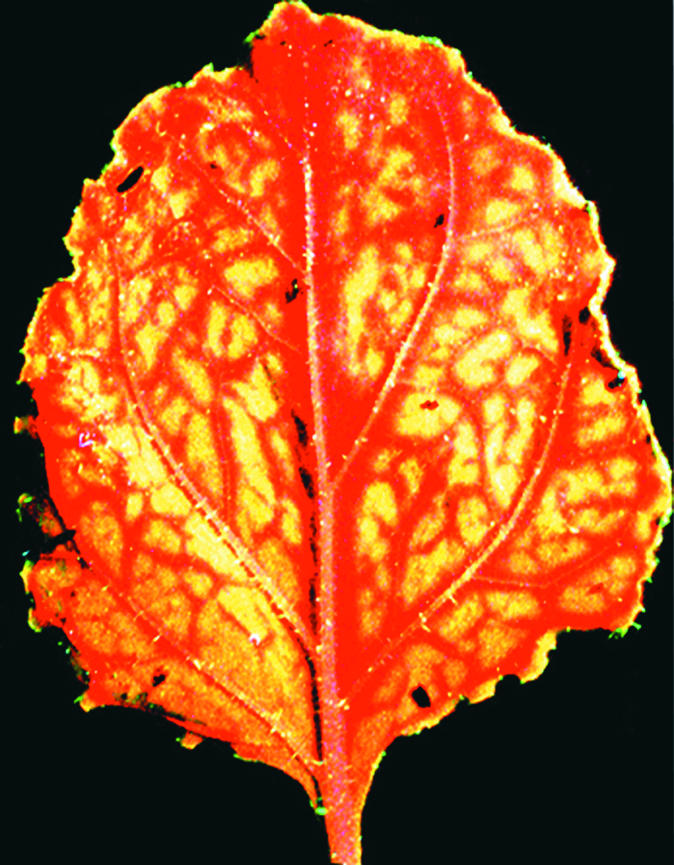
Systemic Silencing of GFP.
Silencing of GFP in response to the mobile silencing signal occurs initially around veins. Systemic silencing (indicated by red florescence produced by chlorophyll) initiates around vascular tissues in response to a mobile silencing signal induced by agro-infiltration. The tissues farther away from veins have not yet silenced and therefore maintain green fluorescence as the result of GFP.
Figure 3.
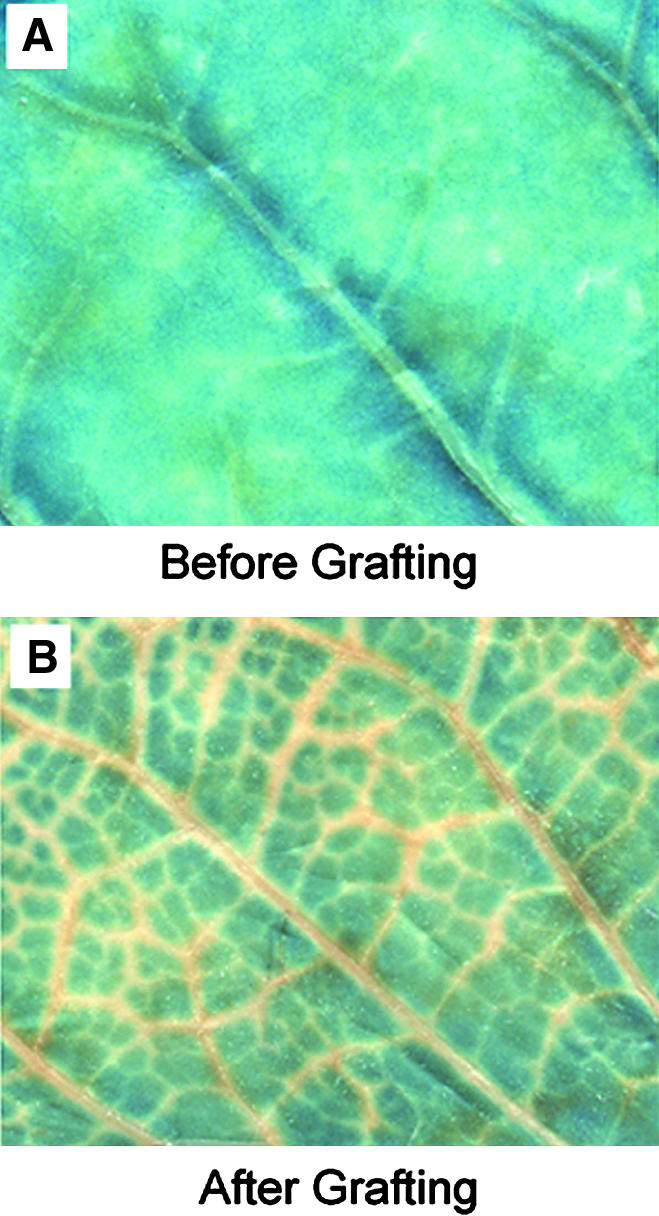
A GUS-Expressing Transgenic Line Is Initially Silenced around Veins When Grafted onto a GUS-Silenced Rootstock.
(A) Histochemical staining of a leaf from a GUS-expressing transgenic tobacco line before grafting. The uniform blue color reflects GUS activity throughout the leaf.
(B) Histochemical staining of a leaf from the transgenic line in (A) after grafting onto a GUS-silenced rootstock. Movement of the signal into the scion is first evidenced by the absence of blue color in the regions along the veins.
Movement of the signal into distant tissues is not sufficient to induce silencing. The same types of experiment that provided direct evidence for the mobile signal have also shown that the nature of the target is important in determining whether—or to what degree—systemic silencing is elicited when signal is provided. In particular, transgenes are much better targets for systemic silencing than are endogenous genes, and the gene must be transcriptionally active. For example, scions expressing a Nia transgene became uniformly silenced for Nia when grafted onto rootstocks silenced for a Nia transgene, whereas the endogenous Nia mRNA in nontransgenic scions and transcriptionally silenced Nia transgenic scions was unaffected (Palauqui et al., 1997). When the level of Nia mRNA in scions lacking a Nia transgene was increased by eliminating feedback inhibition of Nia transcription, the scions became uniformly silenced (Palauqui and Vaucheret, 1998), suggesting that the amount of target RNA is important in establishing systemic silencing in response to the mobile signal. Agro-infiltration experiments using a transgene that produces a replicating RNA were also able to induce systemic silencing of endogenous genes, but—unlike the widespread, persistent silencing observed for a GFP transgene—the silencing was transient and limited to regions near the veins of the affected leaves (Voinnet et al., 2000). This limited degree of systemic silencing was induced whether the targeted RNA was expressed at high levels (small subunit of rubisco) or low levels (phytoene desaturase). Thus, requirements for establishment of systemic silencing by the mobile signal are not well understood, but a favored current hypothesis is that amplification of the signal is necessary for efficient systemic silencing (Palauqui and Vaucheret, 1998; Voinnet et al., 1998).
The maintenance of RNA silencing after an exogenous source of signal is removed also depends on the nature of the target. Grafted scions in which the targeted locus is a transgene that never silences spontaneously (referred to as class I transgenes; Palauqui and Vaucheret, 1998) require a continuous influx of the silencing signal to remain silenced. If such scions are removed from the silenced rootstock and grafted onto a wild-type rootstock, their silencing is lost. However, scions from transgenic lines in which some proportion of the individual plants silence spontaneously (referred to as class II transgenes; Palauqui and Vaucheret, 1998) remain silenced even when the grafted scions are removed from silenced rootstocks or when the bombarded or infiltrated tissues are removed, thereby eliminating the original source of the mobile signal (Voinnet et al., 1998; Palauqui and Balzergue, 1999). These results indicate that RNA degradation in systemic silencing can occur in the absence of an ability to perpetuate systemic silencing and, therefore, possibly independently of production of the mobile signal (Palauqui and Vaucheret, 1998).
WHAT IS THE MOBILE SILENCING SIGNAL?
In principle, the nucleic acid component of the mobile silencing signal(s) involved in transgene silencing could either be DNA or RNA derived from the transgene. Indirect evidence for an RNA component comes from the likely antiviral function of the signal in nontransgenic plants. This antiviral function was suggested in experiments in which RNA silencing was activated in upper leaves of wild-type Nicotiana benthamiana by inoculating lower leaves with movement-defective mutants of Potato virus X (PVX) (Voinnet et al., 2000). Systemic spread of silencing could be monitored in planta because the modified PVX contained fragments of endogenous genes. Thus, the silencing signal generated by localized virus replication was not only primed against the viral genome but also against the corresponding endogenous genes, therefore generating a visual systemic silencing phenotype. Systemic silencing was only apparent when replication-competent PVX was used as an inoculum. Because PVX has an RNA genome that is replicated via RNA intermediates, this result suggests that the signal, or part of it, is an RNA. This suggestion is reinforced by the fact that >90% of plant viruses, many of which are likely to induce systemic silencing, are RNA viruses that replicate without any DNA intermediate.
CANDIDATE RNAS FOR THE MOBILE SILENCING SIGNAL
siRNAs
Small RNAs associated with RNA silencing were discovered in plants (Hamilton and Baulcombe, 1999), and similar RNAs have since been associated with the related RNA degradation processes in animal systems, where they are referred to as siRNAs (Hammond et al., 2000; Parrish et al., 2000; Yang et al., 2000; Zamore et al., 2000; Elbashir et al., 2001b). Indeed, these RNAs are considered a hallmark of RNA silencing. The possibility that siRNAs are involved in systemic signaling is an attractive and popular model (Hamilton and Baulcombe, 1999). The characteristics of siRNAs make them ideal candidates for the mobile signal. They are consistently associated with RNA silencing; they are long enough to convey sequence specificity, yet small enough to move easily through plasmodesmata. In addition, they are sufficient to induce RNA silencing in vitro in Drosophila (Elbashir et al., 2001b) and in vivo in mammalian cell lines and in C. elegans (Caplen et al., 2001; Elbashir et al., 2001a). However, direct evidence that siRNAs are mobile or that they play a role in systemic silencing is lacking.
Two lines of evidence argue against a role for siRNAs in systemic signaling of silencing. One line of evidence deals with the viral suppressor of RNA silencing, helper component–proteinase (HC-Pro; discussed in detail in a later section). When an HC-Pro–expressing tobacco line was crossed to a transgenic line silenced for GUS, the GUS silencing was suppressed, GUS mRNA accumulated, and the GUS siRNAs were eliminated. Strikingly, these plants, despite the absence of detectable levels of GUS siRNA, remained capable of producing and sending the mobile silencing signal when used as rootstocks in grafting experiments (Mallory et al., 2001). Although the simplest interpretation of these data is that siRNAs are not the systemic signal, it remains possible that low quantities of siRNA, below the detection level in these experiments, are actually sufficient to ensure systemic RNA silencing. A second line of evidence against the involvement of siRNAs in systemic silencing comes from work in C. elegans. A mutation in rde-4 interferes with production of siRNAs without interfering with systemic silencing (Tabara et al., 1999; Parrish and Fire, 2001). The evidence for and against the idea that siRNAs are the mobile silencing signal is summarized in Figure 4.
Figure 4.
SiRNAs and the Mobile Signal.
Evidence for and against a role for siRNAs as the mobile silencing signal is shown. Alternate candidates for the signal are also given.
Recent work points to the complexity of small RNAs in eukaryotic cells: the siRNAs may serve roles other than as guides for the RISC complex, and some populations of small RNAs are not involved in RNA silencing. Evidence from animal RNAi systems has raised the possibility that siRNAs are involved in production of new dsRNA to feed into the silencing pathway (Lipardi et al., 2001; Sijen et al., 2001a). Lipardi and co-workers have shown that siRNAs added to an in vitro silencing system become incorporated into long dsRNA via the activity of a cellular RNA-dependent RNA polymerase (RdRP). This result suggests that siRNAs act as primers for the cellular RdRP, resulting in a cycle of dsRNA generation and degradation that amplifies the siRNA (Lipardi et al., 2001). It is not clear if the same siRNA molecules serve dual roles in silencing or if there are subpopulations of siRNA with distinct functions: some serving as guides in the RISC complex and others serving as primers in amplification of siRNA via RdRP. The siRNAs derive from longer dsRNA by the activity of a ribonuclease III (RNase III) called DICER (Hammond et al., 2000; Bernstein et al., 2001b; Knight and Bass, 2001). In Drosphila and C. elegans, DICER also produces another population of small RNAs called small temporal RNAs (stRNAs) (Grishok et al., 2001; Hutvagner et al., 2001; Ketting et al., 2001). Although produced by the activity of the same enzyme, stRNAs differ from siRNAs both structurally and functionally: they are single-stranded (rather than double-stranded), and they interact with the 3′ untranslated region of the target mRNA and inhibit its translation (rather than triggering its degradation). In addition, a novel class of abundant and diverse stRNA-like RNAs, termed microRNAs, have recently been uncovered not only in worms and flies but also in humans (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001), suggesting that stRNA species may be more widespread than previously envisioned. Finally, the small RNAs that accumulate during dsRNA-induced transcriptional gene silencing (TGS) are double-stranded and siRNA-like in size (Mette et al., 2000; Sijen et al., 2001b). However, unlike siRNAs, the TGS-associated small RNAs are not involved in RNA degradation (Mette et al., 2001), and their function, if any, is unknown. Because of the structural and functional diversity of small RNAs, it remains a viable possibility that a subpopulation of small RNAs serves as the mobile silencing signal in RNA silencing.
OTHER CANDIDATES FOR THE RNA SIGNAL
Aberrant RNAs
Another likely candidate for the mobile silencing signal is the RNA transcript from a silenced locus (or a derivative of that mRNA), which is in some way aberrant so that it triggers RNA silencing upon arrival in a new cell. Cell-to-cell and systemic movement of mRNA in plants is not unprecedented, and in fact, some evidence suggests that it is commonplace (reviewed in Jorgensen et al., 1998; Oparka and Santa Cruz, 2000). Thus, although the paradigm for RNA trafficking in plants is virus infection, it is likely that viruses have simply usurped host systems of macromolecular trafficking for their own purposes. Plant viruses encode special proteins to facilitate movement of viral RNA (reviewed in Lazarowitz and Beachy, 1999), but there is good evidence that there are endogenous plant proteins that function as RNA movement proteins. For instance, viroids—plant pathogens with a noncoding RNA genome composed of a circular RNA duplex—move cell to cell and systemically, probably by recruiting specific host factors (Gomez and Pallas, 2001; Owens et al., 2001; Zhu et al., 2001). There are also several examples of endogenous mRNAs, such as the maize KNOTTED1 (Lucas et al., 1995) and SUT1 in tobacco, tomato, and potato (Kuhn et al., 1997), which move through plasmodesmata presumably using endogenous mechanisms for RNA trafficking. Systemic movement of endogenous RNAs across graft junctions also occurs. Pumpkin NACP mRNA moves from a pumpkin rootstock into the apex of cucumber scions (Ruiz-Medrano et al., 1999). In grafting experiments with tomato, an mRNA derived from a dominant negative mutant locus in the rootstock moves into a scion that is wild type for the locus and confers the mutant phenotype in the scion (Kim et al., 2001). The mechanism of RNA trafficking may involve proteins that function in a manner analogous to viral movement proteins such as CmPP16 from Cucurbita maxima (Xoconostle-Cazares et al., 1999). Thus, RNA trafficking, both local and systemic, may be a normal aspect of plant gene expression.
These results raise the possibility that the mobile silencing signal is an mRNA or mRNA/protein complex that moves across the graft junction into the scion via normal pathways for macromolecular trafficking. Upon arrival in the scion, the translocated RNA becomes a template for a cellular RdRP, leading to production of dsRNA and thus initiating RNA silencing. What distinguishes this particular translocated RNA and marks it as a template for RdRP? One possibility is that the RNA is tagged in some way, perhaps by the set of proteins that bind to it. Interestingly, one of the genes required for transgene-induced gene silencing, SDE3 (Dalmay et al., 2001), encodes an RNA helicase related to SMG2 in C. elegans and UPF1 in yeast. In C. elegans, the SMG2-encoded RNA helicase functions in two RNA surveillance pathways: nonsense-mediated decay and RNAi (Page et al., 1999; Domeier et al., 2000). The human ortholog of UPF1 is part of a complex that marks mRNAs carrying premature stop codons and targets them for nonsense-mediated decay (Sun et al., 1998; Bhattacharya et al., 2000; Lykke-Andersen et al., 2001). This result prompts the idea that the SDE3 protein may serve a similar role as a marker protein for aberrant RNAs in RNA silencing. Thus, the mobile silencing signal could be an RNA marked as a template for a cellular RdRP by the binding of the RNA helicase encoded by SDE3. In that case, it would be predicted that SDE3 is required for systemic silencing—a prediction that has not yet been tested.
dsRNA
Another possible candidate for the mobile signal is the larger dsRNA molecules that induce RNA silencing, although evidence for the movement of such dsRNAs is mostly lacking. However, the viroid genome is circular, rod-shaped RNA several hundred bases in length with a high secondary structure content and could conceivably be taken as a model for dsRNA movement. Many viroids are replicated autonomously in the nucleus, presumably by the host RNA–polymerase II, as demonstrated by the α-amanitin sensitivity of the potato spindle tuber viroid replication (Flores, 2001). Neither mature viroids nor their replication intermediates are polyadenylated, yet after replication, viroid RNAs are able to enter a series of transport pathways starting with exit from the nucleus into the cytoplasm; cell-to-cell movement through plasmodesmata; and systemic movement, which requires assistance by special plant proteins in phloem (Gomez and Pallas, 2001; Owens et al., 2001; Zhu et al., 2001).
NO EVIDENCE FOR SIGNALING IN TRANSCRIPTIONAL GENE SILENCING
Early experiments with transgenic plants revealed that gene silencing could be mediated at either the transcriptional level (TGS or transcriptional gene silencing) or at the post-transcriptional level (originally called post-transcriptional gene silencing, or PTGS, but now commonly referred to as RNA silencing) (Vaucheret and Fagard, 2001). Whereas RNA silencing involves sequence-specific RNA degradation in the cytoplasm, TGS involves a block in RNA synthesis in the nucleus. Both types of silencing are correlated with de novo DNA methylation: in transcribed regions in RNA silencing and in promoter regions in TGS. The sequence-specific methylation in silencing suggested the involvement of RNA–DNA or DNA–DNA interactions. Because promoter regions are not normally transcribed, DNA–DNA pairing was a favored model for TGS, and the mechanisms of RNA silencing and TGS were thought to be distinct. In the past few years, however, it has become clear that the two kinds of silencing share additional features that suggest a mechanistic link: both are induced by dsRNA and associated with the accumulation of small RNA species with homology to the inducing dsRNA (Mette et al., 2000; Sijen et al., 2001b). This possibility of a common mechanism raises the question of whether there is also a systemic signal in TGS.
Transgene-Induced TGS
To study whether RNA-mediated TGS spreads systemically, grafting experiments were performed with the H9NP and 271 tobacco lines. Both the H9NP and 271 loci induce TGS in trans, and this induction involves synthesis of promoter dsRNA that is processed to small RNAs of both polarities similar to those involved in RNA silencing (Mette et al., 2000; Sijen et al., 2001b; R. van Blokland, J. Kooter, and H. Vaucheret, unpublished data). Thus, TGS in these lines is induced by dsRNA that is processed to small RNAs, features common to RNA silencing.
The H9NP line was constructed in experiments designed to deliberately transcribe promoter sequences (Mette et al., 1999). In the H9NP system, the nopaline synthase promoter (NOSpro) dsRNA that induces TGS is transcribed from an inverted repeat in the nucleus and is not polyadenylated (Mette et al., 1999). With H9NP, neither silencing nor methylation of the target NOSpro was observed in shoots grafted onto scions of H9NP plants (M.F. Mette, unpublished data). Moreover, dsRNA was not detected in grafted shoots, although minute quantities would not have been detectable with the method used. These results suggest that the dsRNA-induced TGS produced by the H9NP locus is not graft transmissible.
Similar grafting experiments using transgenic line 271 suggest that the inability of TGS to move systemically is a general feature of trans-TGS. The 271 locus contains multiple copies of a plasmid carrying an nptII selectable marker under the control of the Cauliflower mosaic virus (CaMV) 19S promoter and an Nii gene in the antisense orientation under the control of the CaMV 35S promoter (Vaucheret, 1993). Line 271 was not originally designed to transcribe promoter sequences, and how promoter dsRNA is produced in this line remains unknown. In contrast to the H9NP locus, which triggers only TGS, the 271 locus induces both TGS and RNA silencing. It triggers TGS and methylation of unlinked 19S or 35S promoter-driven transgenes and RNA silencing of unlinked Nii endogenous genes or transgenes, but not of unlinked nptII transgenes (Park et al., 1996; Thierry and Vaucheret, 1996). The 271 locus provides an internal control for assessing whether RNA-mediated TGS spreads systemically because RNA silencing of nitrite reductase is graft-transmissible in this line (H. Vaucheret, unpublished data). Grafting experiments demonstrated, however, that only RNA silencing of Nii endogenous genes and transgenes, but not TGS of 35S-driven transgenes, was transmitted from line 271 to scions carrying target Nii or 35S transgenes. These results show convincingly that despite the involvement of dsRNA and small RNAs in both TGS and RNA silencing mechanisms, only RNA silencing spreads systemically.
It is not clear why TGS is not graft-transmissible. One possibility is that dsRNA and small RNAs associated with TGS are confined to the nucleus and thus unable to move cell to cell and into the phloem. A second possibility is that TGS-associated RNAs can move but that they are not amplified in the scion and thus do not induce detectable silencing there. If amplification requires transcription of the target sequence, for example, TGS would not induce amplification because the target is a promoter, and promoter sequences are not ordinarily transcribed. A third possibility is that the dsRNA and the small RNAs that accumulate during TGS are not the mobile signal and that this signal is produced at a step in the RNA silencing pathway that is downstream of any steps in common between TGS and RNA silencing.
Virus-Induced TGS
Transgene-induced TGS relies on promoter dsRNA synthesized in the nucleus. Work with RNA viruses modified to contain promoter sequences has demonstrated that promoter RNAs transcribed in the cytoplasm by viral RNA polymerases can enter the nucleus and induce TGS and RNA-directed DNA methylation of homologous promoters. Virus-induced TGS has been observed in the case of the CaMV 35S promoter (Jones et al., 1999, 2001) but has not yet been reported in the case of endogenous promoters. The efficiency of silencing seems to depend on the virus used; Jones et al. (1999) reported that not all tissues from 35S-GFP N. benthamiana plants infected with PVX-35S are silenced, despite the presence of PVX-35S in nonsilenced tissues, whereas all tissues from 35S-GFP plants infected with Tobacco rattle virus (TRV)–35S are silenced (Jones et al., 2001).
In virus-induced TGS, it is unlikely that a systemic signal for either silencing or methylation is produced from the inactivated transgene. Indeed, Jones et al. (2001) reported that the virus-free progeny of TRV-35S–silenced GFP plants remain initially silenced and progressively revert to expression. In addition, they showed that trans-silencing of an unlinked 35S-GFP locus can occur in the F1 between an infected plant and a noninfected plant carrying the target but not between the virus-free silenced progeny of an infected plant and the target. Taken together, these results suggest that the TG-silenced transgene is unable to produce any signal for systemic propagation or maintenance in the absence of virus, and that the virus itself induces TGS and transgene promoter methylation when it moves systemically through the plant.
VIRAL SUPPRESSORS AND THE MOBILE SIGNAL
The discovery in 1998 that certain plant viruses encode proteins that suppress RNA silencing (Anandalakshmi et al., 1998; Beclin et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998) provided a new approach for dissecting the steps in the RNA silencing pathway. The first indication that a viral suppressor might interfere specifically with systemic RNA silencing came from reversal of silencing assays using the Cucumber mosaic virus (CMV) (Beclin et al., 1998) or the CMV 2b protein (Cmv2b) (Brigneti et al., 1998). In these assays, neither CMV nor Cmv2b was able to reverse established silencing in older tissues, but each completely suppressed the signal-mediated spread of silencing into newly emerging tissues. In contrast, HC-Pro, a viral suppressor encoded by potyviruses, was able to reverse silencing in both types of tissues (Brigneti et al., 1998). These results suggest that these two suppressors interfered with silencing at different points and that each might define a step in the silencing pathway. In this case, it appeared that Cmv2b might interfere specifically with systemic spread of silencing.
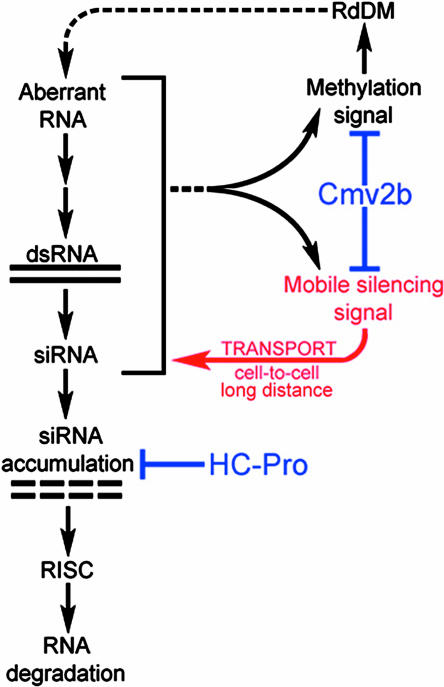
Confirmation that Cmv2b interferes with systemic silencing comes from analyses using a well-characterized transgenic tobacco line silenced for the reporter gene encoding GUS (line 6b5; Elmayan and Vaucheret, 1996). In these experiments, line 6b5 was crossed with a transgenic line expressing Cmv2b (Ji and Ding, 2001). The progeny of this cross (6b5 × Cmv2b plants) were only partially suppressed for GUS silencing, as shown by both the accumulation of GUS mRNA and reduced levels of GUS siRNAs. However, when used as rootstock, these plants completely failed to silence GUS-expressing scions (Guo and Ding, 2002). In contrast, the progeny from a similar cross between 6b5 and HC-Pro–expressing plants showed no signs of GUS silencing, and GUS-specific siRNAs were undetectable (Mallory et al., 2001). However, this efficient suppression of intracellular silencing in the 6b5 × HC-Pro plants did not eliminate their ability to produce and transmit the mobile silencing signal when used as rootstock in grafting experiments (Mallory et al., 2001). Comparing the effect of these two viral suppressors in the same silenced transgenic line allows steps in the silencing pathway to be ordered. Thus, HC-Pro works downstream of the mobile silencing signal at a step that prevents the accumulation of siRNAs, whereas Cmv2b prevents systemic silencing by interfering with the production and/or transmission of a functional silencing signal (Figure 5).
Figure 5.
Contrasting Modes of Action for Two Different Viral Suppressors of RNA Silencing.
This simplified model shows steps in the RNA silencing pathway in transgenic line 6b5. The silenced GUS locus in this line produces aberrant RNA that is used as template for a cellular RdRP to produce dsRNA. The dsRNA is processed through the activity of a DICER-like enzyme to produce siRNAs, which are incorporated into an active RISC complex which effects degradation of specific target RNAs. HC-Pro interferes with silencing at a step that prevents accumulation of the siRNAs without affecting either transgene methylation or the mobile silencing signal. Thus, HC-Pro is shown working at a step downstream of both these signals. In contrast, Cmv2b interferes with both transgene methylation and systemic silencing. The methylation signal and the mobile silencing signal are produced at an unknown point in the silencing pathway indicated by a bracket, including events that occur upstream of HC-Pro action. Because the methylation signal and the mobile silencing signal may be the same molecule, their exit from the pathway is shown as a single dotted line. The systemic portion of the silencing pathway is indicated in red.
The first viral protein shown to suppress systemic silencing was the 25-kD protein (p25) encoded by PVX (Voinnet et al., 2000). Interestingly, PVX-expressing p25 displayed no detectable activity in the reversal of silencing assay described above for Cmv2b and HC-Pro (Brigneti et al., 1998). However, p25 activity became apparent using a newly developed assay called Agrobacterium co-infiltration. In this assay, a GFP-expressing plant is infiltrated with a mixture of two Agrobacterium cultures: one expresses GFP and thereby induces RNA silencing of the GFP transgene; the other expresses a putative suppressor of silencing. In the co-infiltration assay, expression of the putative suppressor occurs prior to or at the same time as initiation of GFP silencing. PVX p25 prevented localized GFP silencing when co-infiltrated with a simple transgene (35S-GFP) but not when co-infiltrated with Agrobacterium expressing a transgene that encodes a replication-competent PVX-GFP vector. Importantly, p25 prevented systemic silencing in both cases (Voinnet et al., 2000). The Cmv2b protein also suppresses systemic silencing in this co-infiltration assay, confirming results obtained in transgenic plants (Guo and Ding, 2002) and in the reversal of silencing assay (Brigneti et al., 1998).
Inhibition of systemic silencing by p25 and Cmv2b might occur either before or after production of the silencing signal in the co-infiltration assay because the silencing inducer and suppressor are expressed in the same cells at the same time. However, in the grafting experiments, persistent export of the GUS-specific silencing signal from 6b5 rootstocks into 6b5 × Cmv2b scions failed to induce degradation of GUS mRNA in scions (Guo and Ding, 2002). Thus, it is unlikely that suppression of silencing in 6b5 × Cmv2b tobacco is due to failure to produce the mobile silencing signal, and this result suggests that Cmv2b suppression occurs after signal synthesis. Additional experiments further illustrate that Cmv2b directly interferes with the activity of the silencing signal rather than with downstream processes (Guo and Ding, 2002). For example, the spread of GUS silencing from a 6b5 rootstock into GUS-expressing scions was blocked when an intergraft from the Cmv2b-expressing tobacco was placed between the scion and the rootstock in double-grafted tobacco plants. Efficient inhibition of systemic GFP silencing by Cmv2b was also observed in the GFP-expressing N. benthamiana line 16c when 35S-Cmv2b was infiltrated at the basal region of a leaf and 35S-GFP infiltrated at the tip of the same leaf; but systemic silencing was not inhibited in 16c plants where the two Agrobacterium strains were infiltrated in a reverse configuration. In these assays, Cmv2b was expressed between the source and recipient tissues of the silencing signal and thus should perturb neither signal production nor signal perception. Together, these findings suggest that Cmv2b interacts directly with the signal or a key component of the signal complex, leading to signal inactivation (Figure 5; Guo and Ding, 2002).
Experiments in the 6b5 silencing system have revealed a correlation between a viral suppressor's effect on systemic silencing and its effect on transgene methylation. HC-Pro blocks RNA silencing in the 6b5 plants without interfering with either systemic signaling or transgene methylation (Mallory et al., 2001). In contrast, Cmv2b interferes with both systemic silencing and transgene methylation in the 6b5 line (Guo and Ding, 2002). These results raise the possibility that the mobile signal also directs methylation. Alternately, systemic silencing and transgene methylation could occur via a common signaling pathway or via partially overlapping pathways. In a simple model (Figure 5), Cmv2b suppresses both transgene methylation and systemic silencing by inactivating the signal molecule(s), whereas HC-Pro works at a step downstream of the mobile silencing/methylation signal loops. Perhaps the different effects of these two suppressors reflect that Cmv2b has a nuclear localization signal (Lucy et al., 2000), whereas HC-Pro is cytoplasmic (Riedel et al., 1998). Furthermore, we expect that the interaction between the different signal loops in silencing is more complex than shown here (Figure 5). For example, our model precludes an effect of HC-Pro on methylation, yet a small reduction in methylation has been reported in a different silencing system after introduction of HC-Pro (Llave et al., 2000).
The differential suppression of intra- and intercellular RNA silencing by HC-Pro and p25/Cmv2b suggests a basis for potyvirus synergism with PVX and CMV (Pruss et al., 1997). Mixed infection with viruses that suppress RNA silencing at different points in the pathway may produce a much more effective interference with this host antiviral defense than infection with either virus alone, leading to the enhanced symptoms and virus accumulation characteristic of synergistic viral disease in plants. Furthermore, the different effects of HC-Pro and p25 on signal movement provide an explanation for the differential ability of plants to recover from infections by PVX and potyviruses. When a transgenic plant expressing a viral sequence is infected by a virus carrying homologous sequences, the transgene is silenced and the virus is eliminated from the plant (Lindbo et al., 1993; Guo and Garcia, 1997; Ruiz et al., 1998). When PVX is the infecting virus in such a system, once the virus is eliminated, the expression of the transgene is reactivated in newly emerging tissues (Ruiz et al., 1998). In contrast, when a potyvirus is the infecting virus, the transgene remains silenced in newly emerging tissues even after the virus is eliminated (Lindbo et al., 1993). Presumably, in the PVX system, p25 interferes with the mobile silencing signal (Voinnet et al., 2000) and prevents the spread of silencing into upper tissues. Thus, in the absence of virus, silencing is not maintained and the transgene is expressed. HC-Pro, in contrast, does not prevent signal export into the apical virus-free tissues (Mallory et al., 2001), thereby enabling a persistent transgene silencing independent of virus accumulation.
PERSPECTIVE
Although the phenomenon of systemic RNA silencing was demonstrated five years ago, the identity of the silencing signal remains largely elusive. To some extent, it is likely that the difficulty in investigating this aspect of RNA silencing lies in the inherent complexity of the silencing process. Hence, what is usually described as “signaling” in transgenic plants does not refer to the physical movement of the silencing factor per se but to the many consequences of its perception in recipient cells located outside the vasculature. This perception leads ultimately to transgene mRNA degradation, which translates into a systemic silencing phenotype. But perception of the signal is also accompanied by the generation of many additional nucleic acid species, including dsRNA and siRNA, that account for the amplification and mRNA degradation steps of RNA silencing. Induction of these new species in each cell makes it very difficult to identify the genuine signaling molecule.
A second difficulty is that signaling of RNA silencing may well involve several types of molecules and transport mechanisms. For instance, a recent report suggests that systemic movement of silencing, but not cell-to-cell spread, is inhibited by nontoxic concentrations of cadmium in tobacco and N. benthamiana (Ueki and Citovsky, 2001). Interestingly, similar concentrations of cadmium were also reported to prevent systemic but not local movement of two tobamoviruses (Citovsky et al., 1998; Ghoshroy et al., 1998). Thus, systemic spread of RNA silencing and of these viruses may share a common step that is cadmium-sensitive and that is dispensable for silencing and localized viral movement. The differential sensitivity to cadmium might prove a useful tool to make a distinction between local and systemic silencing signals.
A fundamental difficulty with understanding systemic silencing is that the mechanisms of intercellular trafficking of macromolecules remain largely unknown. Even if the molecular identity of the signal were known at this point, we still would not understand how and why this molecule moves systemically. Studies of the molecular basis of RNA trafficking should provide a framework for understanding the rules that govern the spread of RNA silencing.
Acknowledgments
We thank B. Roth and M. Endres for providing unpublished images of systemic silencing and B. Roth and A. Mallory for assistance with the figures. We are grateful to D. Baulcombe, M. Zerbini, and L. Bowman for helpful discussions and critical reading of the manuscript. We acknowledge financial support to O.V. from the Gatsby Charitable Foundation, to M.M. from the Austrian Fonds zur förderung der wissenschaftlichen Forschung, to H.V. from the French Ministry of Research, to S.W.D. from the U.S. Department of Agriculture, NRI Competitive Grants Program, and to V.B.V. from the National Institutes of Health and the U.S. Department of Agriculture, NRI Competitive Grants Program.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001677.
References
- Anandalakshmi, R., Pruss, G.J., Ge, X., Marathe, R., Mallory, A.C., Smith, T.H., and Vance, V.B. (1998). A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. (2002). RNA silencing. Curr. Biol. 12 R82–R84. [DOI] [PubMed] [Google Scholar]
- Beclin, C., Berthome, R., Palauqui, J.C., Tepfer, M., and Vaucheret, H. (1998). Infection of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of nonviral (trans)genes. Virology 252 313–317. [DOI] [PubMed] [Google Scholar]
- Bender, J. (2001). A vicious cycle: RNA silencing and DNA methylation in plants. Cell 106 129–132. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. (2001. a). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409 363–366. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Denli, A.M., and Hannon, G.J. (2001. b). The rest is silence. RNA 7 1509–1521. [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya, A., Czaplinski, K., Trifillis, P., He, F., Jacobson, A., and Peltz, S.W. (2000). Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6 1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan, W., Bauw, G., Van Montagu, M., and Inze, D. (1994). Distinct phenotypes generated by overexpression and suppression of S-adenosyl-l-methionine synthetase reveal developmental patterns of gene silencing in tobacco. Plant Cell 6 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti, G., Voinnet, O., Li, W.X., Ji, L.H., Ding, S.W., and Baulcombe, D.C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Caplen, N.J., Parrish, S., Imani, F., Fire, A., and Morgan, R.A. (2001). Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew, R.W. (2001). Gene silencing by double-stranded RNA. Curr. Opin. Cell Biol. 13 244–248. [DOI] [PubMed] [Google Scholar]
- Citovsky, V., Ghoshroy, S., Tsui, F., and Klessig, D. (1998). Non-toxic concentrations of cadmium inhibit systemic movement of turnip vein clearing virus by a salicylic acid-independent mechanism. Plant J. 16 13–20. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (2000). Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 10 638–643. [DOI] [PubMed] [Google Scholar]
- Covey, S.N., Al-kaff, N.S., Langara, A., and Turner, D.S. (1997). Plants combat infection by gene silencing. Nature 385 781–782. [Google Scholar]
- Crete, P., Leuenberger, S., Iglesias, V.A., Suarez, V., Schob, H., Holtorf, H., Van Eeden, S., and Meins, F. (2001). Graft transmission of induced and spontaneous post-transcriptional silencing of chitinase genes. Plant J. 28 493–501. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Horsefield, R., Braunstein, T.H., and Baulcombe, D.C. (2001). SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng, A., Shi, H., Tschudi, C., and Ullu, E. (2001). RNA interference in Trypanosoma brucei: Cloning of small interfering RNAs provides evidence for retroposon-derived 24–26 nt RNAs. RNA 7 1–9. [PMC free article] [PubMed] [Google Scholar]
- Domeier, M.E., Morse, D.P., Knight, S.W., Portereiko, M., Bass, B.L., and Mango, S.E. (2000). A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 289 1928–1931. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001. a). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411 494–498. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. (2001. b). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan, T., and Vaucheret, H. (1996). Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J. 9 787–797. [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans. Nature 391 806–811. [DOI] [PubMed] [Google Scholar]
- Flores, R. (2001). A naked plant-specific RNA ten-fold smaller than the smallest known viral RNA: The viroid. C.R. Acad. Sci. Paris 324 943–952. [DOI] [PubMed] [Google Scholar]
- Ghoshroy, S., Freedman, K., Lartey, R., and Citovsky, V. (1998). Inhibition of plant viral systemic infection by non-toxic concentrations of cadmium. Plant J. 13 591–602. [DOI] [PubMed] [Google Scholar]
- Gomez, G., and Pallas, V. (2001). Identification of an in vitro ribonucleoprotein complex between a viroid RNA and a phloem protein from cucumber plants. Mol. Plant-Microbe Interact. 14 910–913. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Tabara, H., and Mello, C.C. (2000). Genetic requirements for inheritance of RNAi in C. elegans. Science 287 2494–2497. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106 23–24. [DOI] [PubMed] [Google Scholar]
- Guo, H.S., and Ding, S.W. (2002). A viral protein inhibits the long-range signaling activity of the gene silencing signal. EMBO J. 21 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H.S., and Garcia, J.A. (1997). Delayed resistance to plum pox potyvirus mediated by a mutated RNA helicase gene: Involvement of a gene silencing mechanism. Mol. Plant-Microbe Interact. 10 160–170. [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404 293–296. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293 834–838. [DOI] [PubMed] [Google Scholar]
- Ji, L.H., and Ding, S.W. (2001). The suppressor of transgene RNA silencing encoded by Cucumber mosaic virus interferes with salicylic acid–mediated virus resistance. Mol. Plant-Microbe Interact. 14 715–724. [DOI] [PubMed] [Google Scholar]
- Jones, A.L., Thomas, C.L., and Maule, A.J. (1998). De novo methylation and co-suppression induced by a cytoplasmically replicating plant RNA virus. EMBO J. 17 6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Hamilton, A.J., Voinnet, O., Thomas, C.L., Maule, A.J., and Baulcombe, D.C. (1999). RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Ratcliff, F., and Baulcombe, D.C. (2001). RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol. 11 747–757. [DOI] [PubMed] [Google Scholar]
- Jorgensen, R., Atkinson, R., Forster, R., and Lucas, W. (1998). An RNA-based information superhighway in plants. Science 279 1486–1487. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., and Carrington, J.C. (1998). A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 95 461–470. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Haverkamp, T.H., van Luenen, H.G., and Plasterk, R.H. (1999). Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99 133–141. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293 287–289. [DOI] [PubMed] [Google Scholar]
- Knight, S.W., and Bass, B.L. (2001). A role for the Rnase III enzyme DCR-1 in RNA interference and germline development in Caenorhabditis elegans. Science 293 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, C., Franceschi, V.R., Schulz, A., Lemoine, R., and Frommer, W.B. (1997). Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275 1298–1300. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294 853–858. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294 858–862. [DOI] [PubMed] [Google Scholar]
- Lazarowitz, S.G., and Beachy, R.N. (1999). Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R.C., and Ambros, V. (2001). An extensive class of small RNAs in Caenorhabditis elegans. Science 294 862–864. [DOI] [PubMed] [Google Scholar]
- Li, W.X., and Ding, S.W. (2001). Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12 150–154. [DOI] [PubMed] [Google Scholar]
- Lindbo, J.A., Silva-Rosales, L., Proebsting, W.M., and Dougherty, W.G. (1993). Induction of a highly specific antiviral state in transgenic plants: Implications for regulation of gene expression and virus resistance. Plant Cell 5 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipardi, C., Wei, Q., and Paterson, B.M. (2001). RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107 297–307. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., and Carrington, J.C. (2000). Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, W.J., Bouche-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270 1980–1983. [DOI] [PubMed] [Google Scholar]
- Lucy, A.P., Guo, H.S., Li, W.X., and Ding, S.W. (2000). Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19 1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.D., and Steitz, J.A. (2001). Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293 1836–1839. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Ely, L., Smith, T.H., Marathe, R., Anandalakshmi, R., Fagard, M., Vaucheret, H., Pruss, G., Bowman, L., and Vance, V.B. (2001). HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, R.E.F. (1991). Plant Virology, 3rd ed. (San Diego, CA: Academic Press).
- Matzke, M., Matzke, A.J., and Kooter, J.M. (2001). RNA: Guiding gene silencing. Science 293 1080–1083. [DOI] [PubMed] [Google Scholar]
- Mette, M.F., van der Winden, J., Matzke, M.A., and Matzke, A.J. (1999). Production of aberrant promoter transcripts contributes to methylation and silencing of unlinked homologous promoters in trans. EMBO J. 18 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette, M.F., Aufsatz, W., van der Winden, J., Matzke, M.A., and Matzke, A.J. (2000). Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette, M.F., Matzke, A.J., and Matzke, M.A. (2001). Resistance of RNA-mediated TGS to HC-Pro, a viral suppressor of PTGS, suggests alternative pathways for dsRNA processing. Curr. Biol. 11 1119–1123. [DOI] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101 533–542. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J., and Santa Cruz, S. (2000). The great escape: Phloem transport and unloading of macromolecules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 323–347. [DOI] [PubMed] [Google Scholar]
- Owens, R.A., Blackburn, M., and Ding, B. (2001). Possible involvement of the phloem lectin in long-distance viroid movement. Mol. Plant-Microbe Interact. 14 905–909. [DOI] [PubMed] [Google Scholar]
- Page, M.F., Carr, B., Anders, K.R., Grimson, A., and Anderson, P. (1999). SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19 5943–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui, J.C., and Balzergue, S. (1999). Activation of systemic acquired silencing by localised introduction of DNA. Curr. Biol. 9 59–66. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.C., and Vaucheret, H. (1998). Transgenes are dispensable for the RNA degradation step of cosuppression. Proc. Natl. Acad. Sci. USA 95 9675–9680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui, J.C., Elmayan, T., Dorlhac de Borne, F., Crete, P., Charles, C., and Vaucheret, H. (1996). Frequencies, timing, and spatial patterns of cosuppression of nitrate reductase and nitrite reductase in transgenic tobacco plants. Plant Physiol. 112 1447–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui, J.C., Elmayan, T., Pollien, J.M., and Vaucheret, H. (1997). Systemic acquired silencing: Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y.D., Papp, I., Moscone, E., Iglesias, V., Vaucheret, H., Matzke, A.J.M., and Matzke, M.A. (1996). Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J. 9 183–194. [DOI] [PubMed] [Google Scholar]
- Parrish, S., and Fire, A. (2001). Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA 7 1397–1402. [PMC free article] [PubMed] [Google Scholar]
- Parrish, S., Fleenor, J., Xu, S., Mello, C., and Fire, A. (2000). Functional anatomy of a dsRNA trigger. Differential requirement for the two trigger strands in RNA interference. Mol. Cell 6 1077–1087. [DOI] [PubMed] [Google Scholar]
- Pruss, G., Ge, X., Shi, X.M., Carrington, J.C., and Bowman Vance, V. (1997). Plant viral synergism: The potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff, F., Harrison, B.D., and Baulcombe, D.C. (1997). A similarity between viral defense and gene silencing in plants. Science 276 1558–1560. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F.G., MacFarlane, S.A., and Baulcombe, D.C. (1999). Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell 11 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel, D., Lesemann, D.E., and Maiss, E. (1998). Ultrastructural localization of nonstructural and coat proteins of 19 potyviruses using antisera to bacterially expressed proteins of plum pox potyvirus. Arch. Virol. 143 2133–2158. [DOI] [PubMed] [Google Scholar]
- Roberts, A.G., Santa Cruz, S., Roberts, I.M., Prior, D.A.M., Turgeon, R., and Oparka, K.J. (1997). Phloem unloading in sink leaves of Nicotiana benthamiana: Comparison of a fluorescent solute with a fluorescent virus. Plant Cell 9 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, M.T., Voinnet, O., and Baulcombe, D.C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cazares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126 4405–4419. [DOI] [PubMed] [Google Scholar]
- Sijen, T., Fleenor, J., Simmer, F., Thijssen, K.L., Parrish, S., Timmons, L., Plasterk, R.H., and Fire, A. (2001. a). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107 465–476. [DOI] [PubMed] [Google Scholar]
- Sijen, T., Vijn, I., Rebocho, A., van Blokland, R., Roelofs, D., Mol, J., and Kooter, J. (2001. b). Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr. Biol. 11 436–440. [DOI] [PubMed] [Google Scholar]
- Sonoda, S., and Nishiguchi, M. (2000). Graft transmission of post-transcriptional gene silencing: Target specificity for RNA degradation is transmissible between silenced and non-silenced plants, but not between silenced plants. Plant J. 21 1–8. [DOI] [PubMed] [Google Scholar]
- Sun, X., Perlick, H.A., Dietz, H.C., and Maquat, L.E. (1998). A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 95 10009–10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., and Mello, C.C. (1999). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99 123–132. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Sugimoto, K., Kakutani, T., and Hirochika, H. (2001). Linear DNA intermediates of the Tto1 retrotransposon in Gag particles accumulated in stressed tobacco and Arabidopsis thaliana. Plant J. 28 307–317. [DOI] [PubMed] [Google Scholar]
- Thierry, D., and Vaucheret, H. (1996). Sequence homology requirements for transcriptional silencing of 35S transgenes and post-transcriptional silencing of nitrite reductase (trans)genes by the tobacco 271 locus. Plant Mol. Biol. 32 1075–1083. [DOI] [PubMed] [Google Scholar]
- Ueki, S., and Citovsky, V. (2001). RNA commutes to work: Regulation of plant gene expression by systemically transported RNA molecules. Bioessays 23 1087–1090. [DOI] [PubMed] [Google Scholar]
- Vance, V., and Vaucheret, H. (2001). RNA silencing in plants—Defense and counterdefense. Science 292 2277–2280. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H. (1993). Identification of a general silencer for 19S and 35S promoters in a transgenic tobacco plant: 90 bp of homology in the promoter region are sufficient for trans-inactivation. C.R. Acad. Sci. Paris 316 1471–1483. [Google Scholar]
- Vaucheret, H., and Fagard, M. (2001). Transcriptional gene silencing in plants: Targets, inducers and regulators. Trends Genet. 17 29–35. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2001). RNA silencing as a plant immune system against viruses. Trends Genet. 17 449–459. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., and Baulcombe, D.C. (1997). Systemic signalling in gene silencing. Nature 389 553. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., and Baulcombe, D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95 177–187. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Lederer, C., and Baulcombe, D.C. (2000). A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103 157–167. [DOI] [PubMed] [Google Scholar]
- Wassenegger, M. (2000). RNA-directed DNA methylation. Plant Mol. Biol. 43 203–220. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.B., and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411 834–842. [DOI] [PubMed] [Google Scholar]
- Winston, W.M., Molodowitch, C., and Hunter, C.P. (2002). Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295 2456–2459. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cazares, B., Xiang, Y., Ruiz-Medrano, R., Wang, H.L., Monzer, J., Yoo, B.C., McFarland, K.C., Franceschi, V.R., and Lucas, W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283 94–98. [DOI] [PubMed] [Google Scholar]
- Yang, D., Lu, H., and Erickson, J.W. (2000). Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr. Biol. 10 1191–1200. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D. (2001). RNA interference: Listening to the sound of silence. Natl. Struct. Biol. 8 746–750. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101 25–33. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Green, L., Woo, Y.-M., Owens, R., and Ding, B. (2001). Cellular basis of potato spindle tuber viroid systemic movement. Virology 279 69–77. [DOI] [PubMed] [Google Scholar]