Abstract
The androgen receptor (AR) plays a critical role in male sexual development and in normal and malignant prostate cell growth and survival. It has been shown that AR transcriptional activation is regulated through interactions with a variety of transcriptional co-regulators. The Protein Inhibitors of Activated STATs (PIAS) are transcriptional co-regulators, and have been shown to modulate AR-mediated transcription. In this brief, we summarize our recent studies on two novel PIAS-like proteins, Zimp7 and Zimp10. Particularly, we address the functional interactions between the AR and these two proteins, and potential mechanisms by which they regulate AR mediated transcription. In addition, we explore potential roles of Zimp10 in transcriptional regulation in vivo using a recent Zimp10 knockout mouse model. Taken together, our findings thus far suggest that Zimp7 and Zimp10 are functionally non-redundant and share unique characteristics that have not been described for the PIAS family. Further investigation into the functional roles of these two PIAS-like proteins may help to better understand prostate cancer progression, and yield possible new targets for therapeutic intervention.
Introduction
AR-mediated transcription is a complicated process, which is facilitated through interactions with multiple co-regulators. Indeed, it has been suggested that alterations in AR co-regulator expression is a mechanism by which prostate cancers progress to an androgen-insensitive stage [Heinlein and Chang, 2004], highlighting the importance of understanding how transcriptional co-regulators modulate AR activity. Over 200 nuclear hormone receptor co-regulators have been identified in recent years [Chang and McDonnell, 2005]. Among them, the PIAS proteins have been shown to regulate the function of the AR and other nuclear hormone receptors [Schmidt and Muller, 2003].
The PIAS proteins were originally identified as negative regulators of the JAK-STAT cytokine signaling pathway, but have subsequently been shown to regulate the activity of steroid hormone receptors and other transcription factors, such as LEF-1, and p53 [Kotaja et al., 2000; Nelson et al., 2001; Sachdev et al., 2001]. Of particular relevance to AR signaling, the PIAS family member PIASxα was first described as AR-interacting protein 3 (ARIP3) [Moilanen et al., 1999]. In addition to their role as negative regulators of the STAT pathway, the PIAS proteins have been shown to positively and negatively regulate the activity of other transcription factors in different cell contexts [Schmidt and Muller, 2003]. The PIAS proteins possess E3 ligase activity for the ubiquitin-like SUMO pathway, and have been shown to regulate AR activity through sumoylation of the AR and its co-factors [Nishida and Yasuda, 2002; Schmidt and Muller, 2002].
PIAS proteins contain a conserved zinc-binding SP-RING/Miz domain, which is similar to the RING domain in the RING-type E3 ubiqitin ligases [Schmidt and Muller, 2003]. This domain appears essential for PIAS-mediated modulation of transcription because targeted mutations within this region abrogate SUMO-conjugating activity and interactions with target proteins [Kahyo et al., 2001; Kotaja et al., 2002]. Recently, we have identified two novel PIAS-like proteins that contain the SP-RING/Miz domain [Huang et al., 2005; Sharma et al., 2003]. Based on this feature, we have named these proteins Zimp7 and Zimp10 (Zinc finger-containing, Miz-1, PIAS-like protein on chromosome 7 or 10). Like the PIAS proteins, both Zimp7 and Zimp10 interact with the AR and regulate its activity. Importantly, Zimp7 and Zimp10 have been shown to interact with components of the SWI/SNF chromatin remodeling complexes, suggesting a potential mechanism by which the Zimp proteins regulate the activity of the AR or other transcription factors. The following brief outlines our current knowledge of Zimp7 and 10.
Zimp10 is an AR co-activator
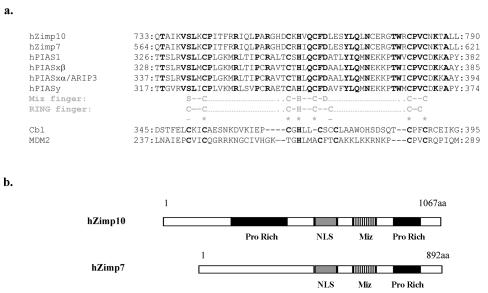
Zimp10 was originally isolated in a yeast two-hybrid assay using a partial AR transactivation domain as bait. Sequence analysis revealed that Zimp10 contains 1067 amino acids and shares a highly conserved SP-RING/Miz domain with members of the PIAS family (Figure 1a). In addition to the SP-RING/Miz domain, Zimp10 also contains a nuclear localization sequence (NLS) and two proline-rich regions (Figure 1b). Interestingly, Zimp10 contains a strong intrinsic transactivation domain within its C-terminus, a function that has also been described for PIAS1 and Miz1 [Kotaja et al., 2000]. Zimp10 mRNA is highly expressed in urogenital tissues, including prostate, testis, and ovary, and in prostate and breast cancer cell lines.
Figure 1. Sequence of the human Zimp7 and Zimp10 Miz domains.
A. Sequence alignment of the Miz domains of hZimp7, hZimp10, and members of the PIAS family. Identical amino acids are indicated in bold. Comparison of the consensus sequences for the Miz finger and RING finger domains are shown in gray. For comparison, the RING finger domains of the E3 ubiquitin ligases Cbl and MDM2 are also shown. B. Comparison of Zimp7 and Zimp10 functional domains. Pro-rich, proline rich region; NLS, Nuclear localization sequence.
Several lines of evidence have demonstrated that Zimp10 physically interacts with the AR and augments AR transcriptional activity [Sharma et al., 2003]. Interestingly, Zimp10 has little or no effect on several other nuclear hormone receptors, including the glucocorticoid receptor (GR), estrogen receptor (ER), thyroid receptor β (TR-β), or vitamin D receptor (VDR). Endogenous Zimp10 protein is primarily localized to the nucleus in all cell lines and prostate tissue samples examined. In addition, immunofluorescent staining has shown that Zimp10 co-localizes with the AR and SUMO-1 at replication foci during S-phase. Consistent with these observations, AR sumoylation is enhanced in the presence of Zimp10. Furthermore, mutation of one (K386R) or both (K386R/K520R) of the major AR sumoylation sites reduces Zimp10-mediated enhancement of AR activity. Interestingly, this is in contrast with reports indicating that PIAS-mediated AR sumoylation represses AR transcriptional activity [Kotaja et al., 2002; Nishida and Yasuda, 2002].
Identification of Zimp7, a Zimp10 homolog
A search for homologs of Zimp10 revealed that the KIAA clone KIAA1886 and Zimp10 share significant sequence similarity. Isolation of the full-length cDNA and translation of the protein product indicated that it is an 892aa protein with a molecular size of approximately 100kDa. A BLAST search mapped the nucleotide sequence to chromosome 7, and we have thus named this protein Zimp7. Like Zimp10, it contains the SP-RING/Miz domain, a NLS, a C-terminal proline-rich region, and a C-terminal transactivation domain. However, Zimp7 lacks the N-terminal proline-rich region present in Zimp10 (Figure 1b). Consistent with their homology, Zimp7 and Zimp10 share many of the same functional characteristics. Zimp7 is a nuclear protein and localizes to replication foci during S phase. Additionally, Zimp7 co-localizes with SUMO-1 and the AR in prostate tissues and several human cell lines. However, the expression profile of Zimp7 is not identical to that of Zimp10, and is most highly expressed in the testis, as well as in prostate, ovary, heart, skeletal muscle, and pancreas. Several lines of evidence have shown that Zimp7 physically interacts with the AR and augments AR-mediated transcription. Although the precise differences between these two proteins are unclear, our recent studies have shown that Zimp7 and Zimp10 may preferentially regulate different subsets of nuclear hormone receptors and other transcription factors [Huang et al., 2005; Sharma et al., 2003].
Understanding the biological role of Zimp10 in vivo
Further insight into the potential mechanisms of Zimp7/10 transcriptional regulation comes from a recent report by Guiterrez et al., which described a Zimp7/10 ortholog in Drosophila called tonalli (tna) [Gutierrez et al., 2003]. Interestingly, tna genetically interacts with brahma, which encodes an ATPase subunit of the SWI2/SNF2 chromatin remodeling complexes. These data suggest that Zimp7 and Zimp10 may regulate transcription through modifications of chromatin structure via interactions with SWI/SNF complexes. Intriguingly, we have observed that both Zimp7 and Zimp10 co-localize with BrdU and/or proliferating cell nuclear antigen (PCNA) in sub-nuclear replication foci during S-phase. In addition, co-immunoprecipitation studies have shown that Zimp7 and Zimp10 interact with Brg-1, a component of SWI/SNF complexes. Zimp7 also associates with the SWI/SNF subunit, BAF57, and increases Brg-1/BAF57-mediated enhancement of AR transcriptional activity [Huang et al., 2005].
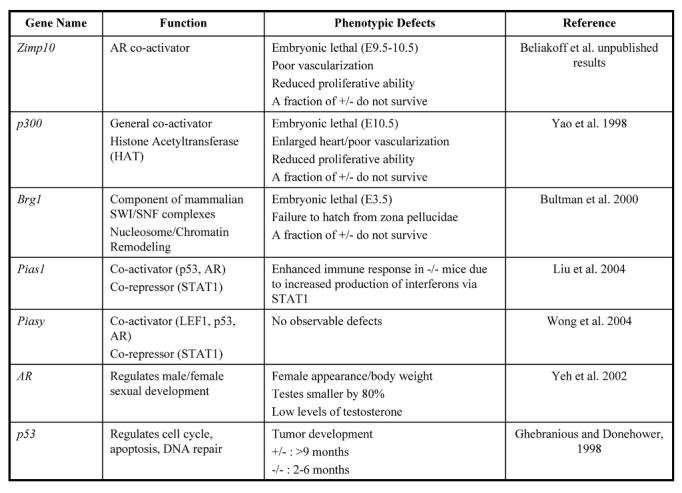
To further characterize the biological roles of Zimp7 and Zimp10, we have used gene-targeting strategies to disrupt the Zimp7 and Zimp10 alleles in mice. Zimp10 knockouts die between 9.5-10.5 days post-coitus. At this stage, Zimp10 knockout embryos are approximately half the size of wild type littermates and display vascular defects. In addition, a fraction of mice heterozygous for the disrupted Zimp10 allele do not survive, suggesting that Zimp10 gene dosage is important for proper development. However, the heterozygotes that do survive have no apparent defects. The embryonic lethal phenotype is in contrast to PIAS knockouts, which are generally viable [Liu et al., 2004; Santti et al., 2005; Wong et al., 2004]. Loss of Zimp10 causes severe defects in cell viability because mouse embryo fibroblasts from Zimp10 knockout embryos show greatly reduced proliferative ability compared to their wild type counterparts. Growth inhibition is also observed in prostate cancer cell lines following knock-down of endogenous Zimp10 protein by interfering RNA. The embryonic lethality observed in Zimp10 knockouts suggests that Zimp7 cannot entirely compensate for loss of Zimp10 function. We are currently in the process of characterizing Zimp7 and Zimp7/Zimp10 knockouts to determine whether Zimp7 and Zimp10 have any functional redundancy. The data from these studies will further advance our knowledge regarding the roles of Zimp7 and Zimp10 in vivo. Of note, mice with a non-functional AR are viable [Quigley et al., 1995], which indicates that Zimp10 may affect AR-independent pathways (Table 1). Indeed, the lethal phenotype of Zimp10 knockouts suggests a critical role for the protein in early development, a requirement that has not been described in knockouts for PIAS family members (Table 1) [Liu et al., 2004; Wong et al., 2004].
Table 1. Comparison of knockout phenotypes for general transcriptional co-regulators, PIAS family members, and specific transcription factors.
The names, functions, and phenotypic defects of knockout mice for various genes are shown. Knockouts for Zimp10 and the general transcriptional co-regulators p300 and Brg-1 display more severe defects than knockouts for PIAS family members, AR and p53.
Conclusions
Zimp7 and Zimp10 are two novel PIAS-like proteins, which share a highly conserved SP-RING/Miz protein-protein interaction domain that is important for PIAS-mediated sumoylation of target proteins. Both Zimp7 and Zimp10 contain strong intrinsic transactivation domains and augment AR activity. The interactions between the Zimp proteins and components of the SWI/SNF chromatin remodeling complexes suggest a possible role for Zimp7 and Zimp10 in chromatin modification. Zimp10 knockout mice exhibit embryonic lethality, a more severe phenotype compared to PIAS knockouts and mice with a non-functional AR, which suggests that Zimp10 may play a more general role in transcriptional regulation and may not share redundancy with Zimp7 or members of the PIAS family. Indeed, the Zimp10 knockout phenotype more closely resembles knockout mouse models for general transcriptional co-factors [Bultman et al., 2000; Yao et al., 1998], rather than knockouts for the PIAS family members [Liu et al., 2004; Wong et al., 2004] or for specific transcription factors [Ghebranious and Donehower, 1998; Yeh et al., 2002]. Further characterization of Zimp7 and Zimp10 knockouts will provide greater insight into the roles of Zimp7 and Zimp10 in vivo.
Acknowledgments
This work was supported by National Institutes of Health Grants CA070297, CA087767, DK061002, Training Grant 5T32 CA09302-27, and the Department of Army Prostate Cancer grant DAMD17-03-1-0090.
References
- Bultman S., Gebuhr T., Yee D., La Mantia C., Nicholson J., Gilliam A., Randazzo F., Metzger D., Chambon P., Crabtree G., Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–95. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Chang C. Y., McDonnell D. P. Androgen receptor-cofactor interactions as targets for new drug discovery. Trends Pharmacol Sci. 2005;26:225–8. doi: 10.1016/j.tips.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Ghebranious N., Donehower L. A. Mouse models in tumor suppression. Oncogene. 1998;17:3385–400. doi: 10.1038/sj.onc.1202573. [DOI] [PubMed] [Google Scholar]
- Gutierrez L., Zurita M., Kennison J. A., Vazquez M. The Drosophila trithorax group gene tonalli (tna) interacts genetically with the Brahma remodeling complex and encodes an SP-RING finger protein. Development. 2003;130:343–54. doi: 10.1242/dev.00222. [DOI] [PubMed] [Google Scholar]
- Heinlein C. A., Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- Huang C. Y., Beliakoff J., Li X., Lee J., Sharma M., Lim B., Sun Z. hZimp7, A Novel PIAS-like Protein, Enhances Androgen Receptor-mediated Transcription and Interacts with SWI/SNF-like BAF Complexes. Mol Endocrinol. 2005 doi: 10.1210/me.2005-0097. [DOI] [PubMed] [Google Scholar]
- Kahyo T., Nishida T., Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell. 2001;8:713–8. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- Kotaja N., Aittomaki S., Silvennoinen O., Palvimo J. J., Janne O. A. ARIP3 (androgen receptor-interacting protein 3) and other PIAS (protein inhibitor of activated STAT) proteins differ in their ability to modulate steroid receptor-dependent transcriptional activation. Mol Endocrinol. 2000;14:1986–2000. doi: 10.1210/mend.14.12.0569. [DOI] [PubMed] [Google Scholar]
- Kotaja N., Karvonen U., Janne O. A., Palvimo J. J. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol Cell Biol. 2002;22:5222–34. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Mink S., Wong K. A., Stein N., Getman C., Dempsey P. W., Wu H., Shuai K. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat Immunol. 2004;5:891–8. doi: 10.1038/ni1104. [DOI] [PubMed] [Google Scholar]
- Moilanen A. M., Karvonen U., Poukka H., Yan W., Toppari J., Janne O. A., Palvimo J. J. A testis-specific androgen receptor coregulator that belongs to a novel family of nuclear proteins. J Biol Chem. 1999;274:3700–4. doi: 10.1074/jbc.274.6.3700. [DOI] [PubMed] [Google Scholar]
- Nelson V., Davis G. E., Maxwell S. A. A putative protein inhibitor of activated STAT (PIASy) interacts with p53 and inhibits p53-mediated transactivation but not apoptosis. Apoptosis. 2001;6:221–34. doi: 10.1023/a:1011392811628. [DOI] [PubMed] [Google Scholar]
- Nishida T., Yasuda H. PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J Biol Chem. 2002;277:41311–7. doi: 10.1074/jbc.M206741200. [DOI] [PubMed] [Google Scholar]
- Quigley C. A., De Bellis A., Marschke K. B., el-Awady M. K., Wilson E. M., French F. S. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr Rev. 1995;16:271–321. doi: 10.1210/edrv-16-3-271. [DOI] [PubMed] [Google Scholar]
- Sachdev S., Bruhn L., Sieber H., Pichler A., Melchior F., Grosschedl R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 2001;15:3088–103. doi: 10.1101/gad.944801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santti H., Mikkonen L., Anand A., Hirvonen-Santti S., Toppari J., Panhuysen M., Vauti F., Perera M., Corte G., Wurst W., Janne O. A., Palvimo J. J. Disruption of the murine PIASx gene results in reduced testis weight. J Mol Endocrinol. 2005;34:645–54. doi: 10.1677/jme.1.01666. [DOI] [PubMed] [Google Scholar]
- Schmidt D., Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci U S A. 2002;99:2872–7. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., Muller S. PIAS/SUMO: new partners in transcriptional regulation. Cell Mol Life Sci. 2003;60:2561–74. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Li X., Wang Y., Zarnegar M., Huang C. Y., Palvimo J. J., Lim B., Sun Z. hZimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. Embo J. 2003;22:6101–14. doi: 10.1093/emboj/cdg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. A., Kim R., Christofk H., Gao J., Lawson G., Wu H. Protein inhibitor of activated STAT Y (PIASy) and a splice variant lacking exon 6 enhance sumoylation but are not essential for embryogenesis and adult life. Mol Cell Biol. 2004;24:5577–86. doi: 10.1128/MCB.24.12.5577-5586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T. P., Oh S. P., Fuchs M., Zhou N. D., Ch'ng L. E., Newsome D., Bronson R. T., Li E., Livingston D. M., Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–72. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- Yeh S., Tsai M. Y., Xu Q., μ X. M., Lardy H., Huang K. E., Lin H., Yeh S. D., Altuwaijri S., Zhou X., Xing L., Boyce B. F., Hung M. C., Zhang S., Gan L., Chang C. Generation and characterization of androgen receptor knockout (ARKO) mice: an in vivo model for the study of androgen functions in selective tissues. Proc Natl Acad Sci U S A. 2002;99:13498–503. doi: 10.1073/pnas.212474399. [DOI] [PMC free article] [PubMed] [Google Scholar]