Short abstract
Long oligonucleotide microarrays are potentially more cost- and management efficient than cDNA microarrays. Unmodified sense and antisense 70-mer oligonucleotides were synthesized and compared with PCR-amplified cDNA clones corresponding to the same genes. The correlation coefficient between oligonucleotide and cDNA probes for identifying differentially expressed genes was 0.80.
Abstract
Background
Long oligonucleotide microarrays are potentially more cost- and management-efficient than cDNA microarrays, but there is little information on the relative performance of these two probe types. The feasibility of using unmodified oligonucleotides to accurately measure changes in gene expression is also unclear.
Results
Unmodified sense and antisense 70-mer oligonucleotides representing 75 known rat genes and 10 Arabidopsis control genes were synthesized, printed and UV cross-linked onto glass slides. Printed alongside were PCR-amplified cDNA clones corresponding to the same genes, enabling us to compare the two probe types simultaneously. Our study was designed to evaluate the mRNA profiles of heart and brain, along with Arabidopsis cRNA spiked into the labeling reaction at different relative copy number. Hybridization signal intensity did not correlate with probe type but depended on the extent of UV irradiation. To determine the effect of oligonucleotide concentration on hybridization signal, 70-mers were serially diluted. No significant change in gene-expression ratio or loss in hybridization signal was detected, even at the lowest concentration tested (6.25 μm). In many instances, signal intensity actually increased with decreasing concentration. The correlation coefficient between oligonucleotide and cDNA probes for identifying differentially expressed genes was 0.80, with an average coefficient of variation of 13.4%. Approximately 8% of the genes showed discordant results with the two probe types, and in each case the cDNA results were more accurate, as determined by real-time PCR.
Conclusions
Microarrays of UV cross-linked unmodified oligonucleotides provided sensitive and specific measurements for most of the genes studied.
Background
The advent of microarray technology has enabled scientists to investigate biological questions in a more global fashion. Instead of studying genes individually, the expression of thousands of genes can be analyzed simultaneously using probes attached to the surface of a microscope slide [1,2,3,4,5,6]. The cDNA microarray represents a popular array type in which double-stranded PCR products amplified from expressed sequence tag (EST) clones are spotted onto glass slides [7,8], allowing gene-expression profiles to be determined with high reproducibility and efficiency. However, construction of cDNA microarrays presents a number of challenges, largely related to costs associated with clone validation, tracking and maintenance. The laborious and problematic tracking of cDNA clones and PCR amplicons may lead to 10-30% misidentification of clones [6]. For all practical purposes, sequence verification of array elements is an ongoing necessity. Other limitations of cDNA microarrays are their difficulty, because of cross-hybridization, in discriminating expression patterns of homologous genes, alternative splice variants and antisense RNAs.
Alternatively, microarrays can be composed of short oligonucleotides (25 bases) synthesized directly onto a solid matrix using photolithographic technology (Affymetrix) [2,9] or constructed from long oligonucleotides (55-70 bases) spotted onto glass slides [10,11,12]. To mimic the Affymetrix design of freely moving probes tethered at one end onto a solid support, in-house manufactured or commercially available long oligonucleotides are modified by the addition of a 5' amino group for covalent attachment onto pre-activated glass slides [5,10]. This oligonucleotide design strategy has been widely viewed as a prerequisite for accurate gene-expression measurements. However, there is no clear evidence that other covalent attachments do not form. With oligonucleotide arrays, problems related to clone tracking, handling of glycerol stocks and failed PCR amplifications are avoided. The completion of numerous microbial, plant and eukaryotic genomes, as well as extensive EST data, provides sufficient sequence information to design unique oligonucleotides capable of distinguishing homologous genes and alternative splice variants. As such, oligonucleotide probes have an added flexibility over PCR amplicons.
Comprehensive studies comparing the Affymetrix approach with cDNA arrays have only recently appeared in the literature [13,14]. Studies comparing long oligonucleotides to cDNA arrays have not been as forthcoming. In the only example to date, 5'-amino-modified 50-mers representing prokaryotic genes were compared to corresponding PCR amplicons [12]. Analysis of the hybridization signals derived from these two probe types, while providing important insights pertaining to sensitivity and specificity, were limited in scope (total of eight genes) and design (interrogation was carried out with complementary targets derived from synthetic RNA as opposed to cellular RNA). A drawback to using modified oligonucleotides is the significant cost associated with the addition of the 5'-amino linker. An alternative strategy is to utilize unmodified oligonucleotides spotted onto glass slides, where attachment is believed to be primarily ionic in nature [11]. However, a comparison of this approach to standard cDNA arrays has yet to be provided. It is imperative that comparisons be carried out on all probe types in the light of conflicting reports regarding the correlation between Affymetrix and cDNA array-based expression measurements [13,14]. Whereas one study shows both approaches correctly identifying 16 out of 17 differentially regulated genes [13], a second study found a correlation of r = 0.328 between matched results from the same two platforms [14]. Discordant results were not resolved in the latter study. Here we test the performance of unmodified 70-mers printed alongside PCR amplicons. Using this unique study design, both probe types can be simultaneously interrogated with a complex target composed of both cellular and synthetic RNA.
Results
Optimal attachment parameters for 70-mers and PCR amplicons on the same slide
The success of microarray assays requires stable binding and retention of probes throughout the entire printing/blocking/hybridization/washing process. Oligonucleotides were spotted alongside PCR amplicons onto TeleChem SuperAmine aminated slides, and immobilized by ultraviolet (UV) cross-linking. To determine the optimal UV cross-linking energy required for efficient oligonucleotide immobilization, a series of spotted arrays from the same printing session were subjected to increasing UV energy (70, 150, 250, and 450 mJ/cm2). Deposition and retention of the probe onto aminated slides were assessed by: staining with Vistra Green solution and subsequent fluorescence scanning at 532 nm; and hybridization with Cy-labeled targets derived from rat brain and heart RNA. Using these two methods, optimal retention of both oligonucleotide and PCR amplicon probes was determined to occur between 250 and 450 mJ/cm2 as described below.
Directly following probe deposition, UV cross-linking at 250 mJ/cm2 and Vistra Green staining, the measured fluorescence intensity from oligonucleotide probes was typically higher than PCR amplicons (Figure 1a). The efficiency of the oligonucleotide immobilization strategy was tested by stringently washing the same slide overnight in 0.2% SDS (at 42°C, SSC absent in wash solution) to remove Vistra Green and any loosely bound nucleic acids, restained with Vistra Green and scanned (Figure 1b). There was essentially no change in the average median intensity values for either the oligonucleotide or PCR amplicon probes after incubation with detergent (Figure 1c).
Figure 1.
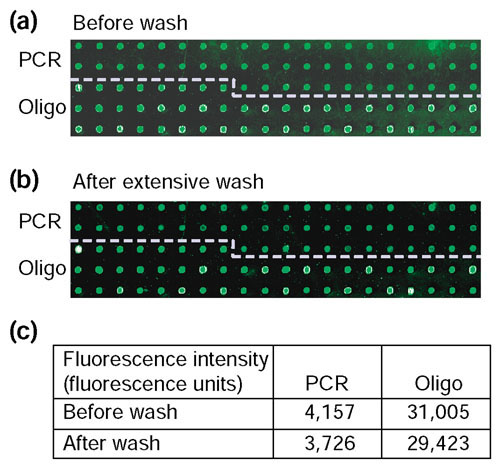
Retention of unmodified rat 70-mer oligonucleotide and cDNA probes printed on the same array. 70-mer oligonucleotides (50 μM) and PCR amplicons (100-200 nM; insert size ranged from 0.5 to 1.5 kb) were printed onto TeleChem SuperAmine slides and immobilized by UV cross-linking at 250 mJ/cm2. The image depicts DNA probes deposited onto slides stained with Vistra Green nucleic acid staining solution. (a) Section of array after staining. (b) The same section of array after the slide was extensively washed overnight in 0.2% SDS at room temperature, restained and scanned. (c) Average DNA fluorescence intensity of target hybridization to PCR amplicon and oligonucleotide probes before and after the extensive wash. Signal intensities were obtained from GenePix 3.0 software (Axon Instruments, CA) after scanning at 532 nm. Results are representative of 10 independent experiments.
These results show that immobilization of unmodified 70-mer oligonucleotides to SuperAmine aminated slides by high-UV cross-linking energy is sufficient and comparable to PCR amplicons. Clearly, our oligonucleotide immobilization protocol should be sufficient to sustain routine microarray hybridization and wash procedures, which are much less stringent than the overnight wash at 42°C with 0.2% SDS and no salt.
The importance of titrating UV cross-linking energy for oligonucleotide immobilization is exemplified by a series of spotted arrays hybridized with Cy-labeled targets derived from rat brain and heart RNA. As seen in Figure 2, there was an increase in the appearance of hybridized spots and signal intensity as the energy of cross-linking was increased. At lower cross-linking energies (for example, 70 and 150 mJ/cm2) it is apparent that oligonucleotide probes were not sufficiently attached to the surface of aminated slides. In contrast, PCR amplicons are sufficiently immobilized onto the aminated surface of glass slides at these intensities [15]. On the basis of the hybridization experiments and in agreement with Vistra Green staining results, optimal attachment of 70-mer oligonucleotides occurred between 250 and 450 mJ/cm2. An improvement in oligonucleotide retention onto aminated slides was not seen at intensities higher than 450 mJ/cm2 (data not shown). Of interest was the finding that different slide chemistries (that is, poly-L-lysine, aldehyde, aminosilane, epoxide) had different UV cross-linking titration curves for optimal attachment of 70-mer oligonucleotides, and increasing the cross-linking intensity in some slide types actually decreased apparent probe deposition (data not shown). Even more surprising was the observation that slides with the same or similar slide chemistry from different vendors exhibited marked differences in the optimal UV cross-linking energy for probe attachment.
Figure 2.

Effect of UV cross-linking intensity on retention of unmodified rat 70-mer oligonucleotides. The 70-mer oligonucleotides (50 μM) were printed onto TeleChem SuperAmine slides that were subjected to different UV cross-linking intensities and hybridized to Cy3- and Cy5-labeled targets derived from reverse transcribed rat heart and brain total RNA. A representative array section is shown for each UV cross-linking energy. (a,d) Experiments in which brain and heart targets were labeled with Cy3 and Cy5, respectively. (b,c) Flip-dye experiments in which brain and heart targets were labeled with Cy5 and Cy3, respectively. Each panel is representative of three to four independent experiments.
Sensitivity of unmodified 70-mers on aminated slides
Defining assay sensitivity is important as the ability to measure gene-expression changes is desired not only for moderate and abundant mRNAs but also for rare transcripts. One method for assessing the sensitivity of microarray assays is to use exogenous spiking controls. These controls also help to identify systematic problems associated with target labeling, slide hybridization and scanning. For this purpose, we developed a set of 10 Arabidopsis control cDNA plasmids. Each of the 10 plasmids was used to synthesize cRNA in vitro. cRNAs were quantitated and differentially spiked into heart and brain RNA samples at specific copy numbers based on the following assumptions: 360,000 mRNA transcripts per cell, 20 pg total RNA per cell and 1 pg mRNA transcript per cell; hence, 100 'spike' copies/cell would be equivalent to about 0.14 ng 'spike' transcript in 10 μg total RNA [12]. To normalize our array data, we spiked five of the ten control cRNAs into the Cy3 and Cy5 labeling reactions at equal copy number, ranging from 40 to 100 copies per cell. The remaining five control cRNAs were spiked into the labeling reactions at concentrations ranging from 1 to 300 copies per cell. We printed Arabidopsis 70-mers and PCR amplicons across six different sectors in order to measure intra-slide variability. Inter-slide variability was evaluated across independent hybridizations and target labeling. Figures 3a and 3b compare the intra- and inter-slide variation of oligonucleotides to discern twofold changes in Arabidopsis targets. Similar results were obtained for the detection of targets spiked at threefold ratios (data not shown). These data show that our oligonucleotide array platform was able to detect two- and threefold changes in transcript number at a sensitivity of one to two copies per cell. Comparing the sensitivity of oligonucleotide probes to PCR amplicons indicates that the two probe types perform equally well at consistently detecting twofold changes in rare transcript levels (Figure 3c).
Figure 3.
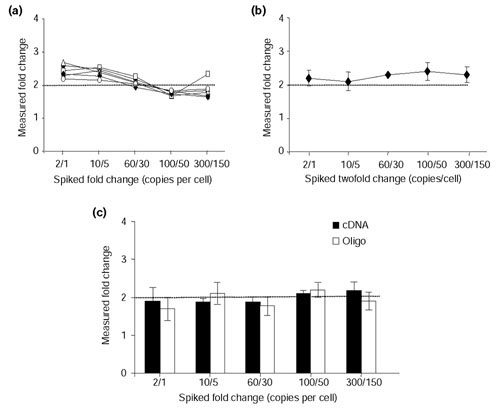
Assessment of sensitivity and reproducibility within and between slides, using the Arabidopsis controls. In vitro transcribed cRNA from a set of five Arabidopsis gene constructs (Ra, Cab, rbcL, Ltp4 and Ltp6) was spiked at varying levels into rat heart and brain RNA (that is, 2/1 represents two copies per cell and one copy per cell of Ra cRNA spiked into heart and brain RNA, respectively). Cy3- and Cy5-labeled targets were hybridized to rat arrays containing Arabidopsis 70-mer and PCR amplicon probes. (a) Intra-slide measurements. Arabidopsis cRNAs were added into the labeling reactions to assess the ability of oligonucleotide probes to discriminate twofold changes. The measured fold-change values are shown for each Arabidopsis target hybridizing to its complementary probe printed across six different sectors of an array. Values derived from a particular sector are represented by an open triangle, open square, open circle, closed circle, closed diamond or closed square. Ra, Cab, rbcL, Ltp4 and Ltp6 cRNAs were differentially spiked at 2/1, 10/5, 60/30, 100/50 and 300/150 copies per cell, respectively. Results are representative of four independent hybridizations. (b) Inter-slide measurements. Arabidopsis cRNAs were spiked into the labeling reactions to evaluate slide-to-slide reproducibility of oligonucleotide probes to discriminate twofold differences. The averaged result from four independent experiments is shown. (c) Comparison of Arabidopsis oligonucleotide and PCR amplicon probes printed on the same array to discriminate twofold changes. Data shown are the mean ± SD of five independent experiments.
Arabidopsis probe elements serve as excellent negative controls when exogenous cRNA is not added to the labeling reaction. In the absence of cRNA spiking, cross-hybridization of Cy-labeled rat targets to Arabidopsis probe elements was negligible (data not shown).
Concordance between probe types when measuring differential gene expression in biological samples
To compare the accuracy of unmodified oligonucleotide arrays to conventional cDNA arrays, we hybridized slides containing both probe types with equal amounts of Cy-labeled target derived from rat heart and brain RNA. Hybridization to 36 out of 38 antisense-strand oligonucleotide probes was negligible to nonexistent under our incubation and wash protocols (data not shown). A positive hybridization signal (threefold above background) was obtained for 65 of the 75 sense-strand oligonucleotide probes interrogated with labeled target. Accordingly, each of the corresponding 65 PCR amplicon probes had a positive hybridization signal (threefold above background). As the intra-slide variation was low (average standard deviation (SD) was around 0.07 and around 0.11 for log2-transformed ratio values derived from PCR amplicon and 70-mer spots, respectively), ratio values from replicate spots within a slide were averaged. Averaged ratios derived from independent experiments were plotted and regression analysis was performed to assess reproducibility of arrays containing oligonucleotide targets (Figure 4a). A similar comparison was performed on PCR amplicon probes (Figure 4b). Both oligonucleotide and PCR amplicon probes showed high reproducibility between replicate experiments (that is, different slides hybridized with targets generated from different batches of RNA samples) with correlation coefficients (r) of 0.95 and 0.96, respectively. Next, we compared oligonucleotide-derived ratios with those obtained from PCR amplicon probes (Figure 4c). A correlation coefficient of r = 0.80 (p < 0.05) and a slope close to unity were obtained, indicating that unmodified oligonucleotide and PCR amplicon probes gave comparable expression ratios. Moreover, there was agreement in the calculated average coefficient of variation (13.4%) for the expression ratios computed from the two probe types.
Figure 4.
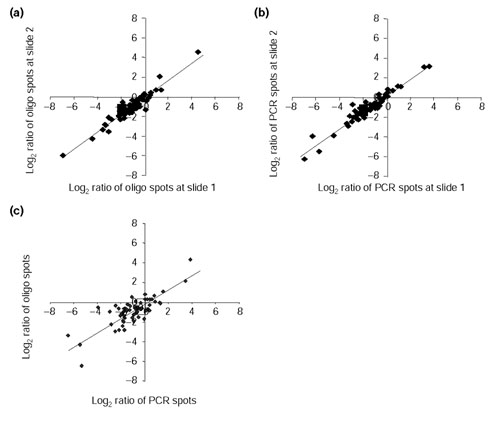
Scatter plots of ratios from replicate experiments for both 70-mer and PCR amplicon probes. (a) Plot of log2 ratios detected by oligonucleotide probes from two independent experiments. The plot shows a high correlation between experiments with a correlation coefficient (r) of 0.95 and a slope of 0.89. Results are representative of five similar comparisons. (b) Plot of log2 ratios detected by PCR amplicons from two independent experiments. The plot shows a high correlation between experiments, with r = 0.96 and a slope of 0.84. Results are representative of five similar comparisons. (c) Correlation of gene-expression ratios detected by 70-mer and PCR amplicon probes. A high r of 0.80 and a slope of 0.73 were obtained when log2 ratio values from oligonucleotide probes were plotted against PCR amplicons. Results are representative of five similar comparisons.
Validation of microarray results with real-time PCR
Of the 65 represented genes that had a positive hybridization signal with both the oligonucleotide and PCR amplicon probe types, 60 were in agreement of each other (Figure 4c). Real-time PCR was used to test the accuracy of our microarray results (Figure 5a). Six genes exhibiting a range of expression differences in heart and brain were selected for validation. These included histone H4, which did not exhibit differential expression in the two tissues (log2 ratio around 0); cytochrome oxidase IV, kynurenine 3-hydroxylase, serine/threonine protein kinase and 14-3-3 protein gamma which exhibited two- to threefold differences (log2 ratio = 1 to 1.6); and desmin which was around 32-fold differentially expressed between the two tissues (log2 ratio approximately 5). In each case, the expression ratios derived from oligonucleotide and PCR amplicon probes were in accord with real-time PCR results.
Figure 5.
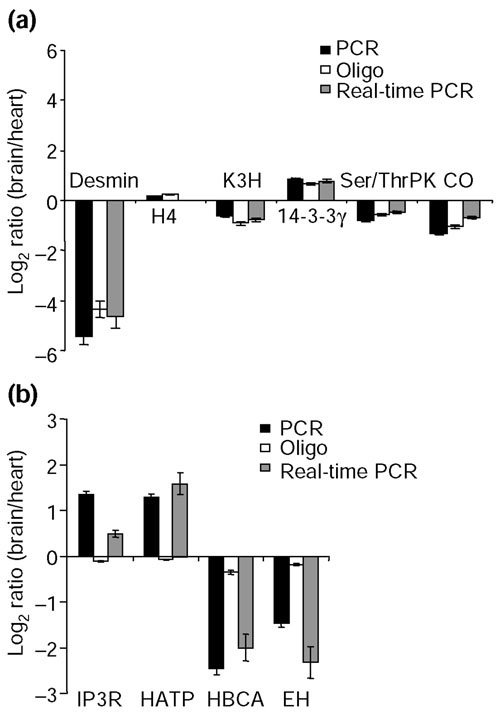
Real-time PCR validation of microarray results. (a) Concordance of gene-expression patterns determined by oligonucleotide and PCR amplicon microarrays and real-time PCR. Data shown are the mean ± SD of four to five independent determinations. H4, histone H4; K3H, kynurenine 3-hydroxylase; 14-3-3γ, 14-3-3 protein gamma Ser/Thr PK, serine/threonine protein kinase; CO, cytochrome c oxidase. (b) Real-time PCR analysis of discordant results from oligonucleotide and PCR amplicon arrays. Data shown are the mean ± SD of four to five independent determinations. IP3R, inositiol-1,4,5-trisphosphate receptor; HATP, H+-ATPase; HBCA, heart branched-chain aminotransferase; EH, cytosolic epoxide hydrolase.
There were five notable discrepancies between the two probe types, as compared to the 60 that were in agreement. A discrepancy was defined as a change equal to or greater than twofold measured with one probe type and no change (or a change in the opposite direction) measured with the other probe type. The resolution of the discordant results for inositol-1,4,5-trisphosphate receptor, H+-ATPase, branched aminotransferase and epoxide hydrolase is presented in Figure 5b. In each case, real-time PCR results were in agreement with the PCR amplicon-derived expression ratios. Interestingly, each of the oligonucleotide-derived expression ratios erroneously suggested that these genes were not differentially expressed in heart and brain tissues.
Effect of oligonucleotide probe concentration on signal intensity
In our initial experiments, unmodified 70-mer oligonucleotides were printed onto TeleChem slides at a relatively high concentration of 50 μM. By comparison, the concentration for printing 5'-amino linker modified 50-mer oligonucleotides was 20 μm [12]. To test the performance of unmodified oligonucleotides at lower printing concentrations, seven rat 70-mer oligonucleotides were serially diluted from 50, 25, 12.5 to 6.25 μM. Each oligonucleotide was chosen on the basis of earlier microarray results showing that both oligonucleotide and cDNA probes could hybridize to heart and brain targets, and that hybridization intensities associated with the seven different gene elements varied by at least an order in magnitude. The selected probes included SCG10 and desmin, which were highly differentially expressed in brain and heart, respectively; 14-3-3-gamma and profilin which were expressed at around twofold higher levels in brain and heart, respectively; and histone H4, 14-3-3-theta and thymosin beta-4, which showed no difference in expression between brain and heart. All diluted oligonucleotides along with their corresponding undiluted PCR amplicons (approximately 100-200 nM) were spotted onto the array at least four times. A stained representative section of an array printed with different starting concentrations of oligonucleotides and a single concentration of the corresponding PCR amplicons is depicted in Figure 6. As the starting oligonucleotide concentration was decreased from 50 μm to 6.25 μM (eightfold dilution), DNA fluorescence decreased on average twofold for the oligonucleotides (data not shown). This suggests that the capacity of the slides to retain 70-mer oligonucleotides in a typical 100 μm diameter spot approached saturation at the higher concentrations.
Figure 6.
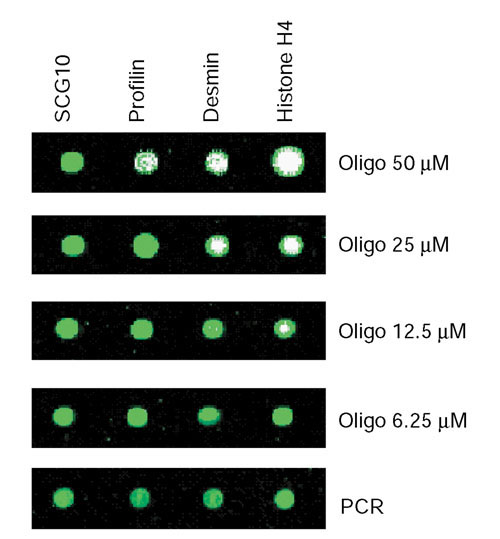
Serial dilution of 70-mer oligonucleotides compared to PCR amplicons. The array was stained with Vistra Green dye. A representative grid of oligonucleotides at each concentration, along with a single concentration of the corresponding PCR amplicon (100-200 ng/μl) is shown. Results are representative of three independent experiments.
Arrays containing diluted oligonucleotides were hybridized with labeled targets derived from brain and heart RNA as before. The median Cy3 and Cy5 hybridization intensities (minus background) were summed for each oligonucleotide concentration along with their corresponding cDNA probe (Figure 7a). A comparison of oligonucleotide and cDNA probes clearly demonstrates that the longer probe length of the latter does not necessarily translate to greater hybridization signal intensities. While the PCR amplicon probes for 14-3-3 protein theta, desmin and thymosin beta-4 generated higher signals than the corresponding oligonucleotide elements, the converse was observed for 14-3-3 protein gamma, histone H4 and profilin. Moreover, there was no correlation (r = 0.06, p > 0.05) between the hybridization signal intensities acquired from PCR amplicon probes and the corresponding oligonucleotide probes. Of interest was the apparent inverse correlation between oligonucleotide concentration and hybridization intensity. Hybridization intensities actually increased with decreasing oligonucleotide concentration for 14-3-3 protein gamma, desmin, SCG10, and to a lesser extent thymosin beta-4 (Figure 7a).
Figure 7.

The role of oligonucleotide concentration on microarray hybridization measurements. (a) Comparison of hybridization intensities of seven genes detected by oligonucleotides printed at different concentrations. Corresponding PCR amplicons at 100-200 ng/μl are included for comparison. The medium background subtracted fluorescent intensities were summed for the Cy3 and Cy5 channels for each gene and plotted. (b) Comparison of the expression ratios of differentially expressed genes detected by oligonucleotides printed at different concentrations. Data are the mean ± SD of four independent experiments.
For the oligonucleotides corresponding to differentially expressed genes (for example, SCG10, desmin, 14-3-3-gamma, profilin), the log2 ratios from four independent hybridizations (including flip dye experiments) were averaged and plotted in Figure 7b. The calculated ratios were highly reproducible and similar across the entire concentration range tested. This suggests that an oligonucleotide concentration as low as 6.25 μM is sufficient for accurate determination of relative expression differences. As the absolute levels of these four transcripts in rat heart and brain are not known with certainty, we repeated these experiments with known concentrations of synthetic Arabidopsis cRNA that were differentially spiked into rat heart and brain RNA. Six Arabidopsis oligonucleotides were accordingly diluted and printed onto aminated slides to test their ability to discriminate twofold differences in synthetic cRNA concentrations ranging from 10 to 300 copies per cell. Our data clearly show that an Arabidopsis oligonucleotide probe concentration as low as 6.25 μM was sufficient to accurately determine twofold differences in cRNA species at a ratio of 20/10 copies per cell (Figure 8).
Figure 8.
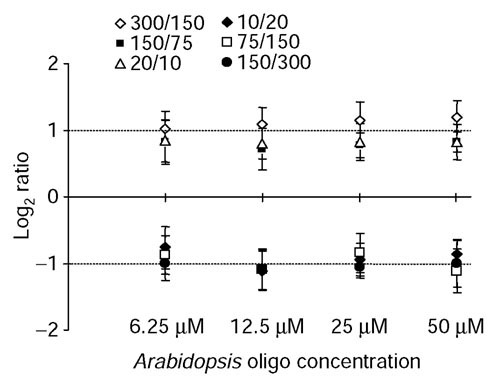
Comparison of the expression ratios of Arabidopsis genes detected by 70-mer oligonucleotides at different printing concentrations. Six Arabidopsis control cRNAs were spiked at varying levels corresponding to twofold differences into two labeling reactions containing rat brain and heart RNA. Another four Arabidopsis control cRNAs were spiked at a 1:1 ratio and served as normalization controls. The measured log2 expression ratios from four independent experiments were plotted as the mean ± SD. The expected twofold change was detectable for all oligonucleotides at the different printing concentrations (twofold difference equals a log2ratio of 1 or -1).
Discussion
In the study reported here, we systematically compared the performance of unmodified 70-mer oligonucleotides to traditional PCR amplicons, both probe types printed and UV cross-linked onto glass slides coated with primary amine groups. Direct comparisons are best accomplished when both probes are printed alongside each other, allowing for simultaneous interrogation with a complex target. Hence, analysis is not confounded by uneven aminosilane coating in different batches of slides, inconsistencies in the array resulting from different print sessions, differences in day-to-day label incorporation, or variations in day-to-day hybridization and wash procedures. A correlation coefficient (r) of 0.80 was obtained from our analysis, indicating that the two probe types gave comparable expression ratios. One variable that was not controlled for in our study was the number of cross-links per DNA molecule. Given a constant UV exposure, many more cross-links per molecule of cDNA probe are presumably formed compared to the shorter oligonucleotide probe. It is possible that the correlation coefficient was not higher as a result of the differential reaction of the two probe types to UV irradiation.
We designed our arrays to contain 75 different probes corresponding to mammalian signal transduction genes with a wide range of expression levels. In heart versus brain comparisons, oligonucleotide probes, like their cDNA probe counterparts, could reproducibly discern differences in mRNA populations as low as twofold (namely, 14-3-3 protein gamma) and as high as around 90-fold (namely, creatine kinase). Hence, the dynamic range of unmodified oligonucleotides is at least two orders of magnitude in fold-change measurements.
In the course of our work, we generated a resource of 10 Arabidopsis spiking control cRNAs along with their corresponding 70-mer oligonucleotide and PCR amplicon probes. As part of our quality-control procedures, all microarray assays routinely incorporate the spiking controls. These reagents will allow the microarray user to add specific concentrations of known transcripts into a complex mix of mammalian target RNA in order to assess, for example, hybridization kinetics, intra-slide variability, inter-slide variability, sensitivity and effectiveness of normalization algorithms. On the basis of experiments with the spiking controls, unmodified oligonucleotides can be used to detect twofold changes in transcript number at a level of 2-20 mRNA copies per cell. It is important to note that our protocol for generating first-strand cDNA target involves the use of random primers. At the outset, the Arabidopsis cRNAs were engineered to contain a 3' poly(A) tail. Hence, alternative protocols using oligo(dT) to prime mRNA for the synthesis of labeled target [15,16] can still take advantage of our spiking control set.
In our initial assessment of cDNA and 70-mer oligonucleotide probe types, the latter was printed at a concentration of 50 μM. Even at a printing concentration as low as 6 μM, oligonucleotide probes were capable of discerning twofold expression differences in complex cellular RNA mixtures and in synthetic spiked cRNAs. In fact, decreasing the oligonucleotide printing concentration from 50 to 6 μM had the effect of increasing the hybridization signal around two- to sixfold for a number of the probes (Figure 7a). The reason is unclear, but it is possible that high-density packing of an oligonucleotide probe within the confines of a small spot interferes with fluorescence emission of the target or hybridization efficiency. Alternatively, the higher spotting concentrations may favor cross-linking of the oligonucleotide probes to each other following UV irradiation. In either case, this phenomenon appears to be sequence dependent as not all probes exhibited this behavior. The present study also demonstrates that longer probes are not necessarily associated with higher hybridization signals, as the hybridization signals from half of the 70-mer oligonucleotide probes were actually higher than or equivalent to their corresponding PCR amplicons, which have an average length of 1 kilobase (kb). Taken together, the combination of unmodified oligonucleotides and low printing concentrations has resulted in an approximately 16-fold reduction in reagent costs. An issue not evaluated in the present study, but one that has significant cost-saving potential, is the effect of reducing the length of unmodified oligonucleotides on microarray sensitivity. Clearly, this is an area for future investigation.
Of the five discordant results found between oligonucleotide and cDNA arrays, real-time PCR data validated the accuracy of the cDNA probe type in every case (Figure 5b). It seems likely that a failure in oligonucleotide probe design was responsible for the discordant data. Analysis of the discordant oligonucleotide sequences (that is, inositol-1,4,5-trisphosphate receptor, H+-ATPase, branched aminotransferase and epoxide hydrolase) did not reveal any obvious secondary structure that might interfere with hybridization. Treatment of spotted arrays with UV light is thought to induce free-radical-based coupling between thymidine residues on the oligonucleotide and carbon atoms on the alkyl amine groups of coated glass slides (Todd Martinsky, TeleChem International, personal communication). The T content of concordant and discordant oligonucleotides was similar, with average values of 26% and 29%, respectively, suggesting that UV cross-linking was not preferentially disrupting hybridization specificity of discordant 70-mers. Moreover, there was a lack of correlation (r = 0.03, p > 0.05) between T content of the oligonucleotides and corresponding hybridization signal intensities. Of interest, however, was the finding that the discordant oligonucleotides had an average GC content of 57% compared to the concordant oligonucleotide average of 50%. Accordingly, the hybridization signal associated with the discordant oligonucleotides was around two- to threefold higher than the concordant oligonucleotides, suggesting that 'non-specific' Cy-labeled targets were cross-hybridizing with the discordant oligonucleotides. This possibility is clearly illustrated for the H+-ATPase gene (Figure 9). Within the H+-ATPase 70-mer sequence is a stretch of 20 contiguous nucleotides perfectly matching a region in the tumor endothelial marker 8 mRNA. It has been shown previously that 15 contiguous nucleotides are sufficient for cross-hybridization of non-target species [12]. There are two important points to note. First, the PCR amplicon for H+-ATPase also contains the same 20 contiguous nucleotides (Figure 9). Regardless of this, this particular probe was still able to distinguish differential expression of the H+-ATPase gene in heart and brain tissue. We postulate that a large fraction of H+-ATPase-specfic Cy-labeled targets (which on average should be 100-200 nucleotides long) were available for hybridization to complementary sequences found on the longer PCR amplicon probe but absent on the shorter 70-mer probe (for example, sequences downstream of the 70-mer). Second, tumor endothelial marker 8 mRNA was identified in mouse. The orthologous rat mRNA has not been cloned yet, which is reflected by the more than 2.5 million mouse EST sequences present in dbEST, compared to only 351,827 ESTs for the rat. On the basis of BLAST searches of human and mouse sequences, contiguous non-target sequences could also be identified in the discordant oligonucleotides for inositol-1,4,5-trisphosphate receptor, branched amino-transferase and epoxide hydrolase. Hence, future oligonucleotide design considerations should include an analysis of mouse and human sequences, because of the relatively small number of available rat sequence for expressed transcripts. In addition, the synthesis of redundant probe sets (for example, two 70-mers per gene) might be warranted to help decrease false negatives by an order of magnitude.
Figure 9.
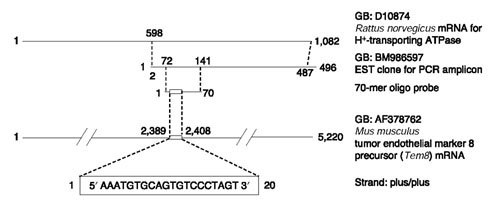
Specificity of 70-mer oligonucleotide probes. An oligonucleotide probe (70-mer) was designed to detect Cy-labeled H+-ATPase cDNA (target) reverse transcribed from rat heart and brain RNA. Nucleotides 1-70 of the oligonucleotide probe correspond to nucleotides 72-141 of an EST clone for H+-ATPase (GenBank accession BM986597). This EST clone serves as the DNA template for generating the PCR amplicon probe. For orientation, the 70-mer oligonucleotide sequence is shown aligned with nucleotides 669-738 of the full-length H+-ATPase mRNA (GenBank accession D10874). Contained within the 70-mer oligonucleotide probe is a stretch of 20 contiguous nucleotides that is also found in the tumor endothelial marker 8 mRNA (GenBank accession AF378762). Other than this stretch of contiguous nucleotides, there is no sequence identity between H+-ATPase and tumor endothelial marker 8.
The mechanism of the adherence of unmodified oligonucleotides to glass slides has been addressed [11]. Attachment involves noncovalent interactions such as electrostatic interactions, where the negatively charged phosphate backbone of the oligonucleotide is attracted to the positively charged surface of the glass slide (for example, a surface containing protonated alkyl amines). Whereas noncovalent interactions appear to be the predominant mechanism for oligonucleotide attachment, covalent linkage is likely to have an important supplementary role in UV-irradiated microarrays. This seems plausible as our stringent overnight washes in strong detergent did not appreciably detach unmodified oligonucleotides from the slide surface. The importance of UV cross-linking cannot be overemphasized. Under-irradiation of cDNA arrays is known to cause insufficient binding of DNA and over-irradiation results in over-nicking of DNA samples [17]. A further complicating factor is our finding that oligonucleotides printed onto different slide chemistries (or slides with similar chemistries from different vendors) will have very different optimal UV titration curves. In our hands, optimal UV cross-linking occurred at 450 and 70 mJ/cm2 for oligonucleotides printed onto TeleChem SuperAmine™ and Corning GAP II™ slides, respectively. For TeleChem slides, under-irradiation (70-150 mJ/cm2) causes insufficient oligonucleotide attachment. For Corning slides, over-irradiation (150-450 mJ/cm2) results in a decrease in the hybridization signal that may reflect excessive covalent attachment of oligonucleotides. As UV cross-linking may adversely affect oligonucleotide accessibility to labeled target during hybridization, we cannot discount the possibility that alternative attachment strategies (for example, 5'-amino-modified oligonucleotides) may provide greater sensitivity and specificity. This issue needs to be explored in the future.
In summary, the present study provides evidence that the performance of unmodified 70-mer oligonucleotides is comparable to cDNAs printed on glass slides. Optimal conditions were identified for oligonucleotide attachment and hybridization/wash conditions, resulting in high assay sensitivity and reproducibility. Our results show that unmodified oligonucleotides can provide an accurate, reproducible and cost-effective means to measure gene-expression profiles. Of interest is the fact that our hybridizations were successfully carried out on slides that simultaneously contained both PCR amplicons and oligonucleotides. Hence, future microarrays can be constructed in a modular fashion, with oligonucleotide-based elements being added to existing PCR amplicons as more genomic sequence information is gathered, in the absence of readily available cDNA clones. Lastly, our findings have broader implications, suggesting that the combination of expression measurements across different platforms (for example, Affymetrix and cDNA arrays, unmodified long oligonucleotides and cDNA arrays) within a single analysis maybe feasible [18].
Materials and methods
Constructing exogenous spiking cRNA controls and a PCR amplicon printing set to assess oligonucleotide sensitivity
Ten Arabidopsis thaliana genes corresponding to chlorophyll a/b-binding protein (Cab), lipid transfer protein 4 (Ltp4), lipid transfer protein 6 (Ltp6), NAC1, ribulose-5-phosphate kinase (PRKase), ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL), rubisco activase (Ra), root cap 1 (RCP1), triosphosphate isomerase (TIM), and papain-type cysteine endopeptidase (XCP2) were chosen for PCR amplification on the basis of their plant-specific expression. PCR amplicons of approximately 500 base-pairs (bp) were amplified from an Arabidopsis cDNA library constructed from leaf tissue or genomic DNA using sense and antisense gene-specific primers flanked by a HindIII and SacI adaptor sites for subcloning, respectively. Primers were synthesized by Invitrogen/Life Technologies or Operon Technologies and purified by PAGE. Primer sequences are as follows: 5'-CCACTGTAGATGGGCTATGC-3' and 5'-AGGGATAACAATATCGCCAA-3' for Cab; 5'-TCACCCAAAAGAGAAGAGCA-3' and 5'-CAAAGCCATCAAGACAAACA-3' for Ltp4; 5'-TCTTATTAGCCGTGTGCCTG-3' and 5'-CAACTAGCAAACCAATGCCC-3' for Ltp6; 5'-CAACATGGGAAGCTGTTTTG-3' and 5'-CAAGCACACGTTATTTCCCC-3' for NAC1;5'-CGGAGAAGAAGAGGAGACCA-3' and 5'-GAGGGTCAAGAAGTCCAGTG-3' for PRKase; 5'-GTTCCACCTGAAGAAGCAGG-3' and 5'-CGCATAAATGGTTGGGAGTT-3' for rbcL; 5'-GAGTGGAAACGCAGGAGAAG-3' and 5'-ACTCCAAGGCTCTCAACGAA-3' for Ra; 5'-TGGTGGACTCTCCGTTCTTC-3' and 5'-CGAGTTGTGACCATAAGCCA-3' for RCP1; 5'-TCAAATCCTCGTTGACAGAC-3' and 5'-CTGTTGCCTCCATTGACAGA-3' for TIM; and 5'-CAAATGGCTCTTTCTTCACC-3' and 5'-TGGTTCTTAACTTCCGCCAC-3' for XCP2. PCR products were digested with the restriction enzymes HindIII and SacI, subcloned into pSP64 poly(A) vector (Promega, Madison, WI) and sequence verified. For printing Arabidopsis genes, inserts in pSP64poly(A) were amplified by PCR with the SP6 (5'-ATTTAGGTGACACTATAG-3') and M13R primers. To generate cRNAs containing a 3' poly(A) tail, pSP64poly(A) constructs were linearized with EcoRI and in vitro transcribed from the SP6 promoter using the MEGAscript™ High Yield Transcription Kit (Ambion, Austin, TX). The Arabidopsis cRNA set was designed to serve as spiking controls to assess a broad range of copy numbers (rare, moderate, abundant) and varying expression ratios (1:3, 1:2, 1:1, 2:1, 3:1). For printing control 70-mers, oligonucleotides were synthesized on the basis of the corresponding 500 bp of each gene in the Arabidopsis control spiking cRNA set (Table 1). Development of our control set of 10 Arabidopsis oligonucleotides and corresponding set of PCR amplicons subcloned into pSP64poly(A) serve as a valuable quality-control resource for cDNA/oligo microarrays. The Arabidopsis control spiking cRNA vector set and protocols will be made freely available to academic investigators upon request.
Table 1.
Arabidopsis 70-mer oligonucleotide probes
| Abbreviation | Complete gene name | GenBank accession number | 70-mer sequence |
| RCP1 | Root cap 1 protein | AF168390 | 5'-AACAGTATCTTGCCTGGGACAACTGCTTTTGGTATTGCTGTGGCAGCTATAATCATGGCTCGAACTGGGA-3' |
| Cab | Photosystem I chlorophyll | X56062 | 5'-TGCTCGCTGTTCCTGGGATTTTGGTACCAGAAGCATTAGGATATGGAAACTGGGTTAAGGCTCAGGAATG-3' |
| A/B-binding protein | |||
| TIM | Triosephosphate isomerase | AF247559 | 5'-GAACAGCTCAAAGACCTTGGCTGCAAGTGGGTCATTCTTGGGCATTCCGAACGGAGACATGTCATCGGAG-3' |
| Ltp4 | Lipid transfer protein 4 | AF159801 | 5'-AGTCCATGTCTAGGCTACCTATCGAAGGGTGGGGTGGTGCCACCTCCGTGCTGTGCAGGAGTCAAAAAGT-3' |
| PRKase | Ribulose-5-phosphate kinase | X58149 | 5'-CAAACCAACCGGAGATTCAACACACTCATCACTTGCGCACAAGAAACCATCGTGATCGGACTAGCTGCTG-3' |
| Ltp6 | Lipid transfer protein 6 | AF159803 | 5'-GCCGTGTGCCTGGTTCTTGCTTTACACTGCGGTGAAGCAGCCGTGTCTTGCAACACGGTGATTGCGGATC-3' |
| rbcL | Ribulose-1,5-bisphosphate carboxylase/oxygenase, large subunit | ATU91966 | 5'-AGCTGCTGAATCTTCTACTGGTACATGGACAACTGTGTGGACCGATGGGCTTACCAGCCT TGATCGTTAC-3' |
| Ra | Rubisco activase | X14212 | 5'-ACGCTGGTGCGGGTCGTATGGGTGGTACTACTCAGTACACTGTCAACAACCAGATGGTTAACGCAACACT-3' |
| XCP2 | Papain-type cysteine endopeptidase | AF191028 | 5'-GCTTCTTCCCACGATTACTCCATCGTTGGATACTCCCCCGAGGATTTGGAATCTCATGACAAACTCATAG-3' |
| NAC1 | NAM-like protein | AF198054 | 5'-GCCTCTGCATCGCTTCCTCCACTGATGGATCCTTACATCAACTTTGACCAAGAACCCTCTTCTTATCTCA-3' |
Rat 70-mer oligonucleotide design
To minimize cross-hybridization, oligonucleotides of 70 bases (unmodified) were designed using the computer program, Pick70 [19]. Oligonucleotide design considerations included uniqueness, avoidance of internal self-annealing structures, narrow Tm range (75-80°C) over the entire oligonucleotide set and masking of low-complexity regions. The TIGR Rat Gene Index containing a non-redundant set of expressed mRNA sequences [20] was used as the 'complete genome source' for selecting 70-mer oligonucleotide sequences with Pick70. Oligonucleotides were chosen to represent 'housekeeping' and signal transduction genes, while other 70-mers were designed to detect tissue-specific transcripts from either brain or heart (for example, those for SCG10, creatine kinase, and desmin). The sequences of the 70-mers and corresponding GenBank accession numbers of the genes are available as and additional data file. Oligonucleotides were synthesized at a 50 nmol scale by Invitrogen/Life Technologies (Carlsbad, CA) or Operon Technologies (Alameda, CA), and resuspended in sterile milliQ water to a final concentration of 100 μM. We selected individual cDNA clones from the TIGR Rat Gene Index whose EST sequences corresponded to the same gene from which the 70-mers were designed. Rat cDNA clone inserts were amplified by PCR with M13F (5'-GTTTTCCCAGTCACGACGTTG-3') and M13R (5'-TGAGCGGATAACAATTTCACACAG-3') primers [15,16]. Insert size ranged from 0.5 to 1.5 kb.
Microarray fabrication
Oligonucleotides (50 μM, except where indicated otherwise) and PCR amplicons (100-200 nM) in 50% DMSO were printed onto SuperAmine slides (TeleChem International, Sunnyvale, CA) using an Intelligent Automation Systems (IAS) arrayer (Cambridge, MA) with a 12-pen print head [16]. The rat 70-mers and PCR amplicons were printed in quadruplicate while the 10 Arabidopsis 70-mers and PCR amplicons were spotted into six different sectors on the slide. After printing, DNA was cross-linked to the slides by UV irradiation with a Stratalinker UV Crosslinker (Stratagene, La Jolla, CA) and stored in a vacuum chamber until use. To assess oligonucleotide retention, slides were UV cross-linked and stained for 10 min in Vistra Green Nucleic Acid staining solution (Amersham Pharmacia, Piscataway, NJ) at a 1:10,000 dilution. Afterwards, slides were washed at least five times, 1 min each, in milliQ water at room temperature, centrifuged to dryness (500 rpm × 5 min), and scanned at 535 nm using a dual laser GenePix 4000B scanner (Axon Instruments, Foster City, CA). Subsequently, slides were gently agitated in 0.2% SDS (no salt) overnight at 42°C, washed extensively with water, scanned to ensure that the dye was completely removed, restained with Vistra Green and rescanned.
Target labeling and array hybridization
To generate labeled single-stranded cDNA target, 10 μg total RNA from rat heart or brain (Clontech, Palo Alto, CA) was reverse transcribed for 2-3 h at 42°C in the presence of 6 μg random primers (Invitrogen/Life Technologies), 1x first-strand synthesis buffer (Invitrogen/Life Technologies), 10 mM DTT, dNTP mix (25 mM dATP, 25 mM dCTP, 25 mM dGTP, 15 mM dTTP, 10 mM amino allyl-dUTP), and 200 units Superscript II reverse transcriptase (Invitrogen/Life Technologies). RNA was hydrolyzed with 200 mM NaOH and 100 mM EDTA for 15 min at 65°C, then neutralized with 200 mM HCl. First-strand cDNA was purified from unincorporated amino allyl-dUTPs on QIAquick PCR purification columns (Qiagen, Valencia, CA) according to manufacturer's instructions, except that QIAquick wash buffer was replaced with 5 mM K+ phosphate buffer (pH 8.5) containing 80% ethanol, and cDNA was eluted with 4 mM K+ phosphate buffer (pH 8.5). Eluted cDNA was lyophilized, resuspended in 4.5 μl 0.1 M Na2CO3 buffer (pH 9), mixed with either Cy3 or Cy5 NHS-ester (Amersham Pharmacia), and incubated for 1 h in the dark at room temperature. Cy3- and Cy5-labeled cDNA targets were then purified on QIAquick PCR purification columns, combined and concentrated by lyophilization, and hybridized to the microarray at 42°C for 16 h in hybridization solution containing 50% formamide, 5x SSC, 0.1% SDS, 20 μg mouse Cot-1 DNA and 10 μg poly(dA). Reverse dye labeling of samples was employed in separate experiments to account for any bias in dye coupling or emission efficiency of Cy dyes. After hybridization, microarray slides were washed by immersion into 2x SSC, 0.2% SDS for 5 min at 42°C, 0.2x SSC, 0.1% SDS for 1 min at room temperature, 0.2x SSC for 1 min at room temperature, and 0.05x SSC twice for 1 min at room temperature, dried by centrifugation, and immediately scanned. Different hybridization and wash conditions were tested (data not shown). The procedures described above have been optimized for both PCR amplicon and oligonucleotide probes regardless of whether the two probe types are printed together or separately. We chose a random primer labeling scheme so that oligonucleotide probe design would not be restricted to any particular region of the mRNA molecule. In contrast, oligo(dT)12-18 priming protocols [16] limit design considerations to the 3' end of the mRNA molecule.
Array image processing and data analysis
Cy3 and Cy5 fluorescence on microarray slides were scanned at 10 μm resolution using a GenePix 4000B scanner and saved as two single TIFF images. The intensities of spots on the two images were subsequently analyzed with GenePix Pro 3.0 software and a dataset was output. We used the following criteria to flag bad or extremely weak spots from the array dataset: spot area < 70 pixels, % saturated pixels > 50%, and sum of the median signal intensity < 1,000. Normalization of the array dataset was based on total median background subtracted intensities from the Cy3 and Cy5 channels and linear regression of the median signal intensities generated from the Arabidopsis control cRNA set spiked into the query RNA samples at a 1:1 ratio [21]. After normalization, expression ratios were calculated for each non-flagged spot and log2 transformed.
Additional data files
The sequences of the 70-mers and corresponding GenBank accession numbers of the genes are available as an additional data file.
Supplementary Material
Sequences of the 70-mers and corresponding GenBank accession numbers of the genes
Acknowledgments
Acknowledgements
We thank Nnenna Nwokekeh for her excellent technical assistance. We also thank members of the TIGR microarray team for technical assistance and helpful comments. This work was supported by a Programs for Genomic Applications grant from the National Heart Lung and Blood Institute.
References
- Harkin DP. Uncovering functionally relevant signaling pathways using microarray-based expression profiling. Oncologist. 2000;5:501–507. doi: 10.1634/theoncologist.5-6-501. [DOI] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Schulze A, Downward J. Navigating gene expression using microarrays - a technology review. Nat Cell Biol. 2001;3:E190–E195. doi: 10.1038/35087138. [DOI] [PubMed] [Google Scholar]
- Schena M, Heller RA, Theriault TP, Konrad K, Lachenmeier E, Davis RW. Microarrays: biotechnology's discovery platform for functional genomics. Trends Biotechnol. 1998;16:301–306. doi: 10.1016/s0167-7799(98)01219-0. [DOI] [PubMed] [Google Scholar]
- Southern E, Mir K, Shchepinov M. Molecular interactions on microarrays. Nat Genet. 1999;21:5–9. doi: 10.1038/4429. [DOI] [PubMed] [Google Scholar]
- Watson A, Mazumder A, Stewart M, Balasubramanian S. Technology for microarray analysis of gene expression. Curr Opin Biotechnol. 1998;9:609–614. doi: 10.1016/s0958-1669(98)80138-9. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Harrington CA, Rosenow C, Retief J. Monitoring gene expression using DNA microarrays. Curr Opin Microbiol. 2000;3:285–291. doi: 10.1016/s1369-5274(00)00091-6. [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Zhao X, Nampalli S, Serino AJ, Kumar S. Immobilization of oligodeoxyribonucleotides with multiple anchors to microchips. Nucleic Acids Res. 2001;29:955–959. doi: 10.1093/nar/29.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call DR, Chandler DP, Brockman F. Fabrication of DNA microarrays using unmodified oligonucleotide probes. Biotechniques. 2001;30:368–372. doi: 10.2144/01302tt06. [DOI] [PubMed] [Google Scholar]
- Kane MD, Jatkoe TA, Stumpf CR, Lu J, Thomas JD, Madore SJ. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 2000;28:4552–4557. doi: 10.1093/nar/28.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen T, Wurmbach E, Pfeffer RL, Ebersole BJ, Sealfon SC. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 2002;30:e48. doi: 10.1093/nar/30.10.e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo WP, Jenssen TK, Butte AJ, Ohno-Machado L, Kohane IS. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics. 2002;18:405–412. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee NH, Dang CV, Liu ET. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J. A concise guide to cDNA microarray analysis. Biotechniques. 2000;29:548–550. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Morley M, Aguilar F, Massimi A, Kucherlapati R, Childs G. Making and reading microarrays. Nat Genet. 1999;21(Suppl):15–19. doi: 10.1038/4439. [DOI] [PubMed] [Google Scholar]
- Malek RL, Irby RB, Guo QM, Lee K, Wong S, Ruffy M, Tsai J, Kwong KY, Frank B, Liu ET, et al. Identification of Src transformation fingerprint in human colon cancer. Oncogene. 2002;21:7256–7265. doi: 10.1038/sj.onc.1205900. [DOI] [PubMed] [Google Scholar]
- Pick70 http://se.osxgnu.org/pub/mirrors/sourceforge/arrayoligosel/
- TIGR rat gene index http://www.tigr.org/tdb/tgi/rgi
- Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of the 70-mers and corresponding GenBank accession numbers of the genes


