INTRODUCTION
Small GTPases, having masses of 21 to 30 kD, are monomeric guanine nucleotide binding proteins related to the α subunit of heterotrimeric G proteins. All small GTPases belong to a superfamily, often named the Ras superfamily because the founding members are encoded by human Ras genes initially discovered as cellular homologs of the viral ras oncogene (Parada et al., 1982). Members of this superfamily share several common structural features, including four guanine nucleotide binding domains and an effector binding domain (Figure 1A) (Zheng and Yang, 2000b; Takai et al., 2001). However, small GTPases also exhibit a remarkable diversity in both structure and function. The Arabidopsis genome is predicted to encode 93 small GTPases, which regulate cellular processes ranging from vesicle trafficking to hormone signaling (V. Vernoud, A. Horton, Z. Yang, and E. Nielson, unpublished data). Given the existence of a relatively small number of heterotrimeric G proteins in plants compared with the number in animals (Assmann, 2002), it is not surprising that small GTPases have emerged as important molecular switches in plant signaling.
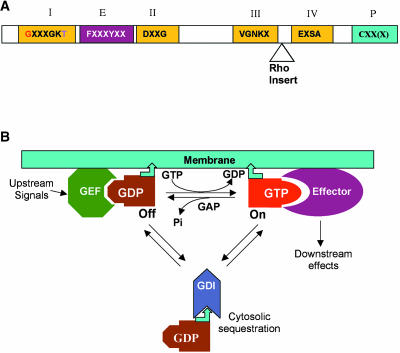
Figure 1.
Conserved Structure and Regulation of Small GTPases.
(A) Conserved structure of small GTPase. All small GTPases contain four conserved domains for guanine nucleotide binding and GTPase activities (I through IV) and an effector domain (E). Residues in red have been used for the generation of CA mutants, and those in purple for DN mutants (see Figure 2). The Rho insert (10 to 12 amino acids) is only found in Rho GTPases. Rab, Ras, and Rho GTPases also contain a C-terminal motif for prenylation (P). The motif can be CAAL (C, cysteine; A, aliphatic amino acid; L, leucine) for geranylgeranylation by geranylgeranyltransferase I (GGTase I), CC/CXC for geranylgeranylation by GGTase II, or CAAX (X indicates any amino acid except for leucine and phenyalanine) for farnesylation. Arf is myristoylated at the N terminus, but Ran has no known modification.
(B) A general scheme for the regulation and action of small GTPases. GAP, GTPase-activating protein; GDI, guanine nucleotide dissociation inhibitor; GEF, guanine nucleotide exchange factor. Bent arrowhead indicates a lipid moiety that becomes attached to membranes.
Small GTPases differ from heterotrimeric G proteins in the mechanisms by which they are regulated by upstream factors as well as those by which they activate downstream targets (Figure 1B). Upon stimulation by an upstream signal, a guanine nucleotide exchange factor (GEF) converts the GDP-bound inactive form into the GTP-bound active form through GDP/GTP replacement. Through its effector domain, the GTP form interacts with one or more specific downstream effector proteins. The GTP form exhibits a weak intrinsic GTPase activity for GTP hydrolysis, requiring a GTPase-activating protein (GAP) for efficient deactivation. In addition, most small GTPases cycle between membrane-bound and cytosolic forms. Because only membrane-associated GTPases can be activated by GEF, their removal by a cytosolic factor called guanine nucleotide dissociation inhibitor (GDI) negatively regulates these GTPases (Figure 1B). These complex modes of regulation and action for small GTPases are apparently conserved in all organisms, including plants. In spite of their small size, the interaction of small GTPases with various regulators and effectors generates functional diversity and creates novel functions in different phyla. Functional innovation of a conserved GTPase together with birth of new members of the small GTPase superfamily endows plants with the capacity to use small GTPases as a key molecular switch for the modulation of many plant-specific signaling pathways and functions.
SMALL GTPASE SUPERFAMILY IN PLANTS: CONSERVED, MISSING, AND NOVEL MEMBERS
Most Small GTPases Are Highly Conserved Regulators of Intracellular Trafficking
The small GTPase superfamily is divided into at least five families, including Ras, Rho, Rab, Arf, and Ran (Bischoff et al., 1999; Takai et al., 2001). The Rab, Arf, and Ran families are conserved in eukaryotes and directly participate in the regulation of eukaryotic hallmark cellular processes (Takai et al., 2001). Arf is required for vesicle budding in the secretory system, whereas different Rabs control transport and docking of specific vesicles. Ran regulates trafficking of RNA and proteins through the nuclear pore. Because of their essential functions in these fundamental processes, they are generally not considered as signaling proteins that transmit extracellular signals.
As expected, Rab, Arf, and Ran are all found in plants and are assumed to regulate the same general processes in plant cells (d'Enfert et al., 1992; Palme et al., 1992; Cheon et al., 1993; Regad et al., 1993; Yoshida et al., 1993; Merke et al., 1996; Haizel et al., 1997; Moore et al., 1997; Bischoff et al., 1999; Steinmann et al., 1999; McElver et al., 2000; Lu et al., 2001; V. Vernoud, A. Horton, Z. Yang, and E. Nielson, unpublished data). The physiological functions of several of these GTPases or their associated proteins have been examined in plants (Bischoff et al., 1999; V. Vernoud, A. Horton, Z. Yang, and E. Nielson, unpublished data). The roles of Ran have not been studied in plants, but the finding that the N terminus of RanGAPs is homologous to nuclear matrix attachment proteins is intriguing (Meier, 2000). Most of our knowledge of Arf function in plants was based on the study of the GNOM gene identified from genetic screen for mutations affecting embryo patterning in Arabidopsis (Steinmann et al., 1999; Geldner et al., 2001). GNOM encodes an Arf GEF (Steinmann et al., 1999). Experiments using gnom mutations, brefeldin A (an Arf GEF inhibitor), and auxin transport inhibitors suggest that an Arf or Arf GEF is critical for polar localization of the PIN1 putative auxin efflux carrier by modulating general protein trafficking between the plasma membrane (PM) and the cytoplasm (Geldner et al., 2001).
Rab is the largest family of small GTPases; 57 distinct Rab GTPases are present in Arabidopsis (Bischoff et al., 1999; Takai et al., 2001; V. Vernoud, A. Horton, Z. Yang, and E. Nielson, unpublished data). Several functional studies of plant Rab homologs seem to conform to their potential roles in vesicle trafficking (d'Enfert et al., 1992; Palme et al., 1992; Cheon et al., 1993; Regad et al., 1993; Yoshida et al., 1993; Merke et al., 1996; Haizel et al., 1997; Moore et al., 1997; Bischoff et al., 1999; Steinmann et al., 1999; Batoko et al., 2000; McElver et al., 2000; Lu et al., 2001). For example, expression of a tomato Rab11 antisense RNA caused defects in the secretion of cell wall–degrading enzymes in ripening fruits (Lu et al., 2001).
Conserved Traffic-Control GTPases May Participate in Plant Signaling
Some surprising observations have been reported regarding the function of plant homologs of Rab11, which may be involved in endocytosis or exocytosis in mammalian cells (Sano et al., 1994; Bischoff et al., 1999; Kang et al., 2001; Lu et al., 2001). The aforementioned tomato antisense rab11 plants also showed pleiotropic phenotypes associated with altered hormone accumulation or responses, for example, reduced apical dominance and ectopic shoots on leaves (Lu et al., 2001). On the other hand, overexpression of a tobacco Rab11 caused an elevated cytokinin level and pleiotropic phenotypes similar to the antisense rab11 phenotypes (Sano et al., 1994). Although these observations could be explained by pleiotropic effects of trafficking defects, further studies should determine whether Rab11 plays any direct roles in the regulation of hormone accumulation or responses. Interestingly, a pea Rab11 (PRA2) has been shown to regulate light-mediated brassinosteroid biosynthesis (Kang et al., 2001). GTP-bound PRA2 directly binds the CPC p450 cytochrome (Kang et al., 2001), suggesting a direct role for PRA2 in signaling. This finding raises an intriguing possibility that plants may have adapted to use some of the “traffic control” proteins for the transmission of extracellular signals.
ROP is the Sole Family of Signaling Small GTPases
Ras and Rho are bona fide signaling proteins known to transmit extracellular signals in yeast and animals. Although Ras has a crucial role in cellular signaling in animals and various lower eukaryotes (Bos, 2000), Arabidopsis genome sequencing reveals no Ras homologs (Arabidopsis Genome Initiative, 2000). Rho GTPases were initially shown to regulate the organization of the actin cytoskeleton and cell polarity development in eukaryotes (Johnson, 1999; Hall and Nobes, 2000; Li and Yang, 2000; Fu and Yang, 2001; Fu et al., 2001, 2002; Takai et al., 2001). But Rho signaling controls many diverse processes, including gene expression, cell wall synthesis, H2O2 production, endocytosis, exocytosis, cytokinesis, cell cycle progression, and cell differentiation in various eukaryotic organisms (Ridley, 2000; Settleman, 2001). Functional diversity of Rho GTPases is reflected by the presence of functionally distinct Rho GTPases in different organisms (Ridley, 2000; Zheng and Yang, 2000b). The Rho family is composed of conserved subfamilies (Cdc42, Rac, and Rho) and members or subfamilies unique to specific phyla (Ridley, 2000; Zheng and Yang, 2000b). Both Cdc42 and Rho are present in yeast and most animals, whereas Rac is animal-specific. Mammals possess several Rho GTPases that do not fall into these subfamilies, as does yeast (Ridley, 2000). Surprisingly, plants do not contain orthologs for any of these fungal and animal Rho-family GTPases.
However, a large number of Rho-related GTPases have been found in plants since their initial identification in pea in 1993 (Yang and Watson, 1993; Winge et al., 2000; Zheng and Yang, 2000b). Phylogenetic analysis of the plant Rho-like GTPases suggests that they all belong to a unique subfamily named ROP (for Rho-related GTPase from plants) that apparently evolved from the ancestor Rho, Rac, or Cdc42 GTPases (Li et al., 1998; Zheng and Yang, 2000b; V. Vernoud, A. Horton, Z. Yang, and E. Nielson, unpublished data). However, the naming of ROP GTPases is confusing in the literature. The Bones group named Arabidopsis Rho-like proteins ARAC (for Arabidopsis Rac) and later renamed ARACs to AtRACs (Winge et al., 1997, 2000). The Chua group also named these GTPases AtRac or At-Rac (Kost et al., 1999; Lemichez et al., 2001). Further confusion came from the use of the same name for different ROPs by different groups, for example, AtRac1 used by the Chua group is ROP6, but was used by Winge et al. for ARAC1 (ROP3) (Winge et al., 1997, 2000; Li et al., 1998; Lemichez et al., 2001). For clarity and consistency, in this review I use ROPs for various members of the ROP subfamily of Rho small GTPases. Table 1 lists all 11 Arabidopsis ROPs and the corresponding names reported. ROPs probably evolved prior to or during the evolution of land plants, because at least three ROP genes are present in the moss Physcomitrella patens (Winge et al., 2000). Consequently, all plant species examined contain multiple ROPs; Arabidopsis has 11 ROPs (Bischoff et al., 1999; Winge et al., 2000; Zheng and Yang, 2000b), whereas maize contains at least nine ROPs (J. Fowler, personal communication). Phylogenetic analysis has placed various ROPs into four distinct groups (Table 2) (Zheng and Yang, 2000b).
Table 1.
Proposed Unifying Nomenclature of Rop Members and Corresponding Names Reported Elsewhere
| ROPa | ROP1 | ROP2 | ROP3 | ROP4 | ROP5 | ROP6 | ROP7 | ROP8 | ROP9 | ROP10 | ROP11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Arac/ AtRACb |
Arac11/ AtRAC11 |
Arac4/ AtRAC4 |
Arac1/ AtRAC1 |
Arac5/ AtRAC5 |
Arac6/ AtRAC6 |
Arac3/ AtRAC3 |
Arac2/ AtRAC2 |
Arac9/ AtRAC9 |
Arac7/ AtRAC7 |
Arac8/ AtRAC8 |
Arac10/ AtRAC10 |
| AtRac/ At-Racc |
AtRac2 | AtRac1 |
Li et al., 1998; this review.
Winge et al., 1997, 2000.
Kost et al., 1999; Lemichez et al., 2001.
Table 2.
Phylogenetic Groups of the Rop-Subfamily Rho GTPases and Known or Potential Functions
| Groups | Arabidopsisa | Cotton (Gh)/Tobacco (Nt) | Maize (Zm)/Rice (Os) |
|---|---|---|---|
| I | ROP8 (function unknown) | — | — |
| II | ROP9/ROP10 (ABA responses–LOF/DN/CA) ROP11 (function unknown) |
— — |
OsRac1 (H2O2 production–DN/CA) OsRac2 to OsRac4, ZmROP6 to ZmROP8 (function unknown) |
| III | ROP7 (inhibition of root hair tip growth–OX) | GhRac9/GhRac13 (H2O2 production–DN/CA) | OsRop5 (function unknown) |
| IV | ROP1 to ROP6 (actin dynamics, polar growth, root hair development, ABA responses–DN/CA/OX) |
GhRac1 (function unknown) | ZmROPB/ZmROPD (function unknown) |
a Approaches used for functional analysis: LOF, loss-of-function; DN, dominant negative mutants; CA, constitutively active mutants; OX, overexpression.
Why do plants only need a single subfamily of signaling small GTPases? An intriguing hypothesis is that ROPs evolved as a unique molecular switch in place of Cdc42, Rac, and Rho as well as Ras in the regulation of actin organization and cell polarity as well as for the transmission of extracellular signals (Li et al., 1998; Winge et al., 2000; Zheng and Yang, 2000b). Hence, ROPs have attracted a great deal of attention over the last few years and are the focus of this review.
FUNCTIONAL VERSATILITY OF ROP GTPASES
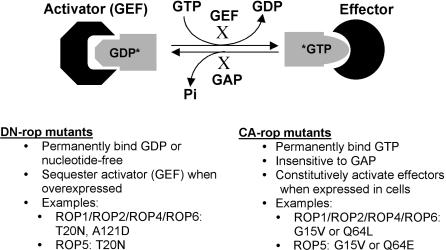
The presence of multiple ROP genes presents a great challenge for the elucidation of their function, because of potential functional redundancy. Therefore, ROP overexpression and gain-of-function rop mutants have been used to investigate the physiological roles of ROP GTPases. These mutants are blocked in the cycling between the GDP and GTP forms (Figure 2). Replacements of specific amino acid residues lock a mutant GTPase either in the inactive form, called a dominant negative (DN) mutant, or in the active form, called a constitutively active (CA) mutant (Figure 2) (Li et al., 1999; Valster et al., 2000; Zheng and Yang, 2000a, 2000b). These rop mutants have been instrumental in understanding the roles of ROPs (Kawasaki et al., 1999; Kost et al., 1999; Li et al., 1999, 2001; Lemichez et al., 2001; Molendijk et al., 2001; Fu et al., 2002; Jones et al., 2002). However, caution should be exercised when interpreting data based on these mutants, as in any other gain-of-function approaches. Furthermore, CA or DN mutants for one ROP gene might interfere with the function of different functionally distinct ROP GTPases, making it difficult to evaluate roles of specific ROP genes. Importantly, recent use of T-DNA insertion mutants in combination with double-stranded RNA interference (RNAi) expression has led to exciting discoveries about the function of specific ROP genes in Arabidopsis.
Figure 2.
Schematics of CA and DN rop Mutants Used for Functional Analysis.
The DN mutant of GTPase is locked in GDP-bound or nucleotide-free form, and the CA mutant is locked in GTP-bound form. Overexpression of DN mutants blocks Rop activation by sequestering activators, whereas CA mutants are insensitive to the action of RopGAPs and thus permanently activate effectors. Examples shown are those used for the investigation of the function of Arabidopsis ROPs (Kost et al., 1999; Li et al., 1999; Lemichez et al., 2001; Li et al., 2001). X indicates the inhibition of the cycling between GDP and GTP forms by the dominant mutations.
ROP Control of Pollen Tube Growth
The examination of the roles of ROPs in plants began with the finding that pea ROP proteins preferentially localize to the apical region of the PM in pollen tubes (Lin et al., 1996). This finding was significant, because pollen tubes elongate by polarized tip growth, a process requiring targeting of Golgi vesicles to the tube apex and their fusion with the apical PM region where ROPs are localized. An essential role for ROPs in pollen tube growth was demonstrated using microinjection of an anti-ROP1 antibody into pea pollen tubes (Lin and Yang, 1997). Three closely related ROPs (ROP1, ROP3, and ROP5) expressed in Arabidopsis pollen were suspected to be functionally redundant in pollen tube growth (Li et al., 1998; Kost et al., 1999). Functional redundancy for ROP1 and ROP5 was supported by the inhibition of pollen tube elongation by pollen-specific expression of either DN-rop1 or DN-rop5 (Kost et al., 1999; Li et al., 1999).
ROPs are also involved in the development of cell polarity in pollen tubes, as suggested by the induction of depolarized pollen tube growth by CA-rop1/CA-rop5 expression or overexpression (OX) of wild-type ROP1 or ROP5 (Kost et al., 1999; Li et al., 1999). A simple explanation for the observed ROP functions in both growth and polarity control is that localized activation of a ROP pathway in the apical PM region leads to pollen tube elongation. This is confirmed by the localization of GTP-bound active ROP to the apical PM region as a tip-high gradient, which was shown using GFP-tagged RIC1 (for ROP-interacting CRIB-containing protein), a protein that binds GTP-ROP1 but not GDP-ROP1 (Figure 3; Wu et al., 2001; G. Wu, Y. Fu, V. Vernoud, and Z. Yang, unpublished data). This is an important observation, not only because it provides evidence for the activation of a signaling protein in a localized PM domain in a plant cell but also because the paradigm of localized ROP signaling may generally apply to ROP-dependent polar growth and cell polarity development in various cell types (Figure 3).
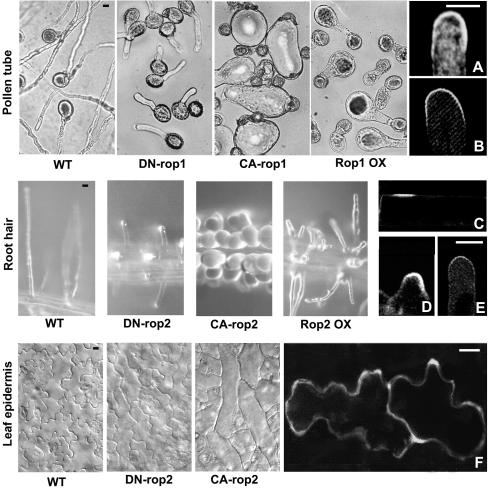
Figure 3.
Rop Regulates Cell Polarity Development and Polar Growth through Recruitment to and Activation at the Site of Action.
Typical cell-shape phenotypes induced by CA-rop, DN-rop, or Rop OX in different cells (Li et al., 1999; Fu et al., 2002; Jones et al., 2002) are shown at left, and Rop localization or its activation is shown at right.
(A) Rop localization to the apical PM region in pea pollen tubes as shown by indirect immunoflurorescence (Lin et al., 1996).
(B) The localization of GFP-tagged RIC1 in tobacco pollen tubes, indicating the localization of GTP-bound active Rop to the apical PM region (Wu et al., 2001).
(C) to (E) The localization of GFP-tagged ROP2 to the future hair forming site in the epidermal cell (C), the tip of swelling prior to the initiation of tip growth (D), and the tip of growing root hair (E) (Jones et al., 2002).
(F) Preferential localization of GFP-ROP2 to the site of lobe formation in expanding leaf epidermal cells (Fu et al., 2002).
All images shown were obtained using confocal microscopy. Scale bars = 10 μm.
Which signals activate the ROP signaling pathway and which factors control the localization of this pathway to the apical PM region in pollen tubes are important and interesting questions. A study by Li et al. (1999) supports the model for an elaborate spatial regulation via a ROP signaling loop (Li et al., 1999; Zheng and Yang, 2000a). A basal level of PM-localized ROPs is activated by an unknown localized cue. Activated ROPs promote the recruitment of more ROP proteins to the site of ROP activation, forming a positive feedback loop of ROP activation–recruitment. Unchecked operation of this loop would cause the activated ROP signaling pathway to spread away from the site of initial activation, as in the case of isotropic growth caused by CA-rop1 (Figure 3); however, the negative regulation of this pathway by GAPs and GDIs restricts the signaling pathway to the tip. This model can explain various observations, including the enhancement of GFP-ROP1 distribution to the apical PM region by ROP1 OX and more severe depolarized growth induced by CA-rop1 expression than wild-type ROP1 OX (Figure 3; Li et al., 1999; Zheng and Yang, 2000a). Given the localization of Rho GTPases to the site of growth and to the leading edge of moving cells in various systems (Symons and Settleman, 2000), this model may provide a paradigm for Rho GTPase control of polar and directional growth or movement in general. However, more work is needed to test this model.
How does the tip-localized ROP signaling pathway regulate pollen tube growth? Current evidence suggests that tip-localized active ROPs modulate both the generation of tip-focused Ca2+ gradients and the assembly of dynamic tip F-actin, both of which are critical for pollen tube growth (Gibbon et al., 1999; Li et al., 1999; Zheng and Yang, 2000a; Feijo et al., 2001; Fu and Yang, 2001). It is proposed that ROPs activate two coordinate downstream pathways respectively, leading to actin assembly and Ca2+ accumulation at the tip (Fu et al., 2001). The former may target vesicles to the site of growth, whereas the latter may regulate vesicle fusion with the apical PM region. This hypothesis is supported by recent identification of two structurally distinct putative ROP1 targets in the control of polar growth in pollen tubes, RIC3 and RIC4 (Wu et al., 2001). The potential coordination of multiple ROP downstream pathways is analogous to Rho1 control of polar growth in yeast via multiple pathways, including recruiting of exocyst (the machinery for exocytosis), activating cell wall assembly, and regulating actin organization (Guo et al., 2001). Unfortunately, RIC3 and RIC4 have no homology to proteins of known function, and thus how ROPs regulate localized F-actin assembly and calcium accumulation remains a mystery. Nonetheless, the study of ROP function in pollen tubes establishes a signaling pathway that controls polarized cell growth in plants.
ROP Regulation of Root Hair Development
Two recent studies demonstrate a pivotal role for ROPs in regulating various aspects of root hair development (Molendijk et al., 2001; Jones et al., 2002). Root hair development involves complex morphogenesis of single epidermal hair-forming cells. It begins with swelling (via diffuse growth) from a site near the basal end of each hair-forming cell. Tip growth, similar to pollen tube growth, is subsequently initiated at the apex of the swelling to form a hair. Localization using an anti-ROP4 antibody and GFP-tagged ROP2 shows that ROPs localize to the tip of elongating Arabidopsis hairs as in pollen tubes (Figure 3) (Molendijk et al., 2001; Jones et al., 2002). Furthermore, expression of CA mutants of ROP2, ROP4, or ROP6 caused either isotropic growth or increased length in Arabidopsis root hairs (Molendijk et al., 2001; Jones et al., 2002), whereas DN-rop2 expression inhibited root hair tip growth (Jones et al., 2002). Interestingly, ROP2 OX causes several morphological changes during root hair development, including increased hair length (Jones et al., 2002). These two studies show that ROP controls tip growth in root hairs as well as in pollen tubes. As in pollen tubes, ROPs control tip growth in root hair apparently via two downstream pathways respectively, regulating tip actin and tip-focused calcium gradients (Baluska et al., 2000; Fu and Yang, 2001; Molendijk et al., 2001; Jones et al., 2002).
Apart from tip growth, ROPs also control the site of swelling formation and the establishment of tip growth sites (Jones et al., 2002). This conclusion came from other ROP2 OX phenotypes, including mislocation of swellings and formation of multiple swellings from a single hair-forming cell, formation of multiple hairs from a single swelling, and continuous branching of root hairs (Jones et al., 2002). The localization of ROPs to the tip of swellings and the future hair site in root hair-forming cells, as revealed by GFP-ROP2 expression (Figure 3) or immunolocalization using anti-ROP4 antibodies (Molendijk et al., 2001; Jones et al., 2002), is consistent with these ROP2 OX phenotypes. The swelling formation is thought to involve diffuse growth independent of F-actin, whereas the establishment of tip growth sites is regulated by microtubules (Bibikova et al., 1999). Thus, ROPs modulate these early processes of root hair development probably through distinct mechanisms different from its control of tip growth during root hair elongation.
Although ROP2, ROP4, and ROP6 in the same phylogenetic group (Table 2) may be functionally redundant in root hair development, to date only ROP2 has been shown to be expressed in root hair-forming cells and in growing root hairs (Jones et al., 2002). Furthermore, the localization patterns for GFP-ROP2 and various ROP2 OX root hair phenotypes suggest that a single ROP may control various stages of cell polarity development in root hairs (Jones et al., 2002). This is in contrast to the control of cell polarity development in yeast, where three distinct G proteins, namely, a Ras-like GTPase or heterotrimeric G protein, Cdc42, and Rho1, respectively, control polar site selection, polarity establishment, and polar growth (Chant, 1999). These observations are consistent with the notion that as the sole subfamily of signaling small GTPases in plants, ROPs have adapted various functions that would otherwise be controlled by distinct types of GTPases in animals and yeast (Fu and Yang, 2001).
ROPs and Cell Expansion in Developing Tissues
Unlike pollen tubes and root hairs, which expand by localized tip growth, cells within developing organs are thought to expand by diffuse growth (Kropf et al., 1998). According to the diffuse growth hypothesis, cell shape formation is determined by the orientation of cellulose microfibrils and cortical microtubules (Kropf et al., 1998). Surprisingly, two recent studies have shown that ROP signaling also plays a general role in the regulation of cell shape formation in developing Arabidopsis tissues (Molendijk et al., 2001; Fu et al., 2002). On the basis of the observation that CA-rop mutants caused isotropic expansion of root and hypocotyl epidermal cells, Molendijk et al. proposed that ROP signaling regulates the polarity of diffuse growth (Molendijk et al., 2001). During organogenesis, polar cell expansion occurs in two phases: an early phase involving both axial and radial or lateral expansion and a late phase involving only elongation (Schindelman et al., 2001; Fu et al., 2002). A series of careful analyses of cell shape changes in transgenic Arabidopsis plants expressing CA-rop2 and DN-rop2 led to the conclusion that ROP2 regulates specifically the early stage of polar cell expansion (Figure 3) (Fu et al., 2002). Furthermore, DN-rop2 expression eliminated a diffuse form of cortical F-actin that is specifically associated with polar expansion in the early phase of leaf epidermal pavement cells, whereas CA-rop2 caused uniform distribution of this F-actin throughout the entire cell cortex (Fu et al., 2002).
The localized assembly of F-actin appears to be a unifying mechanism underlying the ROP control of polar cell growth in different cell types, including tip-growing and non-tip-growing cells (Fu and Yang, 2001; Fu et al., 2001, 2002; Jones et al., 2002). Apart from this unifying mechanism, additional specific ROP-dependent mechanisms likely are required for cell shape formation in different cell types. For example, ROPs appear to regulate the formation of a Ca2+ gradient in tip-growing cells, which has not been found in non-tip-growing cells. The elucidation of mechanisms by which ROP controls cell polarity development and polar growth in different cell systems will require the determination of specific ROP(s) and corresponding ROP targets and regulators involved in each morphogenetic pathway.
ROP Signaling Regulates the Production of the H2O2 Second Messenger
Another well-documented ROP function is regulation of H2O2 production. Interest in this function started with the search for a plant homolog of human Rac2 (one of the regulatory subunits of neutriphil NADPH oxidase) using an anti-human Rac2 antibody (Bokoch, 1994; Xing et al., 1997). In another attempt, Kawaski et al. identified several rice Rac-like GTPases that belong to group II ROPs (Table 1) and showed that expression of CA-OsRac1 and DN-OsRac1 mutants, respectively, activated and inhibited H2O2 production induced by pathogens (Kawasaki et al., 1999). Interestingly, CA-OsRac1-induced H2O2 production was inhibited by the NADPH oxidase inhibitor diphenylene iodonium (DPI). Similar results were obtained when dominant mutants of a cotton ROP (GhRac13) or human Rac1were expressed in Arabidopsis or soybean suspension cultures (Potikha et al., 1999; Park et al., 2000). A role for ROP signaling in the regulation of H2O2 production has also been demonstrated in Arabidopsis responses to oxygen deprivation, which rapidly and transiently activates ROPs in wild-type Arabidopsis seedlings, leading to the ROP-dependent production of H2O2 and H2O2-dependent alcohol dehydrogenase (ADH) gene expression (Baxter-Burrell et al., 2002). The ROP-dependent induction of H2O2 was also inhibited by DPI treatments.
DPI inhibition of ROP-dependent H2O2 production strongly supports the notion that ROP is functionally equivalent to human Rac in its activation of a PM-associated NADPH oxidase. Functional homologs of the gp91phox catalytic subunit of the neutriphil PM NADPH oxidase are present in plants, although no plant homologs of two other regulatory subunits (p47phox and p67phox) have been demonstrated (Sagi and Fluhr, 2001). Novel plant proteins might serve as regulatory subunits of NADPH oxidase. This is in accord with the lack of similarity between the ROP insert region corresponding to residues 126 to 135 of Arabidopsis ROP1 and the Rac insert region known to interact with p67phox. Nonetheless, future studies should determine whether ROP acts as an NADPH regulatory subunit or regulates H2O2 production through other mechanisms. H2O2 has emerged as an important second messenger in plant signaling, such as in the regulation of cotton fiber formation and programmed cell death as well as abscisic acid (ABA) and auxin signaling (Lamb and Dixon, 1997; Potikha et al., 1999; Pei et al., 2000; Joo et al., 2001). Therefore, ROP regulation of H2O2 production may prove to be a very important signaling module in plants.
Negative Regulation of ABA Responses by ROP Signaling
One of the most exciting findings about ROP signaling is the demonstration of its involvement in the negative regulation of ABA responses. A report implicating ROPs in ABA responses describes the respective enhancement and reduction of ABA-inhibited seed germination by DN-rop2 and CA-rop2 expression in Arabidopsis (Li et al., 2001). However, this observation did not demonstrate whether ROP is a direct negative regulator of an ABA pathway or a positive regulator of a pathway that antagonizes ABA responses. A role for ROPs in the negative regulation of ABA responses was more convincingly shown in guard cells by Lemichez et al. (2001). Expression of CA-rop6 in Arabidopsis inhibited ABA-induced stomatal closure in wild-type plants, whereas DN-rop6 expression caused stomatal closure in both the wild type and the abi-1 mutant in the absence of exogenous ABA. This study provided evidence that ABA inactivates one or more ROPs, which apparently act downstream of the ABI1 protein phosphatase, leading to stomatal closure probably through the disruption of actin organization in guard cells (Lemichez et al., 2001). It will be interesting to know whether the ABI1-ROP pathway is guard cell-specific or is also involved in other ABA responses, including those affected by DN-rop2 and CA-rop2 expression.
Studies of loss-of-function mutants have demonstrated ROP9 and ROP10 as more general negative regulators of ABA responses (Z.-L. Zheng and Z. Yang, unpublished data). A rop10 knockout (rop10-1) or a rop9(RNAi) mutant each showed weak enhancement of ABA inhibition of seed germination, seedling greening, and root growth, although rop10-1/rop9(RNAi) double mutations strongly enhanced ABA responses in these aspects. Because both ROP9 and ROP10 contain a putative C-terminal farnesylation motif (see below), one or both of these ROPs could be target of ERA1, the β subunit of protein farnesyltransferase, known to be involved in the negative regulation of ABA responses in both guard cell movement and seed dormancy (Cutler et al., 1996; Pei et al., 1998).
These studies raise an intriguing question: Does the regulation of ABA responses involve multiple ROP-dependent pathways, each controlled by a distinct ROP or a subset of ROPs? The altered ABA responses caused by expression of rop2 or rop6 dominant mutants (Lemichez et al., 2001; Li et al., 2001) could be explained by their interference with the function of ROP9 and/or ROP10. Alternatively, ROP2, ROP6 and ROP9, and ROP10 could respectively regulate distinct ABA pathways. In agreement with the latter is the observation that ROP9 and ROP10 are functionally distinct from ROP2 and ROP6 (Table 1) and that CA-rop10 expression did not interfere with the function of ROP2 and ROP6 in the control of cell expansion and ABA-independent processes (Z.-L. Zheng and Z. Yang, unpublished data). Nonetheless, analysis of various single and multiple rop knockout mutants should help to establish the precise role of ROP GTPases in ABA signaling.
ROP Regulation of Other Hormone Functions and Developmental Processes
ROP regulation of other hormone functions was implicated by analysis of transgenic plants expressing 35S:CA-rop2 or 35S:DN-rop2. 35S:CA-rop2 plants exhibit many morphological phenotypes that resemble auxin- or brassinolide-overproduction plants, whereas DN-rop2 plants exhibit many opposite phenotypes that resemble brassinolide-deficient or -insensitive or auxin-resistant mutants (Li et al., 2001). CA-rop2 expression enhanced exogenous brassinolide-induced hypocotyl elongation of light-grown seedlings, whereas DN-rop2 expression inhibited hypocotyl elongation. Similarly, CA-rop2 expression increased the sensitivity of promotion of lateral root formation induced by exogenous IAA, whereas DN-rop2 inhibited this process. These CA-rop2 and DN-rop2 phenotypes could be explained by the hypothesis that different ROPs are involved in the respective regulation of the responses and/or accumulation of brassinolide and auxin (Li et al., 2001). Alternatively, the transgenic phenotypes could be the result of the primary effect of ROP signaling on the regulation of one hormone and a cross-talk between brassinolide and auxin.
CA-rop2 and DN-rop2 expression also induced other developmental phenotypes, including defects in embryo development, phyllotaxis, and pedicel orientation (Zheng and Yang, 2000b; Li et al., 2001). These phenotypes apparently cannot be explained by the effect of these mutants on hormone functions and cell morphogenesis described above (Zheng and Yang, 2000b; Li et al., 2001). ROP proteins have been shown to localize to developing vacuoles and are preferentially accumulated in the tapetum, microsporogenic cells, and vascular tissues (Li et al., 2001; Lin et al., 2001). Hence, future studies are expected to reveal additional ROP signaling pathways in plants.
Are Different Phylogenetic Groups of ROP GTPases Functionally Distinct?
Does the evolution of ROP GTPases into four phylogenetic groups (Table 2) reflect their functional divergence? As discussed earlier, ROP9 and ROP10 (group II) could be functionally redundant ERA1 targets that negatively regulate ABA responses in Arabidopsis. CA-rop10 expression reduced sensitivity to exogenous ABA but did not cause morphological phenotypes induced by CA-rop2 expression (Z.-L. Zheng and Z. Yang, unpublished data). Similarly, dominant mutants for OsRac1 have been shown to alter pathogen-induced H2O2 production, but no morphological phenotypes were reported for transgenic plants expressing these mutants (Li et al., 2001). In contrast, CA-rop2 expression caused pleiotropic developmental and cell shape phenotypes (Fu and Yang, 2001; Li et al., 2001; Fu et al., 2002; Jones et al., 2002), and CA-rop4 and CA-rop6 expression also caused cell shape phenotypes (Molendijk et al., 2001). ROP2, ROP4, and ROP6 all belong to group IV (Table 2). Furthermore, distinct root hair phenotypes were observed in Arabidopsis plants overexpressing a representative member from each group (Jones et al., 2002). Taken together, these data suggest that group II tends to participate in stress responses, including ABA responses and H2O2 production, whereas group IV seems predominantly to regulate cell polarity and cell expansion through actin organization (Kost et al., 1999; Li et al., 1999; Fu and Yang, 2001; Fu et al., 2001, 2002; Lemichez et al., 2001; Molendijk et al., 2001; Jones et al., 2002).
Different members within each group may regulate distinct processes in different cell types and by distinct mechanisms, depending on their expression pattern and the expression of their functional partners. For example, Arabidopsis ROP1, ROP3, and ROP5 (group II) are expressed in pollen and appear to be functionally redundant in regulating pollen tube tip growth, whereas ROP2, ROP4, and ROP6 appear predominantly to control tip growth and cell expansion in vegetative tissues (Li et al., 1998). ROP regulation of tip growth may differ from that of cell expansion in non-tip-growing cells, as evidence for calcium involvement in the latter is lacking. Elucidation of the interacting partners for different ROPs will provide us with a better understanding of how different ROPs may control distinct signaling pathways.
A DIVERSE ARRAY OF ROP REGULATORS AND TARGETS
ROP GTPases interact with various upstream regulators and downstream effectors, as do Rho GTPases in yeast and animals. The diversity of ROP interactors may account for the functional versatility of ROP as a molecular switch in cell signaling. As expected, some of these ROP interactors are conserved proteins involved in Rho GTPase signaling. Interestingly, many ROP interactors are novel or unique to plants, consistent with the hypothesis that ROPs belong to a plant-specific branch of the Rho family of small GTPases (Li et al., 1998; Zheng and Yang, 2000b).
Regulation of ROP GTPases
Regulation of ROP Subcellular Localization
Like other G proteins, membrane targeting is usually a prerequisite for the activation of ROP GTPases or their interaction of effectors. In some cases, regulated ROP recruitment to a specific subcellular membrane is crucial for the proper function of ROPs (see Figure 3), consistent with their roles in spatial regulation of cellular processes (Kost et al., 1999; Li et al., 1999; Fu and Yang, 2001; Fu et al., 2001, 2002; Lemichez et al., 2001; Molendijk et al., 2001; Jones et al., 2002). As with most small GTPases, membrane association of ROPs is mediated by post-translational modification, including prenylation of a cysteine residue in the C terminus (Kost et al., 1999; Li et al., 1999). All members of groups I, III, and IV contain a signature motif (CAAL) recognized by protein geranylgeranyltransferase I (GGTase I) (Zheng and Yang, 2000b; Li et al., 2001). Possible prenylation sites for group II ROPs are more heterogeneous, with maize ROP6/ROP7 having a putative motif (CAA) for GGTase II (Ivanchenko et al., 2000), Arabidopsis ROP9/ROP10 containing a possible farnesylation motif (CTAA and CGKN), and ROP11 lacking a prenylation motif (Zheng and Yang, 2000b; Li et al., 2001). Interestingly, a C-to-S replacement in the CAA motif of maize ROP6/ROP7 did not significantly affect their localization to the PM (Ivanchenko et al., 2000), and Arabidopsis ROP10 can be localized to the PM in a farnesylation-independent manner (Z.-L. Zheng, D. Crowell, and Z. Yang, unpublished data). This raises the possibility that a different type of modification, for example, palmitoylation, may be crucial for membrane targeting of these ROPs (see below). The final three residues of prenylated Rho GTPases are usually removed by proteolytic cleavage, followed by methylation at the prenylated cysteine residue (Nambara and McCourt, 1999). The methylation appears to stabilize membrane association of prenylated proteins. Prenyly cysteine carboxy methylation transferase is present in plants, suggesting that similar modifications occur in ROP GTPases (Crowell et al., 1998; Rodriguez-Concepcion et al., 2000; Crowell and Kennedy, 2001).
Additional targeting signals are required for ROP GTPase recruitment to specific subcellular compartments. Several subcellular localization patterns have been described for different ROPs: a perinuclear organelle (Arabidopsis ROP4), the entire PM (Arabidopsis ROP9, ROP10, and ROP6; maize ROP6 and ROP7), specific domains of the PM (Arabidopsis ROP1, ROP2, and ROP5), and the tonoplast of developing vacuoles (one or more pea ROPs) (Kost et al., 1999; Li et al., 1999; Bischoff et al., 2000; Lin et al., 2001; Fu et al., 2002). As shown for some animal Rho GTPases, information for specific ROP localization apparently also resides in the C-terminal variable region proximal to the isoprenylaton acceptor site (Bischoff et al., 2000). In mammalian cells, signals for PM localization of prenylated GTPases are either a polybasic domain or palmitoylation of cysteine residues within the C-terminal variable region (Adamson et al., 1992). Group II ROPs contain one or two corresponding cysteine residues that may be palmitoylated, and constitutively localize to the entire PM (Ivanchenko et al., 2000). Group IV ROPs contain a polybasic domain in the C-terminal region, and the members within this group studied to date all show partitioning between the PM and the cytosol (Li et al., 1999; Fu et al., 2002), as mammalian RhoA and RhoC (Adamson et al., 1992). Such a partitioning may allow a dynamic regulation of ROP recruitment to the site of action (Figure 3) (Kost et al., 1999; Li et al., 1999; Fu et al., 2002; Jones et al., 2002). The difference in the regulation of subcellular localization between these two groups of ROPs agrees with their distinct cellular functions as discussed above.
What cellular factors recruit ROP GTPases to specific PM domains and control their partitioning between the cytosol and PM? Interestingly, the localization of ROPs to the hair-forming sites was abolished by brefeldin A treatment, suggesting that vesicle trafficking per se or a secreted protein is involved in recruiting ROPs to these sites (Molendijk et al., 2001). Evidence suggests that active ROP promotes the recruitment of ROP1 to the apical PM region in pollen tubes (G. Wu and Z. Yang, unpublished data). However, it is unknown how ROP promotes its own recruitment to the PM. A ROP-specific GDI plays an important role in the regulation of ROP recruitment to the PM (Bischoff et al., 2000; G. Wu and Z. Yang, unpublished data), because overexpression of Arabidopsis ROP GDI1 removed GFP-tagged ROP1 from the PM and suppressed depolarized growth caused by ROP1 OX (G. Wu, Y. Fu, V. Vernoud, and Z. Yang, unpublished data). In Arabidopsis, three ROP GDIs have been identified (Bischoff et al., 2000). A ROP GDI could also be involved in the regulation of ROP2 localization to specific PM domains (Fu et al., 2002; Jones et al., 2002). It is yet to be determined whether different ROP GDIs have differential effects on the localization of different ROPs.
ROP GTPases Are Not Activated by Conventional GEFs
Fungal and animal Rho GTPases are directly activated by Rho GEFs. These Rho GEFs typically contain a DH domain, the catalytic domain for the GDP to GTP exchange in Rho GTPases (Cerione and Zheng, 1996). Surprisingly, plants lack proteins homologous to the DH domain, although GEFs for other small GTPases such as ARF have been identified in plants (Steinmann et al., 1999). Two possibilities can explain the lack of DH homologs in plants. A novel GEF could have evolved for ROP activation, consistent with the probable early evolution of ROPs prior to the divergence of ancestoral Rho in yeast and animals. Alternatively, a novel mechanism could be involved in ROP activation. For example, ROP could bind GTP with a much higher affinity, and thus GTP-bound ROP could be spontaneously formed as soon as GDP-bound ROP is recruited to the PM. In this case, the rate-limiting step for ROP activation could be the activation of specific ROP-recruiting factor. Such an unconventional Rho activation agrees with the presence of ROP-specific GAPs with high-specific affinity for ROPs (Wu et al., 2000). Interestingly, a ROP-like protein directly associates with the active 450-kD CLV1 receptor-like kinase (RLK) complex but not with the inactive 280 CLV1 complex (Trotochaud et al., 1999). ROPs could be directly regulated by RLKs, although the functional significance of ROP-RLK association remains to be determined. Identification of direct ROP activators is a very important challenge in the study of ROP signaling.
Plant Rho GAPs Are Unique
Like other small GTPases, GAP is required for efficient inactivation of Rho GTPases. A novel class of plant Rho GAPs was identified using the yeast two-hybrid method (Borg et al., 1999; Wu et al., 2000). These GAPs contain a GAP catalytic domain with the greatest similarity to the mammalian Cdc42 GAP (Wu et al., 2000). However, in vitro assays showed that Arabidopsis Rho GAPs strongly promote GTP hydrolysis by ROPs but only weakly affect GTP hydrolysis by Cdc42, suggesting that they act as ROP-specific GAPs. Thus, the plant Rho GAPs are termed RopGAPs (Wu et al., 2000). Importantly, RopGAPs are structurally unique with a Cdc42/Rac-interactive binding (CRIB) motif immediately upstream of the GAP domain. This motif, present in many Cdc42/Rac effectors, allows their specific binding to active Cdc42/Rac GTPases (Aspenstrom, 1999) but has not been found in any animal or fungal Rho/Cdc42/Rac GAPs.
Why do RopGAPs uniquely contain a CRIB motif? The CRIB motif is crucial for ROP-specific RopGAP promotion of GTP hydrolysis, because mutations within the CRIB motif dramatically reduce RopGAP1 activity to a basal level as well as reduce its binding to ROP1 (Wu et al., 2000). Because the CRIB domain from RopGAPs binds the transitional state of ROP, it was proposed that this domain stabilizes or facilitates the formation of the ROP transitional state during GTP hydrolysis (Wu et al., 2000). In vivo studies show that the CRIB motif is critical for the localization of Arabidopsis RopGAP1 to the apical PM region in pollen tubes and for the function of RopGAP1 as a negative regulator in ROP1 signaling to pollen tube tip growth (G. Wu and Z. Yang, unpublished data).
An exciting breakthrough in the study of ROP signaling is the revelation of a role for RopGAP4 in the negative feedback regulation of ROP signaling using a ropgap4 knockout mutant (Baxter-Burrell et al., 2002). This mutant exhibits a high basal level of ROP activity and shows dramatically enhanced ROP-dependent ADH expression induced by oxygen deprivation. Interestingly, oxygen deprivation-activated ROP signaling and H2O2 induce RopGAP4 expression, indicating a RopGAP4-dependent negative feedback loop. These observations establish that ROP acts as a rheostat instead of a switch both to activate the production of H2O2 as a second messenger and to prevent the accumulation of H2O2 to a toxic level. It will be interesting to see whether similar RopGAP-dependent ROP rheostats control other ROP-dependent pathways.
RopGAPs are encoded by a multigene family (Borg et al., 1999; Wu et al., 2000). Arabidopsis contains six RopGAP genes, and RopGAP homologs are present in both monocot and dicot species. N- and C-terminal regions of RopGAPs outside of the conserved CRIB and GAP domains are highly variable and may provide functional specificity for different RopGAPs. Interestingly, the ropgap4 knockout mutant, although having high basal level of active ROPs (Baxter-Burrell et al., 2002), does not exhibit morphological phenotypes that are caused by CA-rop2 expression (Li et al., 2001). Future work should determine whether different RopGAPs differentially interact with ROPs and if knockout mutants for different RopGAPs show distinct phenotypes.
ROP GTPase Downstream Targets
A Class of CRIB-Containing Novel Proteins as Functionally Distinct ROP Targets
In yeast and animals, each subfamily of Rho GTPases activates a plethora of distinct targets with various biochemical functions, ranging from protein kinases through regulators of actin-nucleating complexes to β-1,3-glucan synthase (Aspenstrom, 1999). Surprisingly, no homologs of Rho targets from animals and yeast have been found in plants (Wu et al., 2001). Because effectors normally only interact with the GTP form of small GTPase, a CA-rop1 mutant was used as a bait in a yeast two-hybrid screen to isolate putative ROP effectors from Arabidopsis (Wu et al., 2001). This screen identified a novel protein containing a CRIB motif, termed RIC (for ROP-interacting CRIB-containing protein), which interacted with GTP-bound not GDP-bound ROP1. A database search reveals 10 additional Arabidopsis genes encoding RICs. Regions outside of the CRIB motif are highly variable in the 11 predicted Arabidopsis RICs. Because the CRIB motif is a hallmark structure of many Cdc42/Rac effectors, RICs are attractive candidates for functionally distinct ROP GTPase targets.
Distinct functions for nine of the 11 RIC genes expressed in mature Arabidopsis pollen were tested using a robust transient expression assay in tobacco pollen tubes (Wu et al., 2001). Several categories of RIC OX phenotypes were observed: (1) depolarized growth of pollen tubes as observed in ROP1 OX (RIC3 and RIC4); (2) promotion of tube elongation (RIC10); (3) inhibition of both tube elongation and radial expansion (RIC5); and (4) inhibition of pollen tube elongation (all other RICs). Furthermore, different GFP-tagged RICs displayed different subcellular localization patterns. Finally, the effects of ROP1 OX on RIC OX phenotypes and the subcellular localization of RICs differ among different RICs. For example, ROP1 OX enhanced RIC4 localization to the apical PM region and RIC3 localization to the subcortical region of the tip but had no effect on the localization of RIC2, RIC5, RIC9, and RIC10. On the basis of these results, it was postulated that RIC3 and RIC4 are ROP1 targets in the control of pollen tube growth, whereas RIC9 might be a target of another functionally distinct ROP in pollen tubes (Wu et al., 2001). Most RIC genes are expressed in various Arabidopsis tissues, and RIC homologs are present in different plant species (Wu et al., 2001). These observations strongly support the hypothesis that different RICs act as distinct ROP targets to control various ROP-dependent pathways in plants (Figure 4).
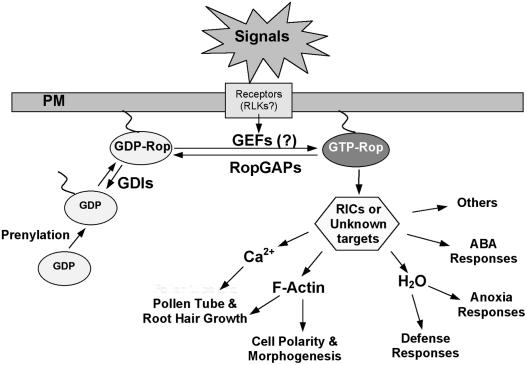
Figure 4.
A Generalized Scheme Illustrating the Functional Diversity of Rop GTPases.
Rop signaling controls many processes and involves many Rop-interacting proteins. Different Rops may interact differentially with different Rop regulators and target proteins to produce functionally distinct Rop signaling pathways. Rop regulation involves not only activation by an unknown mechanism and deactivation by RopGAPs and Rop GDIs, but also compartmentation in specific membranes or membrane domains. In addition to the PM as shown in this figure, Rop is also found in specific PM domains and other compartments such as vacuoles. Targeting to specific compartments could require specific unknown recruiting or docking proteins. Functionally distinct Rop targets including various RICs and possibly other unknown proteins are proposed to control specific downstream effects to achieve functional specificity of each Rop-dependent pathway.
Other Potential ROP Targets
It is possible that some ROP targets, like many yeast and animal Cdc42/Rac effectors, do not use a CRIB motif for their interaction with ROPs (Aspenstrom, 1999). One such potential ROP target is a PIPK that was isolated from tobacco pollen and shown to associate with ROPs in vitro. This PIPK was shown to generate PIP2 (Kost et al., 1999). A GFP-tagged PH domain that specifically binds PIP2 and IP3 was localized to the apical PM region of pollen tubes, and its expression inhibited pollen tube elongation, as did DN-rop (Kost et al., 1999). However, it is not clear whether this PIPK directly binds ROPs and whether it is involved in the regulation of ROP regulation or acts downstream of ROPs in the control of polar growth in pollen tubes. Another possible ROP target is callose synthase. It was shown that GTP- but not GDP-bound ROP1 interacted with UDP-Glucose Transferase 1, a putative subunit of Arabidopsis callose synthase (Hong et al., 2001). This is interesting because yeast Rho1 is known to directly associate with and activate β-1,3-glucan synthase. Further experiments are necessary to determine whether these interactions have functional parallelism.
FUTURE PROSPECTS AND CHALLENGES
Although the study of small GTPases in plants is its early stage, over the last few years rapid progress has been made in our understanding of intracellular signaling pathways mediated by these GTPases, especially by the plant-specific ROP subfamily of Rho GTPases. The surprising finding of Rab11 regulation of brassinolide biosynthesis (Kang et al., 2001) taught us that we cannot assume that the highly conserved Rab, Arf, and Ran GTPases simply control fundamental cellular processes as in yeast and mammals. However, the large number of these GTPases and their potential function in these fundamental processes seemingly present a daunting challenge for investigating their specific roles in signaling. With functional genomic tools available in Arabidopsis, however, it should be possible systematically to analyze their functions using knockout mutants in conjunction with analysis of their subcellular and cellular localization patterns and identification of their functional partners.
ROP, as a sole class of signaling small GTPases in plants, has emerged as an important molecular switch in plant cellular signaling. The use of dominant mutants has revealed a great deal of functional versatility for ROP GTPases (Figure 4), but many outstanding questions about ROP signaling remain. What the precise roles of individual ROPs need to be addressed using loss-of-function rop mutants. To date, only Arabidopsis ROP9/ROP10 have been functionally analyzed using this approach. A systematic functional analysis using this approach may also lead to the identification of many new ROP-dependent pathways. But this approach alone will be insufficient to elucidate complex functions for individual ROPs, for example, overlapping, and multiple functions. A comprehensive study involving biochemical, proteomic, cell biological, and genetic methods in a model system (for example, Arabidopsis) is needed to determine what signals regulate each ROP, how each ROP is localized at cellular and subcellular levels, and how each ROP differentially interacts with different regulators and effectors.
The yeast two-hybrid method has revealed several unique ROP regulators and potential targets (Figure 4). However, what factor recruits ROPs to cellular membranes and what is the molecular mechanism underlying ROP activation remain mysterious. Although a ROP-like protein is known to associate with the CLV1 RLK complex, it is unclear what the functional significance of this association is and whether ROP is associated with other RLK complexes. How ROPs control downstream events to modulate specific processes also requires further exploration. RICs promise to be ROP targets that link ROPs to various downstream pathways, but their functions have yet to be determined using loss-of-function mutants. Because RICs show no homology to functionally known proteins, it will be critical to identify RIC-interacting proteins to understand how ROPs control each downstream pathway. Each ROP-dependent pathway is presumably controlled by a distinct ROP signaling complex. Development of strategies to isolate specific ROP signaling complexes combined with biochemical, cell biological, and genetic analyses of these complexes should ultimately reveal the mechanisms for ROP recruitment and activation as well as those underlying the functional diversity and specificity of ROP GTPases in plants.
Acknowledgments
I am grateful to Sarah Assmann, Keiko Torri, and Zhiliang Zheng for their helpful comments and discussion and to Ying Fu for figure preparation. Work in my laboratory is supported by grants from the National Science Foundation, the U.S. Department of Energy, and the U.S. Department of Agriculture.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001065.
References
- Adamson, P., Paterson, H.F., and Hall, A. (1992). Intracellular localization of the p21rho proteins. J. Cell Biol. 119 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Aspenstrom, P. (1999). Effectors of the Rho GTPases. Curr. Opin. Cell Biol. 11 95–102. [DOI] [PubMed] [Google Scholar]
- Assmann, S.M. (2002). Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell 14 (suppl.), S355–S373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska, F., Salaj, J., Mathur, J., Braun, M., Jasper, F., Samaj, J., Chua, N.H., Barlow, P.W., and Volkmann, D. (2000). Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev. Biol. 227 618–632. [DOI] [PubMed] [Google Scholar]
- Batoko, H., Zheng, H.Q., Hawes, C., and Moore, I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12 2201–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter-Burrell, A., Yang, Z., Springer, P.S., and Bailey-Serres, J. (2002). RopGAP4-dependent Rop GTPase rheostat controls Arabidopsis oxygen deprivation tolerance. Science, in press. [DOI] [PubMed]
- Bibikova, T.N., Blancaflor, E.B., and Gilroy, S. (1999). Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 17 657–665. [DOI] [PubMed] [Google Scholar]
- Bischoff, F., Molendijk, A., Rajendrakumar, C.S., and Palme, K. (1999). GTP-binding proteins in plants. Cell. Mol. Life Sci. 55 233–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, F., Vahlkamp, L., Molendijk, A., and Palme, K. (2000). Localization of AtROP4 and AtROP6 and interaction with the guanine nucleotide dissociation inhibitor AtRhoGDI1 from Arabidopsis. Plant Mol. Biol. 42 515–530. [DOI] [PubMed] [Google Scholar]
- Bokoch, G.M. (1994). Regulation of the human neutrophil NADPH oxidase by the Rac GTP-binding proteins. Curr. Opin. Cell Biol. 6 212–218. [DOI] [PubMed] [Google Scholar]
- Borg, S., Podenphant, L., Jensen, T.J., and Poulsen, C. (1999). Plant cell growth and differentiation may involve GAP regulation of Rac activity. FEBS Lett. 453 341–345. Erratum. FEBS Lett. 458, 82. [DOI] [PubMed] [Google Scholar]
- Bos, J.L. (2000). Ras. In GTPases, A. Hall, ed (Oxford: Oxford University Press), pp. 67–88.
- Cerione, R.A., and Zheng, Y. (1996). The Dbl family of oncogenes. Curr. Opin. Cell Biol. 8 216–222. [DOI] [PubMed] [Google Scholar]
- Chant, J. (1999). Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 15 365–391. [DOI] [PubMed] [Google Scholar]
- Cheon, C., Lee, N., Siddique, A., Bal, A., and Verma, D. (1993). Roles of plant homologs of Rab1p and Rab7p in the biogenesis of the peribacteroid membrane, a subcellular compartment formed de novo during root nodule symbiosis. EMBO J. 12 4125–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell, D.N., and Kennedy, M. (2001). Identification and functional expression in yeast of a prenylcysteine alpha-carboxyl methyltransferase gene from Arabidopsis thaliana. Plant Mol. Biol. 45 469–476. [DOI] [PubMed] [Google Scholar]
- Crowell, D.N., Sen, S.E., and Randall, S.K. (1998). Prenylcysteine alpha-carboxyl methyltransferase in suspension-cultured tobacco cells. Plant Physiol. 118 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S., Ghassemian, M., Bonetta, D., Cooney, S., and McCourt, P. (1996). A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273 1239–1241. [DOI] [PubMed] [Google Scholar]
- d'Enfert, C., Gensse, M., and Gaillardin, C. (1992). Fission yeast and a plant have functional homologues of the Sar1 and Sec12 proteins involved in ER to Golgi traffic in budding yeast. EMBO J. 11 4205–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijo, J.A., Sainhas, J., Holdaway-Clarke, T., Cordeiro, M.S., Kunkel, J.G., and Hepler, P.K. (2001). Cellular oscillations and the regulation of growth: The pollen tube paradigm. Bioessays 23 86–94. [DOI] [PubMed] [Google Scholar]
- Fu, F., and Yang, Z. (2001). The Rop GTPase: A master switch of cell polarity development in plants. Trends Plant Sci. 6 545–547. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Li, H., and Yang, Z. (2002). The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14 777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., Wu, G., and Yang, Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner, N., Friml, J., Stierhof, Y.D., Jurgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413 425–428. [DOI] [PubMed] [Google Scholar]
- Gibbon, B.C., Kovar, D.R., and Staiger, C.J. (1999). Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11 2349–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W., Tamanoi, F., and Novick, P. (2001). Spatial regulation of the exocyst complex by Rho1 GTPase. Nat. Cell Biol. 3 353–360. [DOI] [PubMed] [Google Scholar]
- Haizel, T., Merkle, T., Pay, A., Fejes, E., and Nagy, F. (1997). Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J. 11 93–103. [DOI] [PubMed] [Google Scholar]
- Hall, A., and Nobes, C.D. (2000). Rho GTPases: Molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355 965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Z., Zhang, Z., Olson, J.M., and Verma, D.P. (2001). A novel UDP–glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell 13 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko, M., Vejlupkova, Z., Quatrano, R.S., and Fowler, J.E. (2000). Maize ROP7 GTPase contains a unique, CaaX box–independent plasma membrane targeting signal. Plant J. 24 79–90. [DOI] [PubMed] [Google Scholar]
- Johnson, D.I. (1999). Cdc42: An essential Rho-Type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63 54–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.A., Shen, J.-J., Fu, Y., Li, L., Yang, Z., and Grierson, C.S. (2002). The Arabidopsis Rop2 GTPase is a positive regulator of both root-hair initiation and tip growth. Plant Cell 14 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo, J.H., Bae, Y.S., and Lee, J.S. (2001). Role of auxin-induced reactive oxygen species in root gravitropism. Plant Physiol. 126 1055–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.G., Yun, J., Kim, D.H., Chung, K.S., Fujioka, S., Kim, J.I., Dae, H.W., Yoshida, S., Takatsuto, S., Song, P.S., and Park, C.M. (2001). Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell 105 625–636. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T., Henmi, K., Ono, E., Hatakeyama, S., Iwano, M., Satoh, H., and Shimamoto, K. (1999). The small GTP-binding protein Rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA 96 10922–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., and Chua, N.-H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropf, D.L., Bisgrove, S.R., and Hable, W.E. (1998). Cytoskeletal control of polar growth in plant cells. Curr. Opin. Cell Biol. 10 112–122. [DOI] [PubMed] [Google Scholar]
- Lamb, C., and Dixon, R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 251–275. [DOI] [PubMed] [Google Scholar]
- Lemichez, E., Wu, Y., Sanchez, J.P., Mettouchi, A., Mathur, J., and Chua, N.H. (2001). Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 15 1808–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Lin, Y., Heath, R.M., Zhu, M.X., and Yang, Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to the tip-localized calcium influx. Plant Cell 11 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Shen, J., Zheng, Z., Lin, Y., and Yang, Z. (2001). The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol. 126 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Wu, G., Ware, D., Davis, K.R., and Yang, Z. (1998). Arabidopsis Rho-related GTPases: Differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 118 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and Yang, Z. (2000). Rho GTPase and the actin cytoskeleton. In Actin: A Dynamic Framework for Multiple Plant Cell Functions, C. J. Staiger, F. Baluska, D. Volkmann, and P. Barlow, eds (Dordrecht, The Netherlands: Kluwer Acadmic Publishers), pp. 301–321.
- Lin, Y., Seals, D.F., Randall, S.K., and Yang, Z. (2001). Dynamic localization of Rop GTPases to the tonoplast during vacuole development. Plant Physiol. 125 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., Wang, Y., Zhu, J., and Yang, Z. (1996). Localization of a rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell 8 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y., and Yang, Z. (1997). Inhibition of pollen tube elongation by micro-injected anti-Rop1Ps antibodies suggests a crucial role for Rho-type GTPases in the control of tip growth. Plant Cell 9 1647–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Zainal, Z., Tucker, G.A., and Lycett, G.W. (2001). Developmental abnormalities and reduced fruit softening in tomato plants expressing an antisense Rab11 GTPase gene. Plant Cell 13 1819–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver, J., Patton, D., Rumbaugh, M., Liu, C., Yang, L.J., and Meinke, D. (2000). The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell 12 1379–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, I. (2000). A novel link between ran signal transduction and nuclear envelope proteins in plants. Plant Physiol. 124 1507–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merke, T., Haizel, T., Matsumoto, T., Harter, K., Dallmann, G., and Nagy, F. (1996). Phenotype of the fission yeast cell cycle regulatory mutant pim1–46 is suppressed by a tobacco cDNA encoding a small, Ran-like GTP-binding protein. Plant J. 6 555–565. [DOI] [PubMed] [Google Scholar]
- Molendijk, A.J., Bischoff, F., Rajendrakumar, C.S.V., Friml, J., Braun, M., Gilroy, S., and Palme, K. (2001). Arabidopsis thaliana Rop GTPase are localized to tips of root hairs and control polar growth. EMBO J. 20 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, I., Diefenthal, T., Zarsky, V., Schell, J., and Palme, K. (1997). A homolog of the mammalian GTPase Rab2 is present in Arabidopsis and is expressed predominantly in pollen grains and seedlings. Proc. Natl. Acad. Sci. USA 94 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara, E., and McCourt, P. (1999). Protein farnesylation in plants: A greasy tale. Curr. Opin. Plant Biol. 2 388–392. [DOI] [PubMed] [Google Scholar]
- Palme, K., Diefenthal, T., Vingron, M., Sander, C., and Schell, J. (1992). Molecular cloning and structural analysis of genes from Zea mays (L.) coding for members of the Ras-related ypt gene family. Proc. Natl. Acad. Sci. USA 89 787–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada, L.F., Tabin, C.J., Shih, C., and Weinberg, R.A. (1982). Human EJ bladder carcinoma oncogene is homologue of Harvey carcoma virus Ras gene. Nature 291 474–478. [DOI] [PubMed] [Google Scholar]
- Park, J., Choi, H.J., Lee, S., Lee, T., Yang, Z., and Lee, Y. (2000). Rac-related GTP-binding protein in elicitor-induced reactive oxygen generation by suspension-cultured soybean cells. Plant Physiol. 124 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Z.M., Ghassemian, M., Kwak, C.M., McCourt, P., and Schroeder, J.I. (1998). Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282 287–290. [DOI] [PubMed] [Google Scholar]
- Pei, Z.M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G.J., Grill, E., and Schroeder, J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406 731–734. [DOI] [PubMed] [Google Scholar]
- Potikha, T.S., Collins, C.C., Johnson, D.I., Delmer, D.P., and Levine, A. (1999). The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol. 119 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad, F., Bardet, C., Tremousaygue, D., Moisan, A., Lescure, B., and Axelos, M. (1993). cDNA cloning and expression of an Arabidopsis GTP-binding protein of the ARF family. FEBS Lett. 316 133–136. [DOI] [PubMed] [Google Scholar]
- Ridley, A. (2000). Rho. In GTPases, A. Hall, ed (Oxford: Oxford University Press), pp. 89–136.
- Rodriguez-Concepcion, M., Toledo-Ortiz, G., Yalovsky, S., Caldelari, D., and Gruissem, W. (2000). Carboxyl-methylation of prenylated calmodulin CaM53 is required for efficient plasma membrane targeting of the protein. Plant J. 24 775–784. [DOI] [PubMed] [Google Scholar]
- Sagi, M., and Fluhr, R. (2001). Superoxide production by plant homologues of the gp91(phox) NADPH oxidase. Modulation of activity by calcium and by tobacco mosaic virus infection. Plant Physiol. 126 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, H., Seo, S., Orudgev, E., Youssefian, S., and Ishizuka, K. (1994). Expression of the gene for a small GTP-binding protein in transgenic tobacco elevates endogenous cytokinin levels, abnormally induces salicylic acid in response to wounding, and increases resistance to tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA 91 10556–10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman, G., Morikami, A., Jung, J., Baskin, T.I., Carpita, N.C., Derbyshire, P., McCann, M.C., and Benfey, P.N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settleman, J. (2001). Rac 'n Rho: The music that shapes a developing embryo. Dev. Cell 1 321–331. [DOI] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Gälweiler, L., Palme, K., and Jürgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318. [DOI] [PubMed] [Google Scholar]
- Symons, M., and Settleman, J. (2000). Rho family GTPases: More than simple switches. Trends Cell Biol. 10 415–419. [DOI] [PubMed] [Google Scholar]
- Takai, Y., Sasaki, T., and Matozaki, T. (2001). Small GTP-binding proteins. Physiol. Rev. 81 153–208. [DOI] [PubMed] [Google Scholar]
- Trotochaud, A.E., Hao, T., Wu, G., Yang, Z., and Clark, S.E. (1999). The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valster, A.H., Hepler, P.K., and Chernoff, J. (2000). Plant GTPases: The Rhos in bloom. Trends Cell Biol. 10 141–146. [DOI] [PubMed] [Google Scholar]
- Winge, P., Brembu, T., and Bones, A.M. (1997). Cloning and characterization of rac-like cDNAs from Arabidopsis thaliana. Plant Mol. Biol. 35 483–495. [DOI] [PubMed] [Google Scholar]
- Winge, P., Brembu, T., Kristensen, R., and Bones, A.M. (2000). Genetic structure and evolution of RAC-GTPases in Arabidopsis thaliana. Genetics 156 1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G., Gu, Y., Li, S., and Yang, Z. (2001). A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell 13 2841–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G., Li, H., and Yang, Z. (2000). Arabidopsis RopGAPs are a novel family of Rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for Rop-specific GTPase stimulation. Plant Physiol. 124 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, T., Higgins, V.J., and Blumwald, E. (1997). Race-specific elicitors of Cladosporium fulvum promote translocation of cytosolic components of NADPH oxidase to the plasma membrane of tomato cells. Plant Cell 9 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., and Watson, J.C. (1993). Molecular cloning and characterization of rho, a ras-related small GTP-binding protein from the garden pea. Proc. Natl. Acad. Sci. USA 90 8732–8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K., Nagano, Y., Murai, N., and Sasaki, Y. (1993). Phytochrome-regulated expression of the genes encoding the small GTP- binding proteins in peas. Proc. Natl. Acad. Sci. USA 90 6636–6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z.-L., and Yang, Z. (2000. a). The Rop GTPase switch turns on polar growth in pollen. Trends Plant Sci. 5 298–303. [DOI] [PubMed] [Google Scholar]
- Zheng, Z.-L., and Yang, Z. (2000. b). The Rop GTPase: An emerging signaling switch in plants. Plant Mol. Biol. 44 1–9. [DOI] [PubMed] [Google Scholar]