Abstract
We have systematically isolated a variety of coactivator complexes from HeLa S3 cells using proteomic approaches. In the present report, we have evaluated twelve coactivator complexes involved in nuclear receptor-dependent gene transcription that have been purified by using an immunoprecipitation method. The twelve purified coactivator complexes are SRC-1, SRC-2, SRC-3, CBP, p300, CAPER, E6-AP, ASC-1, CoREST, CRSP3, CRSP2, and CDK7 containing complexes. We have identified 153 protein components associated with these coactivator complexes using mass spectrometry. In order to systematically characterize the functional roles for these components in nuclear receptor-dependent gene transcription and their investigative potential, we have developed a scoring system. This scoring system is comprised of biological and experimental parameters. The biological evaluation considers aspects such as intrinsic enzymatic activity of a protein component, cellular signaling processes in which protein components may be involved, associations with human disease, specific protein motifs, and the known biological roles of other interacting partners of the identified protein. In the experimental evaluation, we include parameters, such as the availability of research materials for the functional study of the identified protein component; such as full-length cDNA clones, antibodies, and commercially available knock-out embryonic stem (ES) cells. Each scoring parameter has been assigned an arbitrary number of points according to perceived relative importance. On the basis of this scoring system, we prioritized each of the protein components in terms of the likelihood of their importance for coactivator complex networking in nuclear receptor-dependent gene transcription.
Introduction
Steroid hormones play important roles in regulating cellular homeostasis and reproductive tissue function through their ligand-dependent interactions with specific nuclear receptors [Anderson, 2004; Lydon et al., 1996]. Coregulators generally mediate nuclear receptor-dependent gene transcription in a ligand-dependent manner [Dong et al., 1997; Halachmi et al., 1994; Hong et al., 1997; Onate et al., 1995; Wang et al., 1998]. A large number of nuclear receptor coactivators have been identified using various methods such as immunoprecipitation and yeast two hybrid analysis [McKenna and O'Malley, 2002]. Currently, ~170 different nuclear receptor coregulators are known and listed on the Nuclear Receptor Signaling Atlas (NURSA) website (www.nursa.org). Nuclear receptor coactivators, such as SRC-1 [Wu et al., 2002], SRC-3 [Wu et al., 2002], CARM1 [Xu et al., 2004], and ASC-2 [Goo et al., 2003] have been known to exist as multi-subunit complexes and this has been substantiated by our additional recent results [Jung et al., 2005]. It is our opinion that virtually all nuclear receptor coactivators function as multi-subunit complexes to dynamically modulate gene transcription.
Different coactivator complexes appear to be sequentially recruited to target promoter regions in a hormone-dependent manner to activate nuclear receptor-dependent gene transcription (www.nursa.org), [Shang et al., 2000; Wang et al., 2005]. These data strongly suggest that this cross-talk between coactivator complexes plays an important role in nuclear receptor-dependent gene transcription. To investigate the coregulator network involved in nuclear receptor-dependent gene transcription, we purified the coactivator complexes listed on the NURSA web site using co-immunoprecipitation, SDS-PAGE separation of the components, and identification of the associated bands by mass spectrometry [Jung et al., 2005].
The rapidly increasing numbers of protein components identified within the purified coactivator complexes are impossible to consider as a whole and require some quantitative approach to determine their relative value for consideration by scientists in the nuclear receptor field. To accomplish this task, we devised a new scoring system to prioritize coactivator complex components. In this way, each protein can be assigned a score that reflects its prospective and investigative value.
Results
Identification of protein components associated with purified coactivator complexes using mass spectrometry
On the basis of coregulators listed on the NURSA web site (www.nursa.org), we have purified an initial series of twelve different coactivator complexes from HeLa S3 cells [Jung et al., 2005]. The purified coactivator complexes are those associated with the SRC-1, SRC-2, SRC-3, CBP, p300, CAPER, E6-AP, ASC-1, CoREST, CRSP3, CRSP2, and CDK7 coactivators. Using mass spectrometry analysis, we identified 153 protein components associated with these coactivator complexes in vivo (Supplementary File 1).
A coactivator component scoring method
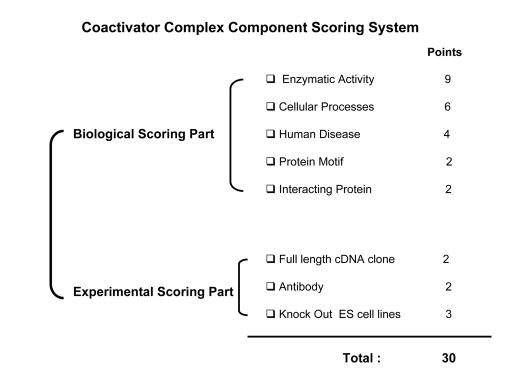
Our coactivator associated component scoring system consists of two parts; one is a biological score and the other is an experimental score (Table 1). Certain components have an intrinsic enzymatic activity and may contain specific protein motifs within their structure. By using these properties of the coactivator associated protein, we can suggest whether they are likely to be involved in specific cellular processes in vivo. Genetic mutations in transcriptional proteins are frequently associated with specific human diseases (such as cancer) due to their importance in cellular processes (Supplementary File 1 and Supplementary File 2). Consequently, analysis of the biological aspects of each protein component is an important step to understand and evaluate its potential physiological role in the regulation of the nuclear receptor mediated gene expression.
Table 1. The nuclear receptor coactivator scoring system.
Our coactivator associated component scoring system consists of two parts; one is a biological score and the other is an experimental score. Each scoring part consists of several subtitles with specific points assigned for the scoring.
Even though each coactivator associated component may have interesting biological properties, if experimental materials, such as a full-length cDNA clone, a commercial antibody against the component, the knockout embryonic stem cells, are not available, it would not be easy to study the function of this component in vivo. Thus, both prospective biological functions and available experimental materials need to be considered before one chooses a protein component for follow-up functional study.
Biological scoring system
Intrinsic enzymatic activity
Intrinsic enzymatic activity of each component is a very important clue to predict the functional role of a given protein component in gene transcription. Using the Human Genome Resources (HGR) database system (www.ncbi.nlm.nih.gov/projects/genome/guide/human), we can assess which enzymatic activity is associated with a given protein component. For example, the HGRD system reveals that both ATP binding activity and damaged DNA binding activity are associated with the MSH2 protein. Consequently, we considered that the CBP complex could be involved in cellular processes requiring both ATP and damaged DNA binding activities since MSH2 was identified as a component of the CBP complex. All enzymatic activities associated with each coactivator associated protein are listed in Supplementary File 1. To determine the priority of each protein component, we assigned nine of thirty points to a protein component containing intrinsic enzymatic activity (all points in our scoring system sum to thirty points ; Table 1 and Supplementary File 1).
On the basis of intrinsic enzymatic activities associated with a protein component, we can determine which biological activity might be associated with a given coactivator complex (Figure 1). As shown in Figure 1, ATP binding (22% of 153 protein components), metal ion binding (12% of 153 protein components), and RNA binding (11% of 153 protein components) activities are most frequently associated with coactivator complexes.
Figure 1. Enzymatic activity associated with protein components of coactivator complexes.
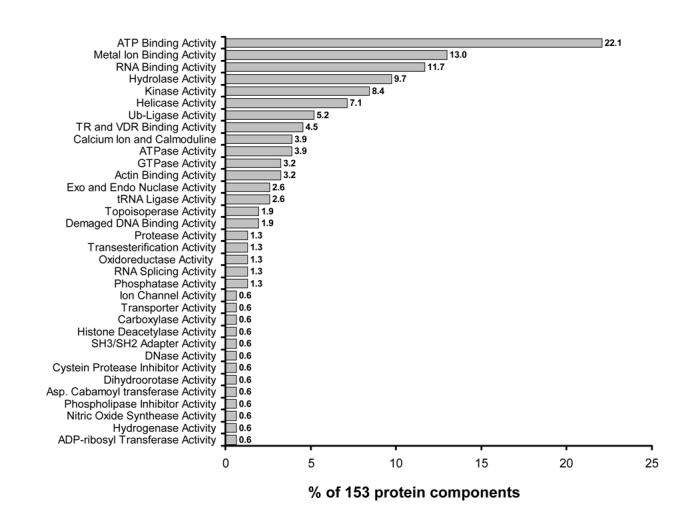
The graphic data reveals the percentage of all (153) protein components identified from 12 coactivator complexes that have a particular enzymatic activity.
Cellular regulation and metabolic processes
Cells dynamically modulate their cellular processes in response to external environmental stimuli. Nuclear receptors and coactivators are required for these adaptation responses. By using the HGRD system (www.ncbi.nlm.nih.gov/projects/genome/guide/human), we can determine which cellular processes are associated with a coactivator associated protein component. For example, the HGRD system revealed that MSH2 is functionally involved in both mismatch repair and in negative regulation of the cell cycle. Thus, one might predict that the CBP complex containing MSH2 could be involved in mismatch repair in cells or mismatch repair activity of MSH2 could be involved in transcription. All cellular processes associated with protein components of coactivators are listed in Supplementary File 1. We assigned six of thirty points to a protein component if it was involved in an identifiable cellular process (Table 1 and Supplementary File 1).
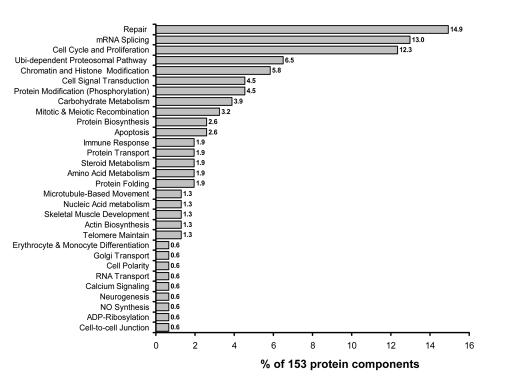
On the basis of the cellular process scores, we collated the main cellular processes associated with our published coactivator complexes (Figure 2). In addition to transcriptional regulation, coactivator complexes may be involved in repair (14% of 154 protein components), mRNA splicing (12% of 154 protein components), cell cycle and cellular proliferation (12% of 154 protein components), cellular metabolism (10.3% of 154 protein components), ubiquitin-mediated proteosomal processes (6% of 154 protein components), and chromatin and histone modifications involved in transcription (6% of 154 protein components).
Figure 2. Physiological processes associated with protein components of coactivator complexes.
The graphic data reveals the percentage of all (153) protein components identified from 12 coactivator complexes that are involved in specific cellular and metabolic processes in vivo.
Human disease
History tells us that human diseases can be closely correlated with genetic mutations in certain coactivator complex components. Therefore, we suggest that human disease related mutations in coactivator complex components are important clues to understanding their physiological roles in cellular processes in vivo. For example, to determine whether genetic mutations in protein components are associated with human cancers, we utilized the Human Genome Mutant Database (HGMD) (http://www.hgmd.cf.ac.uk/hgmd0.html). The HGMD revealed that nucleotide substitutions (157 cases), small deletions (74 cases), and small insertions (31 cases) within the MSH2 gene are associated with colorectal cancer development. A listing of disease related coactivator associated components is provided in Supplementary File 1. We assigned four of thirty points to any coactivator associated protein component that was associated with a human disease (Table 1 and Supplementary File 1).
Protein motif
Since many protein components contain unique protein domains, these motifs can be of value in assigning specific functions to a coactivator associated protein component. Using a Simple Modular Architecture Research Tool (SMRT) database system (http://smart.embl-heidelberg.de), we can predict the protein motif inherent to a specific protein component. For example, the SMRT system shows that MUTSd and MUTSac domains are present in the MSH2 protein. The MUTSd domain is a DNA-binding domain of the DNA mismatch repair MUTS family. MUTSas also is documented within the ATPase domain of DNA mismatch repair MUTS family. On the basis of such protein motif information, we predict that MSH2 is involved in DNA repair processes utilizing ATPase activity. The protein motifs present in coactivator components are listed in Supplementary File 1. We assigned two of thirty points to coactivator associated protein components when they contained specific recognizable protein motifs (Table 1 and Supplementary File 1).
Interacting proteins
To precisely modulate nuclear receptor-dependent gene transcription, coactivator complexes also must physically interact with other factors containing different cellular functions. Using the Biomolecular Interaction Network Database (BIND; http://bind.ca/), we can determine if a given associated protein interacts with other functional proteins. On the basis of such protein-protein interaction information, we can predict an arbitrary molecular mechanism by which a protein within a coactivator complex might act to regulate gene transcription in vivo. We assigned two of thirty points to a coactivator complex protein component if it contained recognizable physical interacting partners (Table 1 and Supplementary File 1).
Experimental scoring system
Availability of a full cDNA clone
Even though a coactivator complex protein component has interesting biological aspects, functional analysis of the protein component is made more difficult if a full length cDNA clone is not already available. Most of the interesting protein components associated with coactivator complexes are over 80 kDa in size. Construction of full length cDNAs for these huge proteins can be a rate limiting step in the functional study of such a protein. To accelerate the functional analysis, we determined availability of the full-length cDNA clone using the Open Biosystem Company Web Site (http://www.openbiosystems.com/). This database system lists availability of a full-length cDNA clone or partial cDNA clone. We assigned two of thirty points to a protein component if a full-length cDNA clone exists (Table 1 and Supplementary File 1).
Antibody availability
Antibodies are useful tools for the functional analysis of protein components. Using an antibody, one can obtain information on protein stability, post-translational modifications, cellular localization under various conditions, and associated proteins using immunoprecipitation. Preparation of an antibody against a specific protein component can be a time consuming and expensive undertaking. Using the Biocompare web site (http://www.biocompare.com/jump/2045/Antibodies.html), we can determine the availability of a commercial antibody. We assigned two of thirty points to a coactivator associated component if a commercial antibody exists (Table 1 and Supplementary File 1).
Commercially available knockout ES cells
After gathering existing information on a protein component from in vitro cellular experiments, we usually assess the physiological role of this protein in vivo. To address this question, we need to generate knockout mice by using gene targeted ES cells. However, the construction of a gene targeting vector and the screening of targeted ES cells can be expensive and labor intensive experiments. BayGenomics Company has inactivated thousands of genes in mouse embryonic stem cells for the purpose of generating knockout mice (by using a gene-trap vector method). Therefore the availability of knockout ES cells for a given protein can be determined using their database (http://baygenomics.ucsf.edu/cgi-bin/BayDbAccess.py). If a genetically deleted ES cell line is available, one can more quickly produce knockout mice to determine the in vivo physiological function of a coactivator associated protein. We assigned three of thirty points to the protein component if commercially available knockout ES cells exist for the protein. (Table 1 and Supplementary File 1).
Discussion
On the basis of our evaluation system, we have assigned scores for each coactivator-associated protein component from 30 (the highest) to zero (the lowest). We would predict that by choosing a high-scoring protein component of a coactivator complex for functional study, one can more rapidly and precisely determine the importance of this protein component in regulation of nuclear receptor-dependent gene transcription. Using this approach, an investigator can prioritize the known coactivator proteins in line with his/her specific interest. Such an evaluation scoring system is likely to be crucial as the increasing numbers of associated proteins are identified for ~200 coactivators, perhaps numbering as many as 2,500 eventually.
Supplementary Material
References
- Anderson E. Cellular homeostasis and the breast. Maturitas. 2004;48 Suppl 1:S13–7. doi: 10.1016/j.maturitas.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Dong H., O'Brien R. J., Fung E. T., Lanahan A. A., Worley P. F., Huganir R. L. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–84. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- Goo Y. H., Sohn Y. C., Kim D. H., Kim S. W., Kang M. J., Jung D. J., Kwak E., Barlev N. A., Berger S. L., Chow V. T., Roeder R. G., Azorsa D. O., Meltzer P. S., Suh P. G., Song E. J., Lee K. J., Lee Y. C., Lee J. W. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–9. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halachmi S., Marden E., Martin G., MacKay H., Abbondanza C., Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–8. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- Hong H., Kohli K., Garabedian M. J., Stallcup M. R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–44. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S. Y., Malovannaya A., Wei J., O'Malley B. W., Qin J. Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes. Mol Endocrinol. 2005;19:2451–65. doi: 10.1210/me.2004-0476. [DOI] [PubMed] [Google Scholar]
- Lydon J. P., DeMayo F. J., Conneely O. M., O'Malley B. W. Reproductive phenotpes of the progesterone receptor null mutant mouse. J Steroid Biochem Mol Biol. 1996;56:67–77. doi: 10.1016/0960-0760(95)00254-5. [DOI] [PubMed] [Google Scholar]
- McKenna N. J., O'Malley B. W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–74. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Onate S. A., Tsai S. Y., Tsai M. J., O'Malley B. W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–7. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu X., DiRenzo J., Lazar M. A., Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Wang Q., Carroll J. S., Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–42. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Stafford J. M., Granner D. K. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. J Biol Chem. 1998;273:30847–50. doi: 10.1074/jbc.273.47.30847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. C., Qin J., Hashimoto Y., Wong J., Xu J., Tsai S. Y., Tsai M. J., O'Malley B. W. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) Coactivator activity by I κ B kinase. Mol Cell Biol. 2002;22:3549–61. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Cho H., Kadam S., Banayo E. M., Anderson S., Yates J. R., 3rd, Emerson B. M., Evans R. M. A methylation-mediator complex in hormone signaling. Genes Dev. 2004;18:144–56. doi: 10.1101/gad.1141704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.