Short abstract
Screening for conditional mutations that increase Drosophila life span has identified genes implicated in membrane transport, phospholipid metabolism and signaling, and actin cytoskeleton organization.
Abstract
Background
A P-type transposable element called PdL has been engineered with a doxycycline-inducible promoter directed out through the 3' end of the element. Insertion of PdL near the 5' end of a gene often yields doxycycline-dependent overexpression of that gene and a mutant phenotype. This functional genomics strategy allows for efficient screening of large numbers of genes for overexpression phenotypes.
Results
PdL was mobilized to around 10,000 new locations in the Drosophila melanogaster genome and used to search for genes that would extend life span when overexpressed. Six lines were identified in which there was a 5-17% increase in life span in the presence of doxyxcycline. The mutations were molecularly characterized and in each case a gene was found to be overexpressed using northern blots. Two genes did not have previously known phenotypes and are implicated in membrane transport: VhaSFD encodes a regulatory subunit of the vacuolar ATPase proton pump (H+-ATPase), whereas Sugar baby (Sug) is related to a maltose permease from Bacillus. Three PdL mutations identified previously characterized genes: filamin encodes the homolog of an actin-polymerizing protein that interacts with presenilins. four wheel drive (fwd) encodes a phosphatidylinositol-4-kinase (PI 4-kinase) and CTP:phosphocholine cytidylyltransferase-l (Cctl) encodes the rate-limiting enzyme in phosphatidylcholine synthesis. Finally, an apparently novel gene (Red herring, Rdh) was found in the first intron of the encore gene.
Conclusions
Screening for conditional mutations that increase Drosophila life span has identified genes implicated in membrane transport, phospholipid metabolism and signaling, and actin cytoskeleton organization.
Background
Drosophila melanogaster has been a leading model for the study of aging for over 80 years [1,2,3,4,5]. The intensive use of Drosophila as a model for developmental biology has produced a wealth of genetic and molecular biological tools that are readily adapted to the study of aging. Aging is associated with characteristic changes at the physiological and molecular level, however organismal life span is still the best measure of aging rate. The most successful studies of aging generally involve manipulations that increase life span. Experimental alterations of environment or genetic makeup that cause decreased life span might create novel diseases or pathologies that do not usually limit life span in a normal individual. In contrast, a manipulation that increases life span is thought to be more likely to identify mechanisms that normally limit the life span of the organism.
Laboratory selection of Drosophila populations for late-life reproduction results in extended life span [6,7,8]. The genetic selection experiments demonstrate that life span has a large genetic component and is highly plastic. Increased life span was generally correlated with increased stress resistance including increased oxidative stress resistance [9,10]. An important question in the study of aging in invertebrates is whether life span can be increased without a trade off with metabolic activity. Certain manipulations that decrease metabolic activity cause increased life span, such as lower culture temperature [11]. In the genetic selection experiments, life span is increased without a reduction in metabolic activity [12,13].
Single gene mutations have been identified that increase Drosophila life span. These mutations identify negative regulators of life span, as mutations expected to decrease activity of the gene lead to increased life span. mth encodes a protein related to G-protein coupled receptors, and appears to negatively regulate life span, stress resistance and body size [14]. Indy is related to a mammalian cotransporter involved in membrane transport of Krebs cycle intermediates, leading to the suggestion that Indy mutations might increase life span by decreasing the availability of nutrients [15].
Certain mutant alleles of either the Inr or the chico genes can increase Drosophila life span [16,17]. Inr is homologous to the mammalian insulin receptor and insulin-like growth factor receptor, while chico is homologous to the mammalian insulin receptor substrate. The data indicate that an insulin-like signaling pathway negatively regulates life span, as had previously been found for the nematode C. elegans [18]. This pathway for life span regulation may be even more highly conserved during evolution, as there are many similarities with life span regulation in mammals and in yeast. The increased life span regulated by the insulin-like signaling pathway in C. elegans is associated with increased resistance to stress, including oxidative stress [18,19,20,21]. This has led to suggestions that increased oxidative stress resistance and decreased amounts of oxidative damage may be the mechanism by which mutation of the insulin-like signaling pathway causes increased life span. Consistent with this idea, life-span extension in C. elegans is associated with increased expression of the gene encoding the mitochondrial antioxidant enzyme manganese-containing superoxide dismsutase (MnSOD) [22]. In Drosophila, mutations of chico that extend life span increase total SOD enzyme activity; however, resistance to oxidative stress was not detectably increased, at least not as assayed by paraquat resistance [16]. Therefore it remains to be determined if mutation of the insulin-like pathway in Drosophila increases life span by increasing SOD activity or by some other mechanism(s).
Increased life span caused by overexpression of a gene by definition identifies a positive regulator of life span. Several genes involved in oxidative stress defense have been found to increase life span when overexpressed in transgenic flies. These include genes for both forms of SOD, the Cu/ZnSOD found in the cytoplasm and outer mitochondrial space, and MnSOD found in the inner mitochondrial space [23,24,25]. These enzymes convert the most common oxygen radical produced by the mitochondria, superoxide, into hydrogen peroxide and water. Abundant catalase enzyme then converts the hydrogen peroxide into molecular oxygen and water. Overexpression of Cu/ZnSOD or MnSOD increases life span up to about 40%, and the amount of life span increase is found to be proportional to the amount of enzyme increase in each case. The life span increase caused by SOD overexpression is not associated with a detectable tradeoff in metabolic activity, as oxygen consumption was normal throughout the extended life span of the flies [24]. Overexpression of the antioxidant enzyme peptide methionine sulfoxide reductase is also associated with increased life span [26]. Finally, several genes implicated in repair and stress responses have been shown to be able to extend Drosophila life span when the flies are cultured at elevated temperatures [27,28,29,30].
Testing specific genes by overexpression in transgenic flies is a candidate gene approach, meaning that the investigator must make an educated guess as to which genes are worth assaying by this relatively labor-intensive method. Recently genetic methods have been developed that allow for the efficient overexpression of random genes in Drosophila. P type transposable elements have been engineered to have transcriptional promoters directed out through the end of the element [31,32]. These elements will often insert upstream of gene coding regions, causing overexpression of the gene and mutant phenotypes. The PdL P element contains an outwardly directed tet-on promoter [33,34]. The transcription of this promoter is activated upon feeding the fly doxycycline, leading to conditional gene overexpression and conditional mutations. Conditional gene overexpression mutations have several potential advantages for study of aging and life span. By waiting until flies are young adults to activate gene overexpression, effects can be identified that are specific to adult aging. The system also provides powerful controls for genetic background effects, because control (no DOX) and over-expressing (+DOX) flies have identical genetic backgrounds and therefore any differences in life span must be due to DOX administration and gene overexpression. The PdL system allows the investigator to search for genes that can increase life span without having to guess ahead of time which genes these might be. Therefore the approach has the potential to identify genes and pathways not previously known or suspected to be involved in life span regulation.
Results
In the rtTA transgenic construct the powerful and tissue-general actin5C promoter drives expression of the rtTA transcriptional transactivator [33]. When flies are fed doxycycline, rtTA binds to target sequences in the synthetic tet-on promoter in the PdL P element. This outwardly-directed promoter often causes overexpression of genes near the PdL insertion site and yields various mutant phenotypes [34]. PdL is 8.4 kilobases (kb) long and contains the P element inverted repeats, the mini-white+ transformation marker gene, and the 500 base-pair (bp) tet-on promoter directed out through the 3' end of the element. The tet-on promoter contains no open reading frame (ORF) or translation initiation sequences. Therefore PdL must insert upstream of an endogenous translation intiation site (ATG) to cause overexpression of a protein. The transcripts initiated within PdL will therefore contain 280 bp of 5' untranslated sequence derived from PdL itself.
To search for genes that might extend life span, new insertions of PdL were created on the second and third chromosomes by appropriate crosses to a strain expressing P element transposase (Figure 1). Approximately 10,000 males bearing a new PdL insert and an rtTA transcriptional transactivator insertion were generated. The males were generated in eight staggered groups, each of around 1,200 individuals. In an attempt to increase the variety of mutations identified, two different rtTA insertion strains were used. The first six cohorts of males contained an insertion of the rtTA construct called rtTA(3)E2 that is associated with moderate levels of target-gene expression and low levels of leaky expression in the absence of DOX [33]. The last two cohorts of males contained an rtTA insertion called rtTA(3)M1 which is associated with a high level of target-gene expression and greater leaky expression. To obtain adult flies of defined age, the second cross was cultured at 25°C in urine specimen bottles (Figure 1). Prior to eclosion of the majority of pupae, bottles were cleared of adults and flies were allowed to emerge over the next 48 hours. The majority of the males will have mated during this time. The males only were then removed and were designated 1 day old, and were maintained at 25°C at 40 per vial in culture vials with food supplemented with 250 μg/ml DOX, and passaged to new vials every 48 hours. The expectation was that some of these flies might live longer due to the overexpression of a beneficial gene. For the first four cohorts, at approximately 30% survival of the cohort, each of the males was individually mated with four third-chromosome balancer stock virgin females. This was so that a stable line could be recovered later if that fly was found to be one of the longest lived. Only about 21% of these males were found to be fertile, meaning that there was selection for males that maintained fertility during aging. After 4 days, the fertile males were removed from the crosses, placed in individual numbered vials, and passaged every two days until they were all dead. For the rest of the cohorts the males were mated at approximately 65% survival of the cohort, which increased the frequency of fertile males to about 32%. Stable lines were recovered for the approximately 1.4% longest-lived flies from each cohort. These 144 lines should therefore be enriched for ones in which PdL is causing overexpression of a gene with a benefit for life span. In addition, lines in which PdL causes overexpression of a gene with large negative effects on life span should have been eliminated by this step.
Figure 1.
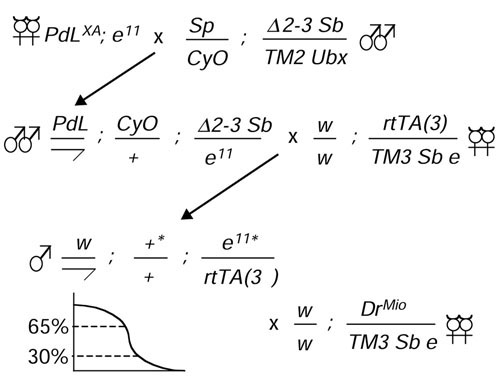
Genetic crossing strategy. The starting PdL insertion on the X chromosome PdL(X)A was mobilized using the Δ2-3 transposase source to generate approximately 10,000 new PdL insertions on the second and third chromosomes. Chromosomes potentially bearing new PdL insertions are indicated by asterisks and were recovered in males that also contained an rtTA transactivator construct. The males were allowed to age in the presence of DOX in staggered cohorts of approximately 1,200 each and were individually mated to balancer virgins. Stable PdL insertion lines were recovered for the longest-lived (approximately 1.4% of the males in each cohort). A total of 110 lines were identified using rtTA insertion rtTA(3)E2, and 34 lines with insertion rtTA(3)M1.
Each of the 144 new PdL insertion lines was crossed to the appropriate rtTA chromosome strain, and males containing both constructs were assayed in large cohorts (around 400 flies) for doxycycline-dependent effects on life span (Figure 2). The mean life spans of the different strains in the absence of DOX varied greatly, from 40 to 84 days. Both increases and decreases were observed in the presence of DOX. For the 110 lines tested with rtTA(3)E2 there was an average 1.8% increase in the presence of DOX (Figure 2a), and for the 34 lines tested with rtTA(3)M1 an average 3.0% increase in the presence of DOX (Figure 2b), consistent with previous results that DOX itself has little to no effect on life span. However, several lines were identified where there was a significantly greater than average increase in life span in the presence of DOX. To confirm these results, these lines were assayed one or more additional times (Table 1). As a control, Oregon-R wild-type flies were repeatedly crossed to the rtTA strains and assayed for life span in parallel. The Oregon-R controls exhibited changes in life span from -5.14% to +4.71%, with an average change of +0.45%. In contrast, six PdL lines were found to yield significant and reproducible increases in life span in the presence of DOX, ranging from 5 to 17%.
Figure 2.
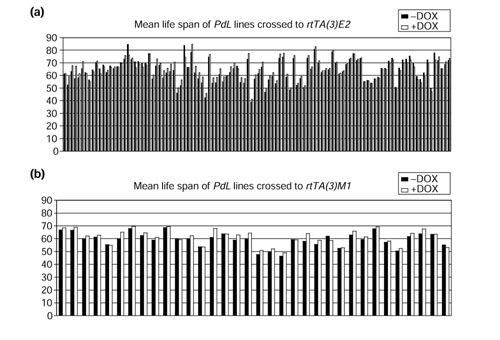
Life-span assay of new PdL insertion lines. The new PdL insertion lines were crossed to the appropriate rtTA insertion line, and male progeny were assayed for life span in the presence (+DOX) and absence of DOX (-DOX). Each line is represented by a pair of bars, black bars for -DOX and white bars for +DOX. (a) Lines crossed to rtTA(3)E2. The average life span -DOX was 64.7 days and the average +DOX was 65.9 days, for an average increase of 1.8%. (b) Lines crossed to rtTA(3)M1. The average life span -DOX was 59.4 days and the average +DOX was 61.2 days for an average increase of 3.0%.
Table 1.
PdL lines
| Life-span phenotype* | |||||
| Line name | Gene (enzyme) | -DOX | +DOX | % Change (p) | Date of assay |
| PdL lines crossed to rtTA(3)E2 | |||||
| PdL(3)3E36 | Red herring/encore | 60.42 ± 0.920 | 64.34 ± 0.864 | +6.50 (0.0021) | 7 October 1999 |
| 55.20 ± 0.729 | 64.55 ± 0.757 | +16.95 (<0.0001) | 6 July 1999 | ||
| 66.91 ± 0.980 | 75.59 ± 0.910 | +13.00 (<0.0001) | 14 February 2001 | ||
| 56.56 ± 0.709 | 62.44 ± 0.854 | +10.38 (<0.0001) | 7 August 2001 | ||
| PdL(2)4G14 | VhaSFD | 76.05 ± 1.039 | 82.71 ± 1.088 | +8.75 (<0.0001) | 14 November 2000 |
| 69.28 ± 1.077 | 76.24 ± 1.538 | +10.6 (0.0002) | 18 July 2000 | ||
| 73.79 ± 1.410 | 77.67 ± 1.251 | +5.27 (0.0404) | 6 March 2001 | ||
| 55.40 ± 0.810 | 59.34 ± 0.887 | +7.11 (0.0011) | 21 August 2001 | ||
| PdL(3)2C33 | Sugar baby CG7334 | 73.91 ± 1.501 | 80.32 ± 1.392 | +8.67 (0.0019) | 15 November 2000 |
| 71.75 ± 1.105 | 77.74 ± 1.169 | +8.35 (0.0002) | 3 July 2000 | ||
| 68.99 ± 1.100 | 72.22 ± 1.034 | +4.68 (0.0329) | 6 March 2001 | ||
| 56.77 ± 0.950 | 57.68 ± 0.617 | +1.59 (0.4241) | 7 August 2001 | ||
| Oregon-R | 62.70 ± 0.963 | 63.81 ± 1.299 | +1.76 (0.4954) | 12 August 2001 | |
| Oregon-R | 68.20 ± 0.978 | 71.41 ± 1.255 | +4.71 (0.0438) | 28 September 2000 | |
| Oregon-R | 71.27 ± 1.407 | 72.46 ± 1.247 | +1.66 (0.5278) | 11 July 2000 | |
| Oregon-R | 76.14 ± 1.498 | 74.45 ± 1.492 | -2.23 (0.4236) | 16 May 2000 | |
| Oregon-R | 68.84 ± 0.903 | 65.30 ± 1.175 | -5.14 (0.0176) | 7 October 1999 | |
| PdL lines crossed to rtTA(3)M1 | |||||
| PdL(3)8S64 | filamin | 58.87 ± 1.295 | 64.11 ± 1.066 | +8.76 (0.0020) | 13 December 2000 |
| 56.00 ± 1.202 | 60.68 ± 1.103 | +8.37 (0.0044) | 3 May 2001 | ||
| PdL(3)8S25 | fwd | 58.90 ± 0.868 | 62.99 ± 0.912 | +6.93 (0.0013) | 22 December 2000 |
| (1-phosphatidylinositol 4-kinase) | 56.25 ± 0.940 | 61.31 ± 1.028 | +9.00 (0.0003) | 3 May 2001 | |
| PdL(3)8R128 | Cct1 | 59.68 ± 1.192 | 64.34 ± 1.052 | +7.80 (0.0035) | 27 February 2001 |
| (cholinephosphate cytidylyltransferase) | 56.54 ± 0.789 | 59.85 ± 0.852 | +5.93 (0.0041) | 10 July 2001 | |
| Oregon-R | 60.44 ± 1.296 | 61.61 ± 1.350 | -1.96 (0.5337) | 13 July 2001 | |
| Oregon-R | 65.79 ± 1.009 | 68.68 ± 0.819 | +4.39 (0.0266) | 2 May 2001 | |
| Oregon-R | 65.45 ± 1.304 | 66.93 ± 1.091 | +2.20 (0.3824) | 30 November 2000 | |
*Mean life span ± SEM, p-value for unpaired, two-sided t-test in parentheses.
Line PdL(3)3E36 was associated with the largest increase in life span (average 12%) (Table 1). The PdL insert in this line was located in the large first intron of the encore gene [35], with the promoter oriented in the sense direction (Figure 3a). PdL caused the expression of a pair of approximately 1.4 kb transcripts that contain a 44 amino acid ORF identified by the Drosophila genome project as CG14975, as determined by northern blot with a CG14975 probe. These transcripts were not detected in the absence of DOX in the adult males and this putative gene was named Red herring (Rdh). Northern analysis using a probe from encore exon 3 indicated that levels of the normal sized encore transcript were not detectably altered by DOX, as indicated by the arrowhead (Figure 3a). However, DOX appeared to induce expression of both larger and smaller RNAs containing encore coding-region sequences, resulting in a smear of hybridization in the upper part of the lane, as indicated by the asterisk. Representative survival curves are presented (Figure 3a).
Figure 3.
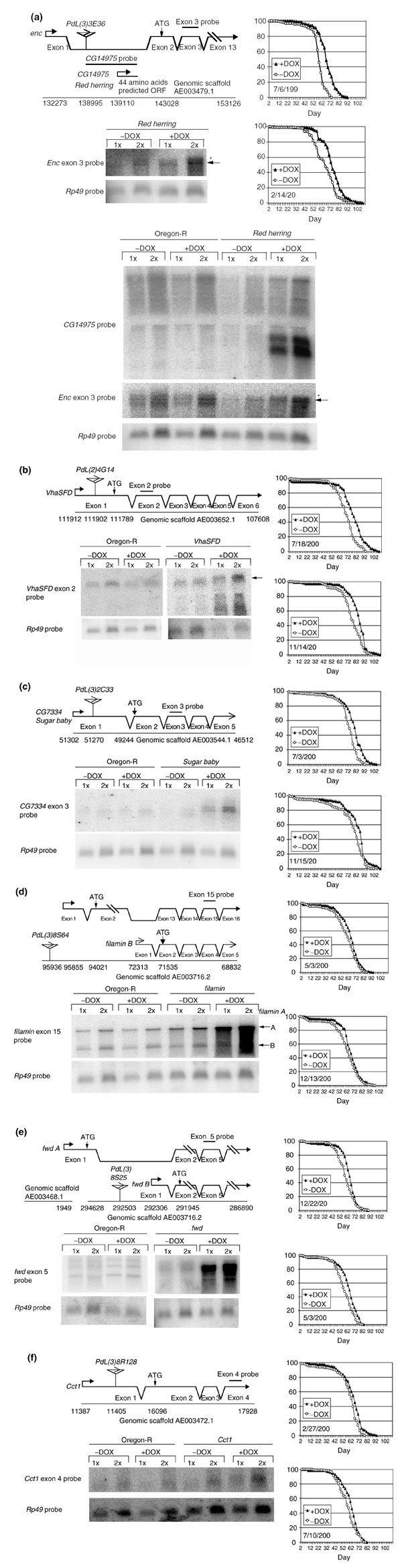
Characterization of new PdL mutations. (a) PdL(3)3E36 (Red herring and encore (enc)); (b) PdL(2)4G14 (VhaSFD); (c) PdL(3)2C33 (Sugar baby); (d) PdL(3)8S64 (filamin); (e) PdL(3)8S25 (four wheel drive (fwd)); (f) PdL(3)8R128 (CTP:phosphocholine cytidylyltransferase-1 (Cct1)). The molecular organization of each mutated gene is diagrammed. The PdL insertion is indicated by a triangle, with an arrow indicating the direction of transcription of the PdL tet-on promoter. Transcript start sites and ATG translation start codons are marked by arrows. The location of PdL insertions, transcription start sites, ATG translation start sites, and transcript termination sites are indicated by genomic scaffold numbers. mRNA was isolated from whole adult male flies cultured ±DOX and analyzed by northern blot. Hybridization with ribosomal protein gene Rp49 probe was used as a loading control. The locations of gene-specific DNA probes used for northern analyses are indicated. Transcript sizes were estimated using RNA markers run in adjacent lanes (data not shown). Two representative survival curves are presented for each line.
Line PdL(2)4G14 contains an insert at the 5' end of the VhaSFD gene, encoding the vacuolar proton pump ATPase (H+-ATPase) SFD subunit [36]. DOX caused an approximately fourfold overexpression of the VhaSFD transcript and an average 8% increase in life span (Figure 3b). Representative survival curves for the PdL(2)4G14 line are presented (Figure 3b). Line PdL(3)2C33 contains an insert at the 5' end of a gene with homology to a maltose permease from Bacillus stearothermophilus [37], which was named Sugar baby (Figure 3c). DOX caused an approximately 8.5-fold overexpression of the Sugar baby transcript, and was associated with an average 6% increase in life span.
Three genes with previously characterized mutant phenotypes were also identified by PdL insertions. Line PdL(3)8S64 contained an insert at the 5' end of the filamin gene 'A' transcript (Figure 3d). Filamin is an actin-binding protein involved in cytokinesis and ring canal formation [38,39]. filamin A was overexpressed approximately fivefold in the presence of DOX and associated with an average 8.5% increase in life span. Expression of the filamin 'B' transcript was not detectably altered. Line PdL(3)8S25 contained an insert at the 5' end of the 'B' transcript of the four wheel drive (fwd) gene (Figure 3e). fwd encodes a 1-phosphatidylinositol 4-kinase (PI 4-kinase) that regulates actin organization and ring canal formation during germline cytokinesis [40]. fwd B was overexpressed approximately 10-fold and associated with an average 8% increase in life span. Finally, line PdL(3)8R128 had an insert at the 5' end of the CTP:phosphocholine cytidylyltransferase 1 (Cct1) gene (Figure 3f). Cct1 encodes a homolog of the rate-limiting enzyme in phosphatidylcholine biosynthesis [41]. Cct1 was overexpressed approximately 1.7-fold and was associated with an average 7% increase in life span.
Discussion
The genetic screen described here had several unusual aspects, and the potential to identify a novel set of genes involved in life-span regulation. First, the screen was designed to identify positive regulators of life span, that is, genes that will increase life span when overexpressed. Most genetic screens for life-span regulators in Drosophila and other organisms, such as C. elegans, have involved loss-of-function mutations and therefore have identified negative regulators of life span [14,15,42,43]. Second, mutants were identified on the basis of the phenotype of increased life span under normal conditions, as opposed to some surrogate phenotype expected to correlate with life span, such as increased stress resistance. Because the PdL mutations generated here were screened directly for extended life span under normal conditions, the screen had the potential to identify genes and pathways not previously suspected to have a role in aging and life-span regulation.
Results were reported recently for a screen for positive regulators of life span using a different P-element-based gene overexpression strategy [29]. In that study a heat-stress inducible system was used, and lines were screened for life span at the moderate heat stress condition of 30°C. Several genes were identified that increased survival when overexpressed, including genes involved in heat stress and oxidative stress responses. It is interesting to note that there is no overlap with the genes identified here with PdL at 25°C. This may be due to the small fraction of the genome that has been tested so far, or it may indicate that different mechanisms limit life span under normal and moderate heat-stress conditions.
It is difficult to estimate the number of genes that were tested in the PdL screen, but it is certain to have been only a tiny fraction of the genome. In previous experiments approximately 7% of PdL insertions were found to cause lethal and visible phenotypes when the PdL promoter was activated during larval and pupal development [34]. Seven percent is certainly a large underestimate of the frequency at which genes are overexpressed, as embryogenesis was not tested, and not all genes will be lethal or have an obvious visible phenotype when overexpressed. The original 10,000 PdL insertions generated here went through an initial enrichment step of unknown efficiency, to yield the 144 lines that were tested in large cohorts. Lines in which a gene was overexpressed that had a positive effect on life span should have been enriched in the final 144 lines, whereas ones with a significant negative effect should have been eliminated. The enrichment step appears to have been at least partially successful, as the largest negative effect on life span observed among the 144 lines tested was -7.5%, and PdL is known to be able to create mutations with negative effects of up to -30% [34]. The molecular characterization of the mutations suggests a low frequency of false positives. If the six PdL insertion strains were a random set, then PdL would be expected to sometimes be in the wrong orientation and/or downstream of the ATG translation start. However, all six lines were found to have PdL insertions at the 5' end of a gene and oriented in the sense direction, and in all six lines a transcript was overexpressed that was expected to contain a complete ORF.
The magnitude of life-span increases caused by the PdL mutations was generally small, ranging from 5 to 17%. This is in contrast to overexpression of SOD genes or certain single gene mutations such as mth, where life-span increases range from 20 to 50% [14,23,24,25]. There are at least two possible reasons for the relatively small life span increases observed here. First, as only a fraction of the genome has been tested, it may be that genes with larger effects exist but have not yet been identified. Second, the tet-on system is sometimes leaky, meaning that at certain genomic locations there can be significant expression of the tet-on promoter in the absence of DOX [33]. It is possible that in some of the lines the increase in life span between -DOX and +DOX is reduced because there is already some gene overexpression and life-span extension in the -DOX flies. Such a line might be expected to have an unusually high -DOX life span relative to the other lines, and it is notable that this appears to be the case for the lines bearing mutations in the VhaSFD and Sugar baby genes. When this difference is included in the calculations, the life span increases for VhaSFD and Sugar baby are approximately 20% and 17%, respectively.
The largest average increase in life span caused by DOX (12%) was obtained for the PdL(2)3E36 insertion in the large first intron of the encore gene. A pair of approximately 1.4 kb transcripts was expressed containing a 44 amino acid ORF with no detectable homologies. While there are a number of Drosophila genes encoding functional ORFs of this size, it is also possible that these transcripts do not encode a functional protein but rather function at the RNA level to affect life span. There are no cDNAs in the databases containing these intronic sequences, and we were unable to detect these transcripts in wild-type adult males using northern blots. The data suggest that these transcripts are not expressed under normal conditions, or are expressed at very low levels. Further experiments will be required to determine the mechanism of life-span extension in this line, and the elusive nature of the intronic gene suggested the name Red herring. The northern analyses also indicated induction of a small amount of a large transcript containing the encore coding region. Therefore it is also possible that the life span extension in this line is caused by an increase in the expression of encore, encore is a member of a novel family of proteins with multiple functions during Drosophila oogenesis [44].
Two of the genes identified appear to be involved in membrane transport. The VhaSFD gene encodes the Drosophila homolog of the vacuolar H+-ATPase SFD subunit. The vacuolar ATPase is an ATP-driven proton pump found in the endomembranes of eukaryotic cells [36,45,46,47]. It is composed of a cytoplasmic ATPase domain called V1 that contains VhaSFD and other subunits, and a multisubunit membrane-channel component called V0. The vacuolar ATPase is involved in the acidification and energization of organelles such as Golgi-derived vesicles, clathrin-coated vesicles, synaptic vesicles and lysosomes. The proton pumping is required for processes such as protein trafficking, receptor-mediated endocytosis, neurotransmitter release and intracellular pH regulation and waste management. The free membrane V0 domain has also been implicated in membrane fusion events. In higher organisms the vacuolar H+-ATPase is also found in the plasma membrane of specialized epithelial cells where it functions in bulk transport. In Drosophila this is the Malpighian (or renal) tubule. The VhaSFD subunit is required for the ATPase activity and assembly of the vacuolar H+-ATPase and may function as a dissociable regulatory subunit. Therefore, overexpression of VhaSFD may affect life span by increasing activity of the vacuolar H+-ATPase.
The Sugar baby gene is related to a maltose permease from Bacillus stearothermophilus [37], suggesting that it may function in transport of maltose or other sugars across a membrane. Recently, loss-of-function mutations in another putative membrane transporter, Indy, have been found to increase life span [15]. It is possible that overexpression of the putative transporter encoded by Sugar baby could increase life span by increasing uptake of a particular sugar and altering energy metabolism pathways.
Filamin is an actin-binding protein with a homolog in humans called actin-binding protein 280 (ABP280) [38,39]. Filamin binds to several cell-membrane proteins and intercellular ligands involved in signal transduction, and appears to act as a downstream effector in remodeling of the actin cytoskeleton. During Drosophila oogenesis, filamin localizes to the ring canals, which are membrane-associated actin cytoskeletal structures that create intercellular bridges between germline cells. Loss-of-function mutations of filamin disrupt the normal function of these actin cytoskeletal structures. Filamin has several additional interactions with potential relevance to aging studies. Presenilins are membrane proteins found in both humans and Drosophila, and mutations in human presenilins are associated with early onset familial Alzheimer's disease (FAD) [48]. Drosophila filamin binds to both Drosophila and human presenilins, and filamin interacts genetically with presenilin in Drosophila [49]. Dominant adult phenotypes produced in Drosophila by overexpression of presenilin can be suppressed by overexpression of filamin. Filamin also interacts specifically with Drosophila Toll and Tube proteins [50]. Toll and Tube are members of a signal transduction pathway that activates the Rel transcription factor Dorsal and is involved in developmental patterning and in the adult immune response. Several of the immune-response genes regulated by Rel transcription factors are dramatically induced during Drosophila aging (G.L., J. Carrick and J.T., unpublished data).
fwd encodes a phosphatidylinositol 4-kinase (PI 4-kinase) that converts phosphatidylinositol (PI) to phosphatidylinositol 4-phosphate (PIP) [40]. This is the first step in the synthesis of the key regulatory membrane phospholipid PIP2, which is generated from PIP by a PI 4,5-kinase. PIP2 interacts directly with a number of proteins, regulating both their sub-cellular localization and their activity. Data from yeast and mammalian systems implicates homologous PI 4-kinases and PIP2 in regulation of secretion and membrane vesicle trafficking [51,52]. In Drosophila, fwd gene function is required for the formation of ring canals and cytoplasmic bridges during cytokinesis in male meiosis. PIP2 is also the precursor to the important second messengers PIP3 and diacylglycerol. PIP3 is involved in the insulin-like signaling pathway that affects life span in Drosophila and C. elegans [53], and it will be of interest to determine if fwd overexpression is somehow affecting this pathway.
CTP:phosphocholine cytidylyltransferase (Cct) catalyzes the conversion of phosphocholine to CDP-choline and is the rate-limiting enzyme in the phosphatidylcholine biosynthetic pathway [41]. Phosphatidylcholine is a major membrane phospholipid and is a precursor to second messengers involved in signal transduction at the membrane, including the PIP, PIP2 and PIP3 discussed above. There are two genes encoding Cct in Drosophila (Cct1 and Cct2), and mutations in the Cct1 gene disrupt patterning during oogenesis (T. Gupta and T. Schupbach, personal communication). Overexpression of Cct1 might affect life span by increasing synthesis of phosphatidylcholine and altering membrane structure and/or phospholipid signaling pathways.
It is striking that most of the genes identified in this screen as positive regulators of Drosophila life span are implicated in functions at the membrane, including regulation of transport, phospholipid metabolism, signal transduction and actin cytoskeleton organization. It is tempting to speculate that some of these genes may be acting in the same pathway(s) for life-span regulation. For example, both filamin and fwd are involved in the function of ring canals, which are membrane-associated actin cytoskeletal structures that create intercellular bridges between germline cells during early mitotic divisions [38,39,40]. encore is also required for normal early mitotic division of the germline cells [44]. In C. elegans, germline cells have recently been shown to send signals that regulate life span of the adult organism [54,55,56], and it will be of interest to determine if any of the genes identified here increase life span through effects on the germline. Both fwd and Cct1 are involved in phospholipid metabolism, and phospholipid signaling pathways are implicated in life-span regulation in Drosophila and other organisms [53]. The conserved insulin-like life-span regulatory pathway includes a PI 3-kinase that converts PIP2 into PIP3. The enzymes encoded by fwd and Cct1 are both in the biosynthetic pathway for PIP2 and therefore might affect the insulin-like pathway by altering the availability of this substrate. Further experiments will be required to confirm the role of these genes in life-span regulation, and to determine their interactions with each other and in known or novel life-span regulatory pathways.
Materials and methods
Drosophila strains
Drosophila culture and life-span assays
Drosophila were cultured on standard agar/molasses/corn meal/yeast media [59]. To obtain adult flies of defined age, the indicated PdL lines and Oregon-R control strain were crossed to an rtTA stock and cultured at 25°C in urine specimen bottles. Prior to eclosion of the majority of pupae, bottles were cleared of adults and newly eclosed flies were allowed to emerge over the next 48 h. The majority of the males will have mated during this time. The males only were then removed and were designated 1 day old, and were maintained at 25°C at 40 per vial in culture vials with food. At 4 days of age the males were split into control and experimental groups of 200 males each, with experimentals (+DOX) placed on culture media supplemented with 250 μg/ml DOX. Dead flies were counted at each passage, and the number of vials was progressively reduced to maintain approximately 40 flies per vial. To calculate mean life spans for the experimental (+DOX) and control (-DOX) cohorts, each fly's life span was tabulated and their life spans were averaged and the standard error of the mean (SEM) calculated. Statistical significance of differences in mean life span was calculated for each experiment using unpaired two-sided t tests.
ANOVA
Two-way analysis of variance (ANOVA) was performed for each life-span dataset. The first dataset was the 110 lines crossed to rtTA(3)E2 (Figure 2a). Sources of variance were line, treatment (DOX), and line × treatment. Results are presented in Table 2. A P-value ≤ 0.05 was considered statistically significant. The significant line × treatment interaction for dataset 1 indicated that DOX affects life span only in certain lines. The significance and 95% confidence interval were calculated for each line with experiment-wise error rate for multiple comparisons reduced using the Bonferroni method (see supplemenetary table in Additional data files). DOX decreased life span in 6 lines and increased it in 34 lines. The second dataset was the 34 lines crossed to rtTA(3)M1 (Figure 2b). Results were line F = 57.168, p < 0.001; treatment F = 48.131, p < 0.001; line × treatment F = 1.898, p = 0.001. The significant line × treatment interaction again indicates that DOX affects life span only in certain lines. The significance and 95% confidence interval were calculated for each line, with experiment-wise error rate reduced using the Bonferroni method (see supplementary table in Additional data files). DOX decreased life span in one line, and increased it in seven lines.
Table 2.
ANOVA results for 110 lines crossed to rtTA(3)E2
| Source | Degrees of freedom | Mean square | F | p |
| Line | 109 | 30,407.071 | 134.854 | < 0.001 |
| Treatment | 1 | 19,763.341 | 87.650 | < 0.001 |
| Line × treatment | 109 | 720.007 | 3.193 | < 0.001 |
| Error | 40,129 | 225.481 |
Each line in which DOX increased life span was crossed to the appropriate rtTA strain and assayed one to three additional times. For six lines there was a reproducible increase in life span with DOX (Table 1). As an additional control, Oregon-R wild-type was repeatedly crossed to rtTA(3)E3 and to rtTA(3)M1 strains and assayed for life span in the presence or absence of DOX. These datasets were analyzed using two-way ANOVA with date of assay, treatment (DOX), and date × treatment as the sources of variance. Dataset 3 was the four independent assays of line PdL(3)3E36 (Red herring/encore) (Table 1). Results were date F = 100.690, p < 0.001; treatment F = 129.780, p < 0.001; and date × treatment F = 5.387, p = 0.001. Dataset 4 was the four independent assays of line PdL(2)4G14 (VhaSFD). Results were date F = 145.348, p < 0.001, treatment F = 43.294, p < 0.001, date × treatment F = 1.057, p = 0.366. Dataset 5 was the four independent assays of line PdL(3)2C33 (Sugar baby). Results were date F = 121.648, p ≤ 0.001; treatment F = 24.728, p ≤ 0.001; date × treatment F = 2.283, p = 0.077. Dataset 6 was the five independent assays of Oregon-R crossed to rtTA(3)E2 (Table 2). Results were date F = 28.171, p < 0.001, treatment F = 0.005, p = 0.944, date × treatment F = 2.313, p = 0.056. Dataset 7 was the two independent assays of line PdL(3)8S64 (filamin). Results were date F = 7.192, p = 0.007, treatment F = 17.871, p < 0.001, date × treatment F = 0.057, p = 0.812. Dataset 8 was the two independent assays of line PdL(3)8S25 (fwd). Results were date F = 5.319, p = 0.021, treatment F = 23.784, p < 0.001, date × treatment F = 0.271, p = 0.603. Dataset 9 was the two independent assays of line PdL(3)8R128 (Cct1). Results were date F = 15.405, p < 0.001, treatment F = 17.177, p < 0.001, date × treatment F = 0.455, p = 0.500. Dataset 10 was the three independent assays of Oregon-R crossed to rtTA(3)M1. Results were date F = 16.611, p < 0.001, treatment F = 3.777, p = 0.052, date × treatment F = 0.315, p = 0.730.
Southern analysis of PdL copy number
DNA was isolated from PdL lines and restriction digested with XbaI, HindIII, PstI and TaqI in separate reactions. DNA was transferred to a Southern blot and hybridized with a radiolabeled 172 bp fragment from the 3' P end of PdL. This probe fragment was generated by PCR amplification with primers located within the 3' P end, IRREV (ATGATGAAATAACATAAGGTGGTCCCG) and P3MCSREV (ATGAGTTAATTCAAACCCCACGGACAT).
Inverse PCR amplification of PdL flanking sequences
Protocols were as previously described [60,61]. Briefly, DNA equivalent to one fly was restriction digested with TaqI. The DNA was extracted with phenol/chloroform, precipitated with ethanol, resuspended and treated with T4 ligase overnight at 16°C. PCR amplification was performed using primers Pry1 (CCTTAGCATGTCCGTGGGGTTTGAAT) and IR (CGGGACCACCTTATGTTATTTCATCATG) located in the 3' P end. PCR protocol was as follows: step 1, 95°C for 5 min; step 2, 95°C for 30 sec; step 3, 51°C for 1 min; step 4, 72°C for 1 min; step 5, repeat steps 2-4 40 times; step 6, 72°C for 10 min. The PCR product was subcloned into the pCR2.1-TOPO cloning vector (Invitrogen). Dideoxy sequencing was carried out using the Sequenase version 2.0 DNA sequencing kit (US Biochemical) and the T7 and M13 reverse sequencing primers. Each inverse PCR product will contain 284 bp of 3' P-element sequences. The sizes of the products were: PdL(3)3E36 (Red herring/encore) 534 bp; PdL(2)4G14 (VhaSFD) 465 bp; PdL(3)2C33 (Sugar baby) 464 bp; PdL(3)8S64 (filamin) 362 bp; PdL(3)8S25 (fwd) 345 bp; PdL(3)8R128 (Cct1) 298 bp.
DNA sequence analyses
PdL-flanldng DNA sequences were used to query GenBank databases using BLASTN with default settings as provided at the National Center for Biotechnology Information (NCBI) website [62]. Genomic scaffold sequences and numbering and ORF designations are according to the NCBI databases and Drosophila genome sequence [63].
Northern blot analyses
RNA was isolated from adult Drosophila using the RNAqueous kit (Ambion), fractionated on 1.0% agarose gels and transferred to GeneScreen membranes (DuPont/NEN). 1x = 4 μg, and 2x = 8 μg, except for the analysis of Sugar baby where 1x = 8 μg. The DNA probe for exon 5 of the fwd gene was generated by PCR amplification from Drosophila genomic DNA using primers FWDFWD (TGCTTCCTCCATTTGGCGAAC) and FWDREV (ATCATCTGTGGCTCAGAGTCG). The probe for exon 2 of the VhaSFD gene was generated using primers VhaFWD (CCAGCTGATCCTTCAGGAACTGC) and VhaREV (ACCAGGACGATCAACTGGGCTTC). The probe for exon 15 of the filamin A gene was generated using primers AMINAFWD (GCCAATGTAGGCCTTCTTCAG) and FILAMINAREV (ATCCATGCCATCCACGTCAAG). The probe for exon 1 of the CG1049 gene was generated using primers CG1049EX1FWD (AGTTGTGTTTGTGTCCGACG) and CG1049EX1REV (ATAAACAGAGCAGAGCAGAGC). The probe for exon 4 of the CG1049 gene was generated using primers CG1049EX4FWD (TCTGTCCGATGAATTCATCGCC) and CG1049EX4REV (ATGATTCAGGTTCTCACGTCCG). The CG14975 ORF probe was generated using primers CG14975FWD (TCAGCCGAGAGATTCTAGAGAG) and CG14975REV (CATCGACCATTGTTCTCTCTCC). The 3E36 intergenic region probe was generated using primers 3E36-140120FWD (TTTCCATTTCCCTTCCACTGCC) and 3E36-1406038REV (TTACAGCTGCTCACTCACTCAC). The probe for exon 3 of the encore gene was generated using primers ENCFWD (AATGAAGCGGAGTTCCCAAAGC) and ENCREV (ATAAAGCCCGAGGTGTTGTTGC). The probe for exon 3 of the Hr39 gene was generated using primers Hr39FWD (ACATGTCCAGCATCAAAGCGG) and Hr39REV (TATCGGTTGTAGTGCGCAGAC). The 8R96 intergenic region probe was generated using primers 8R96-225683FWD (CAAGTGGGCTCCATAATAGC) and 8R96-226069REV (TGGAGCTCTCGGTCTGTTAG). The probe for exon 4 of the CG8677 gene was generated using primers CG8677FWD (ATCCGTACCAGTGGCTAAAAGG) and CG8677REV (TTCTTCAACAGCACCACTCGTC). The loading control was ribosomal protein gene Rp49 [64]. DNA probes were 32P-labeled using the Prime-It II DNA labeling kit (Stratagene). Hybridization was carried out in Church-Gilbert solution at 65°C overnight. Hybridization signals were visualized and quantitated using the phosphoimager and ImageQuant software (Molecular Dynamics), and exposure times were 4 days. Transcript size was determined by comparison with 1 kb RNA ladder (Gibco-BRL) according to the manufacturer's instructions.
Relative RNA levels and the fold induction of transcripts in the presence of DOX were estimated as follows. A box was drawn around the band for each gene transcript and intensity measured in arbitrary units using the phosphoimager and ImageQuant. An equal sized box was drawn around a region of the lane containing no bands and that value was subtracted as background. Rp49 loading control was quantitated in the same way for each lane. Each Rp49 intensity value was divided by the median Rp49 intensity value to generate a loading correction factor for each lane. A normalized intensity value for each gene transcript was then calculated by multiplying by the Rp49 correction factor for that lane. This quantitation was done twice for each phosphoimage. The 1x and 2x northern lanes for each RNA sample were quantitated and the numbers were averaged. The resulting relative expression levels are presented in arbitrary units ± standard deviation (SD). VhaSFD: -DOX = 4.8 ± 1.2; +DOX = 20 ± 5.5; fold induction approximately 4. Sugar baby: -DOX = 1.5 ± 0.5; +DOX = 12.8 ± 2.4; fold induction approximately 8.5. filamin: -DOX = 59 ± 11; +DOX = 318 ± 64; fold induction approximately 5. fwd: -DOX = 240± 7; +DOX = 2,303± 28; fold induction approximately 10. Cct: -DOX = 2.3 ± 0.4; +DOX = 4.0 ± 0.2; fold induction approximately 1.7.
Additional data files
A table showing the calculation of 95% confidence intervals for all the datasets is available.
Supplementary Material
A table showing the calculation of 95% confidence intervals.
Acknowledgments
Acknowledgements
We thank the following undergraduates and graduate rotation students for helping with the genetic screen: Yasmin Alt, Rapee Boomplueang, Briana Chavez, Shama Currimbhoy, Sherif Hanna, Mark Heller, Michael Hsu, Hnin Htun, Armine Karapetian, Jimmy Lee, John Lee, Paymann Moin, Viet Nguyen, Neil Rampal, Deepali Shinde, Vanessa Yu and Yvette Yeung. This research was supported by a grant from the Department of Health and Human Services to J.T. (AG11644). We thank Beth E. Rabin for expert assistance with statistical analyses.
References
- Arking R, Dudas SP. Review of genetic investigations into the aging process of Drosophila. J Am Geriatr Soc. 1989;37:757–773. doi: 10.1111/j.1532-5415.1989.tb02240.x. [DOI] [PubMed] [Google Scholar]
- Baker GT, Jacobsen M, Mokrynski G. Aging in Drosophila. In: Cristofalo V, editor. In Cell Biology Handbook in Aging. Boca Raton, FL: CRC Press; 1989. pp. 511–578. [Google Scholar]
- Rose MR. Evolutionary Biology of Aging. New York: Oxford University Press; 1991. [Google Scholar]
- Tower J. Aging mechanisms in fruit flies. BioEssays. 1996;18:799–807. doi: 10.1002/bies.950181006. [DOI] [PubMed] [Google Scholar]
- Tower J. Transgenic methods for increasing Drosophila life span. Mech Ageing Dev. 2000;118:1–14. doi: 10.1016/s0047-6374(00)00152-4. [DOI] [PubMed] [Google Scholar]
- Luckinbill LS, Arking R, Clare M, Cirocco W, Buck S. Selection for delayed senescence in Drosophila melanogaster. Evolution. 1984;38:996–1003. doi: 10.1111/j.1558-5646.1984.tb00369.x. [DOI] [PubMed] [Google Scholar]
- Partridge L, Fowler K. Direct and correlated responses to selection on age at reproduction in Drosophila melanogaster. Evolution. 1992;46:76–91. doi: 10.1111/j.1558-5646.1992.tb01986.x. [DOI] [PubMed] [Google Scholar]
- Rose M, Charlesworth B. A test of evolutionary theories of senescence. Nature. 1980;287:141–142. doi: 10.1038/287141a0. [DOI] [PubMed] [Google Scholar]
- Arking R, Buck S, Berrios A, Dwyer S, Baker GT. Elevated paraquat resistance can be used as a bioassay for longevity in a genetically based long-lived strain of Drosophila. Dev Genet. 1991;12:362–370. doi: 10.1002/dvg.1020120505. [DOI] [PubMed] [Google Scholar]
- Service PM, Hutchinson MD, MacKinley MD, Rose MR. Resistance to environmental stress in Drosophila melanogaster selected for postponed senescence. Physiol Zool. 1985;58:380–389. [Google Scholar]
- Miquel J, Lundgren PR, Bensch KG, Atlan H. Effects of temperature on the life span, vitality and fine structure of Drosophila melanogaster. Mech Ageing Dev. 1976;5:347–370. doi: 10.1016/0047-6374(76)90034-8. [DOI] [PubMed] [Google Scholar]
- Arking R, Buck S, Wells RA, Pretzlaff R. Metabolic rates in genetically based long lived strains of Drosophila. Exp Gerontol. 1988;23:59–76. doi: 10.1016/0531-5565(88)90020-4. [DOI] [PubMed] [Google Scholar]
- Ross RE. Age-specific decrease in aerobic efficiency associated with increase in oxygen free radical production in Drosophila melanogaster. J Insect Physiol. 2000;46:1477–1480. doi: 10.1016/s0022-1910(00)00072-x. [DOI] [PubMed] [Google Scholar]
- Lin Y-J, Seroude L, Benzer S. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science. 1998;282:943–946. doi: 10.1126/science.282.5390.943. [DOI] [PubMed] [Google Scholar]
- Rogina B, Reenan RA, Nilsen SP, Helfand S. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science. 2000;290:2137–2140. doi: 10.1126/science.290.5499.2137. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu M-P, Yin C-M, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Kenyon C. A conserved regulatory system for aging. Cell. 2001;105:165–168. doi: 10.1016/s0092-8674(01)00306-3. [DOI] [PubMed] [Google Scholar]
- Larsen PL. Aging and resistance to oxidative stress in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1993;90:8905–8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci USA. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Johnson TE. Life extension and stress resistance in Caenorhabditis elegans modulated by the tkr-1 gene. Curr Biol. 1998;8:1091–1094. doi: 10.1016/s0960-9822(98)70448-8. [DOI] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Parkes TL, Elia AJ, Dickson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nature Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- Sun J, Folk D, Bradley TJ, Tower J. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics. 2002;161:661–672. doi: 10.1093/genetics/161.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Tower J. FLP recombinase-mediated induction of Cu/Zn-superoxide dismutase transgene expression can extend the life span of adult Drosophila melanogaster flies. Mol Cell Biol. 1999;19:216–228. doi: 10.1128/mcb.19.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Tang XD, Chen M-L, Joiner MA, Sun G, Brot N, Wiessbach H, Heinemann SH, Iverson L, Wu C-F, Hoshi T. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc Natl Acad Sci USA. 2002;99:2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavous DA, Jackson FR, O'Connor CM. Extension of the Drosophila life span by overexpression of a protein repair methyltransferase. Proc Natl Acad Sci USA. 2001;98:14814–14818. doi: 10.1073/pnas.251446498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong K-H, Matsuo T, Fuyama Y, Aigaki T. Neural-specific over-expression of Drosophila Plenty of SH3s (DPOSH) extends the longevity of adult flies. Biogerontology. 2001;2:271–281. doi: 10.1023/a:1013249326285. [DOI] [PubMed] [Google Scholar]
- Seong K-H, Ogashiwa T, Matsuo T, Fuyama Y, Aigaki T. Application of the gene search system to screen for longevity genes in Drosophila. Biogerontology. 2001;2:209–217. doi: 10.1023/a:1011517325711. [DOI] [PubMed] [Google Scholar]
- Tatar M, Khazaeli AA, Curtsinger JW. Chaperoning extended life. Nature. 1997;390:30. doi: 10.1038/36237. [DOI] [PubMed] [Google Scholar]
- Hay BA, Maile R, Rubin GM. P element insertion-dependent gene activation in the Drosophila eye. Proc Natl Acad Sci USA. 1997;94:5195–5200. doi: 10.1073/pnas.94.10.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci USA. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieschke ET, Wheeler JC, Tower J. Doxycycline-induced transgene expression during Drosophila development and aging. Mol Gen Genet. 1998;258:571–579. doi: 10.1007/s004380050770. [DOI] [PubMed] [Google Scholar]
- Landis G, Bhole , Lu L, Tower J. High-frequency generation of conditional mutations affecting Drosophila development and life span. Genetics. 2001;158:1167–1176. doi: 10.1093/genetics/158.3.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk CV, Hawkins NC, Schupbach T. Encore is a member of a novel family of proteins and affects multiple processes in Drosophila oogenesis. Development. 2000;127:4753–4762. doi: 10.1242/dev.127.22.4753. [DOI] [PubMed] [Google Scholar]
- Dow JAT. The multifunctional Drosophila melanogaster V-ATPase is encoded by a multigene family. J Bioenerg Biomembr. 1999;31:75–83. doi: 10.1023/a:1005400731289. [DOI] [PubMed] [Google Scholar]
- Liong EC, Ferenci T. Molecular cloning of a maltose transport gene from Bacillus stearothermophilus and its expression in Escherichia coli K-12. Mol Gen Genet. 1994;243:343–352. doi: 10.1007/BF00301070. [DOI] [PubMed] [Google Scholar]
- Li M, Serr M, Edwards K, Ludmann S, Yamamoto D, Tilney LG, Field CM, Hays TS. Filamin is required for ring canal assembly and actin organization during Drosophila oogenesis. J Cell Biol. 1999;146:1061–1073. doi: 10.1083/jcb.146.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Cooley L. Drosophila Filamin encoded by the cheerio locus is a component of ovarian ring canals. Curr Biol. 1999;9:1221–1230. doi: 10.1016/s0960-9822(99)80502-8. [DOI] [PubMed] [Google Scholar]
- Brill JA, Hime GR, Scharer-Schuksz M, Fuller MT. A phospholipid kinase regulates actin organization and intercellular bridge formation during germline cytokinesis. Development. 2000;127:3855–3864. doi: 10.1242/dev.127.17.3855. [DOI] [PubMed] [Google Scholar]
- Cornell RB, Northwood IC. Regulation of CTP:phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biochem Sci. 2000;25:441–447. doi: 10.1016/s0968-0004(00)01625-x. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch A, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk C, Hawkins NC, Schupbach T. Encore is a member of a novel family of proteins and affects multiple processes in Drosophila oogenesis. Development. 2000;127:4753–4762. doi: 10.1242/dev.127.22.4753. [DOI] [PubMed] [Google Scholar]
- Forgac M. Structure and properties of the clathrin-coated vesicle and yeast vacuolar V-ATPases. J Bioenerg Biomembr. 1999;31:57–65. doi: 10.1023/a:1005496530380. [DOI] [PubMed] [Google Scholar]
- Graham LA, Stevens TH. Assembly of the yeast vacuolar proton-translocating ATPase. J Bioenerg Biomembr. 1999;31:39–47. doi: 10.1023/a:1005455411918. [DOI] [PubMed] [Google Scholar]
- Nishi T, Forgac M. The vacuolar (H+)-ATPases - nature's most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- Hardy J. Amyloid, the presenilins and Alzheimer's disease. Trends Neurosci. 1997;20:154–159. doi: 10.1016/s0166-2236(96)01030-2. [DOI] [PubMed] [Google Scholar]
- Guo Y, Zhang SJ, Sokol N, Cooley L, Boulianne GL. Physical and genetic interaction of filamin with presenilin in Drosophila. J Cell Sci. 2000;113:3499–3508. doi: 10.1242/jcs.113.19.3499. [DOI] [PubMed] [Google Scholar]
- Edwards DN, Towb P, Wasserman SA. An activity-dependent network of interactions links the Rel protein Dorsal with its cytoplasmic regulators. Development. 1997;124:3855–3864. doi: 10.1242/dev.124.19.3855. [DOI] [PubMed] [Google Scholar]
- Balla T. Phosphatidylinositol 4-kinases. Biochim Biophys Acta. 1998;1436:69–85. doi: 10.1016/s0005-2760(98)00134-9. [DOI] [PubMed] [Google Scholar]
- Hsuan JJ, Minogue S, dos Santos M. Phosphoinositide 4- and 5-kinases and the cellular roles of phosphatidylinositol 4,5-bis-phosphate. Adv Cancer Res. 1998;74:167–216. doi: 10.1016/s0065-230x(08)60767-8. [DOI] [PubMed] [Google Scholar]
- Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals for the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signalling. Nature Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Flybase http://flybase.bio.indiana.edu
- Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Ashburner M. Drosophila: A Laboratory Handbook. Plainview, NY: Cold Spring Harbor Laboratory Press. 1989.
- Tower J, Karpen GH, Craig N, Spradling AC. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;113:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J, Kurapati R. Preferential transposition of a Drosophila P element to the corresponding region of the homologous chromosome. Mol Gen Genet. 1994;244:484–490. doi: 10.1007/BF00583899. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- O'Connell P, Rosbash M. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A table showing the calculation of 95% confidence intervals.


