Abstract
LXRα and -β are nuclear receptors that regulate the metabolism of several important lipids, including cholesterol and bile acids. Previously, we have proposed that LXRs regulate these pathways through their interaction with specific, naturally occurring oxysterols, including 22(R)-hydroxycholesterol, 24(S)-hydroxycholesterol, and 24(S),25-epoxycholesterol. Using a ligand binding assay that incorporates scintillation proximity technology to circumvent many of the problems associated with assaying extremely hydrophobic ligands, we now demonstrate that these oxysterols bind directly to LXRs at concentrations that occur in vivo. To characterize further the structural determinants required for potent LXR ligands, we synthesized and tested a series of related compounds for binding to LXRs and activation of transcription. These studies revealed that position-specific monooxidation of the sterol side chain is requisite for LXR high-affinity binding and activation. Enhanced binding and activation can also be achieved through the use of 24-oxo ligands that act as hydrogen bond acceptors in the side chain. In addition, introduction of an oxygen on the sterol B-ring results in a ligand with LXRα-subtype selectivity. These results support the hypothesis that naturally occurring oxysterols are physiological ligands for LXRs and show that a rational, structure-based approach can be used to design potent LXR ligands for pharmacologic use.
Nonsteroidal nuclear hormone receptors are ligand-activated transcription factors that regulate an array of signaling pathways. Several members of this protein family, including the vitamin D receptor, retinoic acid receptor, and peroxisome proliferator-activated receptor, are primary regulators in nutrient metabolism and are known to be associated with a variety of metabolic disorders (1). These receptors function by forming obligate heterodimers with the retinoid X receptor (RXR) and binding specific DNA sequences (response elements) within the promoters of the genes they regulate. Upon binding ligand, the heterodimerized receptor undergoes a conformational change that results in enhanced transcription of the target gene. Thus far, all known native ligands for nuclear receptors are small hydrophobic compounds, many of which are the products of lipid metabolism (1).
The liver X receptors (LXRs) were originally identified as orphan members of the nuclear receptor superfamily because their ligands were unknown. Like other receptors in the family, LXRs heterodimerize with RXR and bind to specific response elements (LXREs) characterized by direct repeats separated by four nucleotides (2–4). Two genes (α and β) are known to encode LXR proteins. LXRα is expressed most highly in the liver and to a lesser extent in the kidney, small intestine, spleen, and adrenal gland (2, 4). In contrast to the restricted expression pattern of LXRα, LXRβ is ubiquitously expressed (5).
In a comprehensive screen for LXR ligands we observed that the naturally occurring oxysterols 24(S)-hydroxycholesterol [24(S)-HC], 22(R)-hydroxycholesterol [22(R)-HC], and 24(S),25-epoxycholesterol [24(S),25-EC] are potent transcriptional activators of LXRα and -β (6–8). These oxysterols exist at concentrations that activate LXR in tissues (e.g., liver, brain, and placenta) where both cholesterol metabolism and LXR expression are high (9–11). Furthermore, we have shown that these oxysterols can drive LXR-dependent transactivation through the promoter for cholesterol 7α-hydroxylase (Cyp7a) (7, 8), the rate-limiting enzyme in bile acid synthesis (12). Conclusive evidence supporting the hypothesis that LXRs are key regulators of cholesterol homeostasis has come from the characterization of LXRα knockout mice (8). The loss of LXRα results in the rapid accumulation of cholesterol esters in the liver, because of the inability to stimulate the metabolic clearance of cholesterol through the synthesis of bile acids. As expected, a major defect in these mice is the inability to up-regulate Cyp7a expression. Interestingly, LXRβ, which is also expressed in the liver, does not compensate for the loss of LXRα, suggesting that the two receptors may have different biological functions.
One of the key remaining questions of LXR action is whether the oxysterol activators are bona fide ligands. In this report we have used a recently developed ligand-binding assay (13) to unequivocally demonstrate that oxysterols bind LXRs at physiological concentrations, supporting the hypothesis that LXRs mediate signal transduction through their direct interaction with oxysterols. We also report the design and synthesis of potent LXRα and -β ligands.
MATERIALS AND METHODS
Ligands.
Purchased ligands include the following: 9-cis-[20-methyl-3H]retinoic acid (72 Ci/mmol; 1 Ci = 37 GBq) and [26,27-3H(N)]-24(S),25-EC (76 Ci/mmol) (DuPont/NEN); 22(R,S)-HC, 25-HC, 7α-HC, 24,25-dehydrocholesterol, cholic acid, pregnenolone, 9-cis-retinoic acid (9cRA) (Steraloids, Research Plus, and Sigma). Efficient stereocontrolled syntheses for the following ligands are described elsewhere (14): 24(S),25-EC; 22(R)-hydroxy-24(S),25-EC; 24(R),25-EC; 22(S)-hydroxy-24(R),25-EC; 7-keto-, 7β-hydroxy-, and 7α-hydroxy-24(S),25-ECs; 24(S),25-iminocholesterol; and 22(R),24(S)-dihydroxycholesterol. The methyl ester and dimethylamide of cholenic acid were prepared from cholesterol trisnorcarboxylic acid via acid chloride by reaction with methanol and dimethylamine, respectively.
Cell Cotransfection Assays.
CV-1 cell conditions and transient transfections were as described (4). Receptor expression plasmids encoding full-length human LXRβ (CMX-hLXRβ) or LXRα (CMX-hLXRα) (4) were cotransfected with a luciferase reporter plasmid [TK-CYP7a-LXRE(X3)-LUC] (8) containing three tandem copies of the sequence (gcttTGGTCActcaAGTTCAagtta) from the rat Cyp7a gene (15). Increasing concentrations of ligand (0.1 to 40 μM) were added to cells in media containing 5% lipid-depleted calf bovine serum. Transfection data were normalized to a β-galactosidase internal standard. Data are presented as mean relative light units (RLU) from triplicate assays ± SEM. EC50 values (Table 1) generated from duplicate assays were determined by fitting the data to a sigmoidal dose–response curve (GraphPad Prism, GraphPad Software, San Diego). Efficacy values represent the fraction of maximal fold activation of each compound relative to 24(S),25-EC. Fold activation was determined by dividing the maximal activation of each compound by the activation observed in the absence of compound.
Table 1.
Structure–activity relationships of LXR ligands
| Compound | Wild-type LXRα
|
Wild-type LXRβ
|
||||
|---|---|---|---|---|---|---|
| Ki, nM | EC50, μM | Efficacy | Ki, nM | EC50, μM | Efficacy | |
| 1 24(S),25-EC | 200 ± 20 | 4 | 1.0 | 200 ± 10 | 3 | 1.0 |
| 2 24(R),25-EC | 1,200 ± 300 | 10 | 0.47 | 710 ± 60 | 10 | 0.50 |
| 3 22(R)-ol-24(S),25-EC | PC | ∗ | — | 300 ± 40 | ∗ | — |
| 4 22(S)-ol,24(R),25-EC | 440 ± 70 | — | — | 920 ± 90 | — | — |
| 5 24(S),25-IC | 990 ± 90 | ∗∗ | — | 1,000 ± 110 | ∗∗ | — |
| 6 Methyl-H-cholenate | 110 ± 20 | 8 | 0.74 | 170 ± 10 | 8 | 0.83 |
| 7 Dimethyl-HCA | 130 ± 10 | 2 | 0.60 | 100 ± 10 | 2 | 0.50 |
| 8 24(S)-HC | 110 ± 10 | 4 | 0.40 | 100 ± 5 | 3 | 0.70 |
| 9 24(R)-HC | PC | 7 | 0.20 | PC | 4 | 0.38 |
| 10 22(R)-HC | 380 ± 50 | 5 | 0.40 | 130 ± 30 | 3 | 0.57 |
| 11 22(S)-HC | 150 ± 10 | — | — | 160 ± 10 | — | — |
| 12 22(R),24(S)-diHC | 950 ± 40 | — | — | 710 ± 90 | — | — |
| 13 25-HC | 180 ± 30 | 7 | 0.16 | 300 ± 30 | ∗ | — |
| 14 24(S),25-diHC | 1,200 ± 240 | — | — | PC | — | — |
| 15 24(R),25-diHC | PC | — | — | 1,700 ± 390 | ∗ | — |
| 16 24,25-DC | — | No sat | 0.16 | — | No sat | 0.31 |
| 17 7(α)-ol,24(S),25-EC | 1,400 ± 300 | 8 | 0.40 | 2,300 ± 180 | 6 | 0.50 |
| 18 7(β)-ol,24(S),25-EC | 1,200 ± 200 | ∗ | — | 4,500 ± 630 | ∗ | — |
| 19 7k,24(S),25-EC | 1,800 ± 590 | ∗ | — | 2,000 ± 280 | ∗ | — |
| 20 7(α)-HC | PC | ∗ | — | PC | ∗ | — |
| 21 7-KC | >5,000 | — | — | >5,000 | — | — |
| 22 Cholesterol | — | — | — | — | — | — |
| 23 5,6-24(S),25-diEC | 390 ± 70 | 7 | 0.87 | 1,700 ± 250 | ∗ | — |
Ki values are presented ± SEM. EC50, effective concentration for 50% maximal activation; Efficacy, maximal fold activation relative to 24(S),25-EC; EC, epoxycholesterol; H, hydroxy; HC, hydroxycholesterol; diHC, dihydroxycholesterol; IC, iminocholesterol; KC, ketocholesterol; HCA, hydroxycholenamide; DC, dehydrocholesterol; ∗, efficacy ≤ 10% at 40 μM; ∗∗, toxic; —, below detection; No sat, no saturation; PC, poor competitor (competes <70% at 50 μM).
Receptor Protein Purification.
Polyhistidine human RXRα (His10-hRXRα) (16), LXRαLBD or LXRβLBD fusion proteins (His6-hLXRα-LBD, His6-hLXRβ-LBD) were expressed in Escherichia coli strain BL21(DE3) (Novagen). Cultures grown in Luria-Bertani medium were induced with 0.5 mM isopropyl β-d-thiogalactoside (IPTG) for 4 h at 25°C. Pellets were suspended in lysis buffer (250 mM NaCl/16 mM Na2HPO4/4 mM NaH2PO4/1% Triton X-100/10 mM imidazole/200 μg of lysozyme per ml). The supernatant was incubated with Ni2+-NTA agarose (Qiagen). The resin was washed twice with 20 vol of 50 mM Hepes, pH 7.5/250 mM NaCl supplemented first with 50 mM then 75 mM imidazole. Protein was eluted with a linear 75–500 mM imidazole gradient. Peak fractions were tested for purity by SDS/PAGE, pooled, and cleared of imidazole over a PD-10 column (Pharmacia) equilibrated with 20 mM Hepes, pH 7.5/200 mM NaCl/2.5 mM EDTA/5 mM DTT. Concentrations were determined by UV spectral analysis.
Ligand-Binding Assay.
Scintillant-filled beads precoated with polylysine to permit protein binding (Amersham) were diluted in scintillation proximity assay (SPA) buffer [10 mM K2HPO4/10 mM KH2PO4/2 mM EDTA/50 mM NaCl/1 mM DTT/2 mM CHAPS/10% (vol/vol) glycerol, pH 7.1; CHAPS is 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate] to a final concentration of 10 mg/ml. Binding assays were performed in 96-well plates (Packard) in a total volume of 100 μl containing beads (0.2 mg per well) and His6-hLXRα-LBD (600 ng per well), His6-hLXRβ-LBD (250 ng per well), or His6-hRXRα (250 ng per well). The amount of protein used did not deplete ligand concentrations. [3H]-24(S),25-EC (Fig. 1A) or [3H]-9cRA was diluted in SPA buffer and added to wells for a final concentration of 25 nM or 5 nM, respectively. Competition binding assays using a single concentration of unlabeled competitor (Fig. 1B) contained 25 μM 24(S),25-EC or 5 μM 9cRA. In other competition binding assays, unlabeled ligands were serially diluted in SPA buffer, then added at final concentrations ranging from 3 nM to 50 μM. Plates were shaken at 25°C for 3 h, and then radioactivity was measured with a Packard Topcount at 1 min per well. All concentrations were assayed in triplicate and the results were averaged. Values from wells void of competitor represented 100% binding.
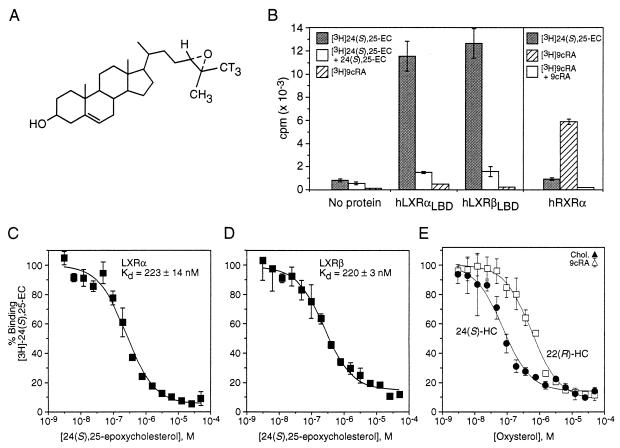
Figure 1.
Naturally occurring oxysterols bind LXRα and LXRβ. (A) Structure of [3H]-24(S),25-EC, showing the incorporation of tritium atoms (T). (B) Binding specificity for LXRs and RXR by their cognate ligands. Receptor protein (His6-hLXRαLBD, His6-hLXRβLBD, or His10-hRXRα) immobilized on SPA beads was incubated with [3H]-24(S),25-EC (shaded bars) or [3H]-9cRA (hatched bars) in the absence or presence of 1000-fold excess of nonradiolabeled 24(S),25-EC (open bars in left panel) or 9cRA (open bar in right panel). (C and D) Competition curves for 24(S),25-EC binding His6-hLXRαLBD or His6-hLXRβLBD. LXRLBD protein immobilized on SPA beads was incubated with 25 nM [3H]-24(S),25-EC and increasing concentrations (3 nM to 50 μM) of nonradiolabeled 24(S),25-EC. (E) Binding of naturally occurring oxysterols to LXRα. Competition of 25 nM [3H]-24(S),25-EC with either nonradiolabeled 24(S)-HC (•) or 22(R)-HC (□) at concentrations ranging from 3 nM to 50 μM. Values for cholesterol (▴) and 9cRA (▵) are shown at the highest tested concentration, 50 μM.
Generation of Ki Values.
Competition curves were generated by nonlinear regression analysis with GraphPad Prism, and apparent equilibrium dissociation constants (Ki values) were determined by using a method described by DeBlasi and colleagues (17) based on the Cheng–Prusoff equation (18). Ki values for compounds that served as weak competitors in the assay (competition <70% at the highest concentrations tested) are described as poor competitors (PC) (Table 1). All Ki and SEM values reported are averages generated from duplicate or triplicate assays. Compounds that did not compete in the assay are listed as being below the level of detection.
RESULTS
Scintillation Proximity Binding Assay.
Understanding the mechanism by which oxysterols regulate specific aspects of cholesterol metabolism through LXRα and LXRβ requires analysis of direct ligand binding to receptor and quantitation of binding affinities. One of the technical difficulties associated with using oxysterols in solution is their hydrophobic nature, which promotes nonspecific interactions and micelle formation at high concentrations. For these reasons, traditional saturation binding assays that require separation of free from bound radioligand (e.g., using dextran-treated charcoal or hydroxylapatite) have not yielded interpretable results. To circumvent these problems, we used a binding assay based on SPA technology (13), which maintains true equilibrium conditions and does not require separation of bound from free radioligand. For this assay, receptor protein is immobilized on SPA beads through electrostatic interactions (see Materials and Methods). Upon radioligand binding, the scintillant in the beads is activated, and the formation of radioligand/receptor complexes is measured with a scintillation counter. Ki values are determined by using competition assays in which a single concentration of radioligand is displaced by increasing concentrations of unlabeled competitor (see Materials and Methods).
Naturally Occurring Oxysterols Bind LXRα and LXRβ at Physiological Concentrations.
The SPA was validated for nuclear hormone receptors by first examining the binding of RXRα to its natural ligand, 9cRA. Using this assay, we found that [3H]-9cRA binds RXRα with a Kd of 10 nM (data not shown), which correlates exactly with values previously determined from a variety of ligand-binding assays (19–21). To show the binding specificity of oxysterols for LXRα and LXRβ, we tested the ability of the best reported LXR oxysterol activator, 24(S),25-EC, as well as 9cRA, to bind LXRα, LXRβ, or RXRα proteins (Fig. 1B). Significant specific binding with 25 nM [3H]-24(S),25-EC was detected only in the presence of LXRα or LXRβ. Conversely, 5 nM [3H]-9cRA specifically bound RXRα, but not LXRα or LXRβ, further confirming the selective binding properties of these receptors. In the absence of protein, binding of either radiolabeled ligand to the SPA beads was negligible.
Homologous competition binding assays (Fig. 1 C and D) demonstrate that both LXRα and LXRβ subtypes bind 24(S),25-EC with a Kd of approximately 200 nM. This value corresponds well with reported liver concentrations of 24(S),25-EC (ref. 10; T. Spencer and D.J.M., unpublished observations), and is below the concentration required for activation of LXRα and LXRβ in cultured CV-1 cells (Table 1). The observation that cell-based activation requires a higher concentration of compound than in vitro ligand binding is characteristic of nuclear receptor ligands and reflects the difference of measuring Kd values in vitro and EC50 values in cells. Two other potent LXR activators, 24(S)-HC and 22(R)-HC, which are also present in vivo (11, 22) bind LXRα with Ki values of 110 nM and 380 nM, respectively (Fig. 1E, Table 1). Similar binding affinities were observed with LXRβ (Table 1). Neither cholesterol nor 9cRA acted as a competitor in the assay (Fig. 1E), confirming specific binding. The observation that 24(S),25-EC, 24(S)-HC, and 22(R)-HC can bind LXRα and LXRβ at concentrations similar to those found within cells supports the hypothesis that these oxysterols are effective endogenous ligands of both LXRs.
Structure–Activity Relationships of LXR Ligands.
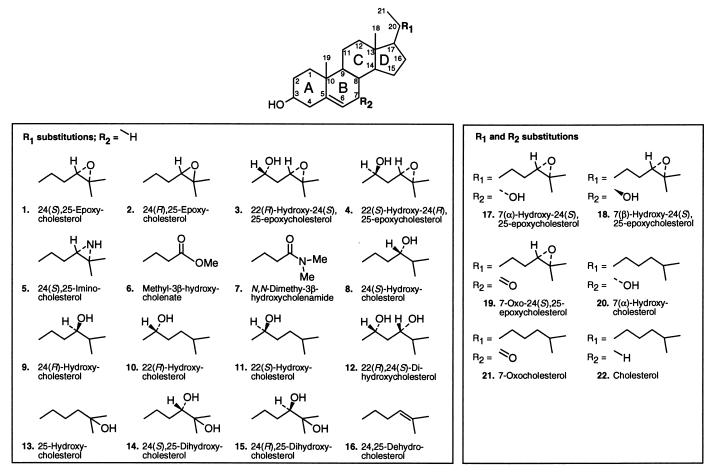
Cholesterol neither activates (6) nor binds LXR (Fig. 1E), yet the introduction of an epoxide, hydroxyl, or keto group on the side chain renders this compound biologically active (6, 7). To better understand the specificity of these oxidized cholesterol metabolites for LXR, we performed a structure–activity relationship study on a series of potential oxysterol ligands. Compounds with several stereospecific modifications to the side chain or the B-ring of the cholesterol structure were synthesized (Fig. 2). Each compound was evaluated with LXRα and LXRβ for its binding affinity by using the SPA, and for transactivation potential (EC50 and efficacy) by using a cell-based cotransfection assay (Table 1). All compounds were also analyzed for transcriptional activation by using chimeric receptors in which the yeast Gal4 DNA-binding domain is fused with the ligand-binding domain of either LXRα or LXRβ (data not shown). Similar results were observed with chimeric and wild-type LXR proteins.
Figure 2.
Structures of naturally occurring and synthetic oxysterols, with chemical substitutions on the side chain carbons 21–27 (R1) or the B-ring (R2) of the sterol scaffold.
Stereochemistry.
If a chiral center of a molecule is involved in biologically important interactions, alteration of stereochemistry at one or more stereocenters generally decreases biological activity. Changing the spatial orientation of the oxygen at carbon 24 on the sterol side chain from the naturally occurring S arrangement to yield 24(R),25-EC and 24(R)-HC (2 and 9; Fig. 2) resulted in a 50% reduction of transcriptional activation (Table 1) and a 6-fold and 4-fold decrease in affinity of 24(R),25-EC for LXRα and LXRβ, respectively (Table 1). In vivo, the nonnatural compounds 24(R)-HC and 24(R),25-EC are more rapidly metabolized than the natural ligand 24(S),25-EC (23–25). Switching the stereochemistry of 22(R)-HC (10; Fig. 2) from the naturally occurring R to synthetic S configuration (11; Fig. 2) resulted in a complete loss of LXRα and LXRβ transactivation (Table 1). Surprisingly, 22(S)-HC binds both LXR subtypes competitively and with high affinity (150 nM for LXRα, 160 nM for LXRβ; Table 1), yet does not function as an antagonist in cells (data not shown). The most plausible explanation for the lack of activity in cells is that 22(S)-HC never reaches its nuclear receptor target. As has been shown to be the case for many pharmacophores, 22(S)-HC may specifically bind cellular proteins that prevent its localization in the nucleus, may be pumped out of the cells by means of a multidrug resistance (MDR)-related mechanism, or may be rapidly metabolized in cell culture. In support of this latter hypothesis, we note that the unnatural stereoisomers of several oxysterols are much better substrates for esterification by acyl-CoA:cholesterol acyltransferase enzymes than are their LXR-active counterparts (26). Alternatively, the binding potential of LXR may be different in vivo, where it exists as part of a multimeric complex bound to RXR, other cofactor proteins, and DNA. Thus, the in vivo state of LXR may impart a second level of ligand binding specificity that is not seen with the purified receptor protein alone. These results suggest that LXRs are able to maintain ligand specificity through several mechanisms, including differential binding, selective access to the nuclear receptor, and ligand metabolism.
Effects of Mono- or Multiple Oxidation.
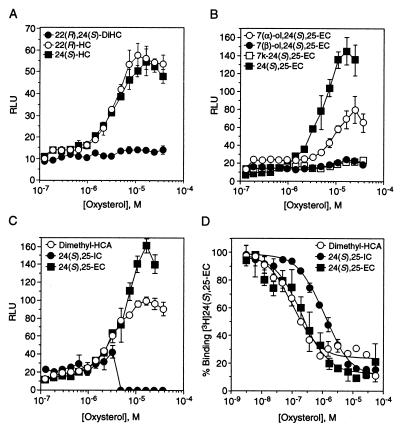
To further understand the specific determinants of LXR ligands for binding and gene activation we combined the functional chemical groups of the compounds that exhibited the highest affinity and activation for LXRs and compared them to their monooxidized parent compounds. Introduction of multiple hydroxyl groups at positions 22, 24, or 25 on the side chain of cholesterol, (e.g., 12, 14, 15; Fig. 2) yielded compounds that were completely inactive in gene transcription, which correlates with the low binding affinities observed for both LXRα and LXRβ (Fig. 3A, Table 1). 24,25-Dehydrocholesterol, which is unsaturated at carbons 24 and 25, did not bind, but weakly activated in culture at the highest concentration, possibly because of lability of the double bond and enzymatic hydroxylation (23, 25). We have also studied the effect of adding the 22-hydroxyl group to the side chain of 24(S),25-EC, using the diastereomers 22(S)-hydroxy-24(R),25-EC (4; Fig. 2) and 22(R)-hydroxy-24(S),25-EC (3; Fig. 2). The results were strikingly similar to those observed with 22(S)-HC; 22(S)-hydroxy-24(R),25-EC bound weakly, but did not activate either LXR subtype (Table 1) or act as an antagonist when tested in cells. Also, 22(R)-hydroxy-24(S),25-EC binds LXRβ with a Ki of 300 nM (Table 1), but does not activate or antagonize receptor function. As mentioned above, the ability of some oxysterols to bind and not activate is likely a reflection of the receptor context in vitro versus in vivo, where ligand metabolism and association of other factors may play a key role in ligand specificity. The above results demonstrate that oxygens at more than one carbon on the side chain of cholesterol diminish LXR binding and activation as compared with the monooxygenated analogs. Thus, there may be insufficient space for both oxygens or a disruption in the contact between the side-chain oxygen and the proton donor in the binding pocket of LXR.
Figure 3.
Multiple oxidation decreases binding and gene activation, whereas single hydrogen bond acceptors are potent ligands and activators of LXR. (A–C) Transcriptional activation profile of LXRα with increasing concentrations of 22(R),24(S)-diHC (•), 22(R)-HC (○), or 24(S)-HC (■) (A); 7α- (○) or 7β- (•) -hydroxy-24(S),25-EC, 7-oxo-24(S),25-EC (□), or 24(S),25-EC (■) (B); dimethylhydroxycholenamide (○), 24(S),25-iminocholesterol (•), or 24(S),25-EC (■) (C). Compounds were tested for their ability to activate the LXR-responsive element in the Cyp7a gene in CV-1 cells. Activation is represented as RLU. (D) Competition binding profile of His6-hLXRαLBD with dimethylhydroxycholenamide (○), 24(S),25-iminocholesterol (•), or 24(S),25-EC (■). LXRLBD protein, SPA beads, and 25 nM [3H]-24(S),25-EC were incubated with increasing concentrations of nonradiolabeled competitors (as in Fig. 1). Similar results were observed with LXRβ.
Since monooxidation of the cholesterol side chain is required for strong binding and activation of LXRs, we tested whether monooxidation at other positions that are biologically relevant would produce LXR ligands. 7α-Hydroxycholesterol (7α-HC; 20; Fig. 2) was tested because it is the product of the Cyp7a enzyme, and therefore, an intermediate in bile acid synthesis. 7α-HC (Table 1), and its isomer 7β-HC (data not shown) are poor ligands for LXRα and LXRβ; however, the combination of the 7α-hydroxyl group and the 24(S),25-epoxide (17; Fig. 2) resulted in a ligand with moderate LXRα and LXRβ transcriptional activity (Fig. 3B; Table 1). Interestingly, the isomer 7β-hydroxy-24(S),25-EC is transcriptionally inactive (Fig. 3B). The finding that only the 7α-hydroxy metabolite is active and that the 7α, but not the 7β, hydroxylation is enzymatically regulated suggests that the 7α-hydroxy metabolite may be an endogenous ligand.
Introduction of Amide, Imino, or Ester Moieties.
As demonstrated above, position, stereochemistry, and functionality of the oxygen on the side chain of the sterol are important parameters for LXR/ligand interactions. Thus, stereospecific epoxidation and hydroxylation result in strong binding to LXRα and LXRβ. It is known from crystal structures of other nuclear receptors that binding of an agonist induces a conformational change in the ligand-binding domain, thus encapsulating the ligand in the hydrophobic binding pocket of the receptor (27–29). This ligand-binding paradigm emphasizes the requirement for crucial contacts between ligand and receptor. The similarity in binding of 22(S)-HC, 24(S)-HC, and 24(S),25-EC to LXR may be due to the fact that both hydroxyl groups and the epoxide can act as hydrogen bond acceptors, potentially forming a single strong contact with a specific proton-donating amino acid in the ligand binding pocket of LXR.
Since placement of a nucleophilic oxygen on carbon 24 yielded the most efficacious LXR ligands, we extended the study by introducing an ester (6, Fig. 2) or dimethylamide carbonyl (7, Fig. 2) at carbon 24. Selection of these potential ligands was guided by the prediction that the carbonyl group would serve as a better hydrogen bond acceptor than hydroxyl or epoxy subunits. Presence of the methyl ester moderately enhanced the binding affinity above 24(S),25-EC (Ki = 110 nM for LXRα, 170 nM for LXRβ; Table 1), even though the EC50 values remained high. The dimethylamide (7, Fig. 2), however, exhibited the best binding and activation profile yet observed for both LXR subtypes (EC50 = 2 μM, Ki = 130 nM LXRα; 100 nM LXRβ; Fig. 3 C and D; Table 1). Replacement of the 24(S),25-epoxide subunit with an imino group (5; Fig. 2) that is less able to be a hydrogen bond acceptor, since it would be protonated at neutral pH, decreased the affinity of the compound for both receptors, and it also rendered the compound toxic to cells (Fig. 3 C and D; Table 1). These results confirm the idea that hydrogen bonding plays an important role in ligand binding and suggest that further modification of the side chain may result in higher-affinity ligands.
Discovery of a Ligand Selective for LXRα.
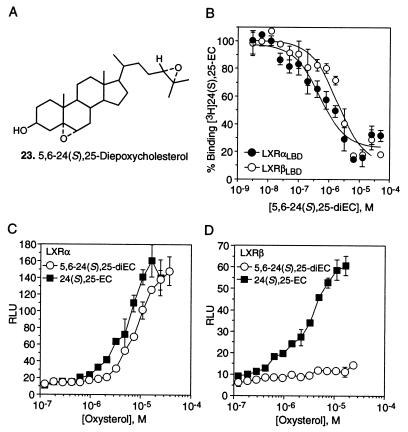
To further delineate the biological roles of the LXR subtypes we sought to identify ligands that selectively activate either LXRα or LXRβ. The level of identity that these receptors share in their ligand-binding domains (77%) has been shown to be amenable to ligand selectivity for subtypes of other nuclear hormone receptors (30, 31). Our survey of prospective LXR ligands shows that the introduction of a second epoxide function at the 5,6 position on 24(S),25-EC (23; Fig. 4A) converts an equipotent LXRα and LXRβ ligand structure into one with LXRα selectivity. 5,6–24(S),25-diEC and 24(S),25-EC bind and activate LXRα with similar affinities (Ki = 390 nM and 200 nM, respectively) and efficacies (Table 1; Fig. 4 B and C). In sharp contrast, 5,6–24(S),25-diEC has a lower affinity for LXRβ (Ki = 1700 nM, Table 1; Fig. 4B) and is a weak LXRβ activator (Fig. 4D). It is not clear whether LXRα selectivity by 5,6–24(S),25-diEC results from a specific interaction between the 5,6-epoxide and an amino acid in the LXRα ligand-binding pocket, or whether alteration in the B-ring geometry produces a better fit between ligand and receptor. Regardless, the results show that the ligand-binding pockets of LXRα and LXRβ differ, and that these differences may be exploited to design potent LXR subtype-selective ligands.
Figure 4.
Identification of a LXRα-selective ligand. (A) Structure of 5,6-α-24(S),25-diEC. (B) Binding profile of 5,6–24(S),25-diEC for His6-hLXRαLBD (•) or His6-hLXRβLBD (○). LXRα or LXRβ protein, SPA beads, and 25 nM [3H]-24(S),25-EC were incubated with increasing concentrations of nonradiolabeled 5,6–24(S),25-diEC. (C and D) Activation profiles with increasing concentrations (3 nM to 5 μM) of either 5,6–24(S),25-diEC (○) or 24(S),25-EC (■) for LXRα or LXRβ on the LXR-responsive element in the Cyp7a gene. Transcriptional activation is represented in RLU.
CONCLUSIONS
The present study demonstrates that oxysterols are the physiological ligands for LXRs and further supports the existence of a LXR-mediated pathway for oxysterol signaling. The key involvement of this pathway in the regulation of cholesterol homeostasis (6–8) suggests that LXRs may be exploited as drug targets for the treatment of disease states associated with hypercholesterolemia. We conclude from our structure–activity relationship analysis that potent LXR ligands require a single stereoselective oxygen on the sterol side chain that functions as a hydrogen bond acceptor. This functionality may be enhanced by introduction of a chemical moiety that increases electron density on the oxygen, possibly strengthening the contact with a proton-donating amino acid in the ligand-binding pocket of LXR. This analysis has also led to the discovery of an LXR subtype-selective ligand that should provide a useful tool for further evaluating the biological role of LXRα. Furthermore, this discovery suggests that the differences between LXRα and LXRβ may be used to design more potent and subtype-selective LXR ligands for pharmacologic application.
Acknowledgments
We thank Dr. David Corey for thoughtful discussions and help in the preparation of this manuscript, Dr. Joseph Goldstein for reviewing the manuscript, and Dr. Joyce Repa for critical comments. The SPA was developed by S.A.J. and S.A.K.; the LXR bacterial expression constructs were provided by G.B.W. Design and synthesis of oxysterol ligands were performed by M.J.G. and E.J.C. D.J.M. is an Associate Investigator with the Howard Hughes Medical Institute. This work was funded by the Howard Hughes Medical Institute, the Robert A. Welch Foundation, and the National Institutes of Health.
ABBREVIATIONS
- RXR
retinoid X receptor
- LXR
liver X receptor
- HC
hydroxycholesterol
- EC
epoxycholesterol
- 9cRA
9-cis-retinoic acid
- Cyp7a
cholesterol 7α-hydroxylase
- RLU
relative light units
- SPA
scintillation proximity assay
References
- 1.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apfel R, Benbrook D, Lernhardt E, Ortiz M A, Salbert G, Pfahl M. Mol Cell Biol. 1994;14:7025–7035. doi: 10.1128/mcb.14.10.7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song C, Kokontis J M, Hiipakka R A, Liao S. Proc Natl Acad Sci USA. 1994;91:10809–10813. doi: 10.1073/pnas.91.23.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willy P J, Umesono K, Ong E S, Evans R M, Heyman R A, Mangelsdorf D J. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- 5.Song C, Hiipakka R A, Kokontis J M, Liao S. Ann NY Acad Sci. 1995;761:38–49. doi: 10.1111/j.1749-6632.1995.tb31367.x. [DOI] [PubMed] [Google Scholar]
- 6.Janowski B A, Willy P J, Rama-Devi T, Falck J R, Mangelsdorf D J. Nature (London) 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann J M, Kliewer S A, Moore L B, Smith-Oliver T A, Oliver B B, Su J L, Sundseth S S, Winegar D A, Blanchard D E, Spencer T A, Willson T M. J Biol Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 8.Peet D J, Turley D, Ma W, Janowski B A, Lobaccaro J M, Mangelsdorf D J. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 9.Lavy U, Burstein S, Gut M, Javitt N B. J Lipid Res. 1977;18:232–238. [PubMed] [Google Scholar]
- 10.Spencer T A, Gayen A K, Phirwa S, Nelson J A, Taylor F R, Kandutsch A A, Erickson S K. J Biol Chem. 1985;260:13391–13394. [PubMed] [Google Scholar]
- 11.Lütjohann D, Breuer O, Ahlborg G, Nennesmo I, Sidén Å, Diczfalusy U, Björkhem I. Proc Natl Acad Sci USA. 1996;93:9799–9804. doi: 10.1073/pnas.93.18.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell D W, Setchell K D. Biochemistry. 1992;31:4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- 13.Nichols J S, Parks D J, Consler T G, Blanchard S G. Anal Biochem. 1998;257:112–119. doi: 10.1006/abio.1997.2557. [DOI] [PubMed] [Google Scholar]
- 14.Corey, E. J. & Grogan, M. J. Tetrahedron. Lett., in press.
- 15.Chiang J Y L, Stroup D. J Biol Chem. 1994;269:17502–17507. [PubMed] [Google Scholar]
- 16.Petty K J. Mol Cell Endocrinol. 1995;108:131–142. doi: 10.1016/0303-7207(94)03466-7. [DOI] [PubMed] [Google Scholar]
- 17.DeBlasi A, O’Reilly K, Motulsky H J. Trends Pharmacol Sci. 1989;10:227–229. doi: 10.1016/0165-6147(89)90266-6. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Y, Prusoff W H. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 19.Heyman R A, Mangelsdorf D J, Dyck J A, Stein R B, Eichele G, Evans R M, Thaller C. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 20.Levin A A, Sturzenbecker L L, Kazmer S, Bosakowski T, Huselton C, Allenby G, Speck J, Kratzeisen C I, Rosenberger M, Lovey A, Grippo J F. Nature (London) 1992;355:359–361. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]
- 21.Peet D J, Doyle D F, Corey D R, Mangelsdorf D J. Chem Biol. 1998;5:13–21. doi: 10.1016/s1074-5521(98)90083-7. [DOI] [PubMed] [Google Scholar]
- 22.Dixon R, Furutachi T, Lieberman S. Biochem Biophys Res Commun. 1970;40:161–165. doi: 10.1016/0006-291x(70)91060-0. [DOI] [PubMed] [Google Scholar]
- 23.Steckbeck S R, Nelson J A, Spencer T A. J Am Chem Soc. 1982;104:893–895. [Google Scholar]
- 24.Taylor F R, Kandutsch A A, Gayen A K, Nelson J A, Nelson S S, Phirwa S, Spencer T A. J Biol Chem. 1986;261:15039–15044. [PubMed] [Google Scholar]
- 25.Saucier S E, Kandutsch A A, Clark D S, Spencer T A. Biochim Biophys Acta. 1993;1091:63–67. doi: 10.1016/0005-2760(93)90291-g. [DOI] [PubMed] [Google Scholar]
- 26.Cases S, Novak S, Zheng Y, Myers H M, Lear S R, Sande E, Welch C B, Lusis A J, Spencer T A, Krause B R, Erickson S K, Farese R V. J Biol Chem. 1998;273:26755–26764. doi: 10.1074/jbc.273.41.26755. [DOI] [PubMed] [Google Scholar]
- 27.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 28.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 29.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. Nature (London) 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 30.Willson T M, Wahli W. Curr Opin Chem Biol. 1998;1:235–241. doi: 10.1016/s1367-5931(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 31.Charpentier B, Bernardon J, Eustache J, Millois C, Martin B, Michel S, Shroot B. J Med Chem. 1995;38:4993–5006. doi: 10.1021/jm00026a006. [DOI] [PubMed] [Google Scholar]