Abstract
Large-scale genomics analyses have grown by leaps and bounds with the rapid advances in high throughput DNA sequencing and synthesis techniques. Nuclear receptor signaling is ideally suited to genomics studies because receptors function as ligand-regulated gene switches. This review will survey the strengths and limitations of three major classes of high throughput techniques widely used in the nuclear receptor field to characterize ligand-dependent gene regulation: expression profiling studies (microarrays, SAGE and related techniques), chromatin immunoprecipitation followed by microarray (ChIP-on-chip), and genome-wide in silico hormone response element screens. We will discuss each technique, and how each has contributed to our understanding of nuclear receptor signaling.
Introduction
Given that they function as gene switches, signaling by nuclear receptors is ideally suited to analyses using genomics approaches. The last few years have seen the development of a number of techniques for analysis of gene expression and regulation on chromosome-wide or genome-wide scales. We will assess below the strengths and limitations and the complementarity of three general approaches for genome-wide identification of nuclear receptor target genes; expression profiling, ChIP-on-chip experiments, and in silico identification and mapping of hormone response elements, and discuss how these techniques have dramatically enhanced our understanding of the mechanisms and physiological consequences of nuclear receptor signaling.
Genome-wide expression profiling of nuclear receptor-regulated gene expression
Microarray analysis
Microarray technology was developed in the early 1990s to monitor the expression of many genes simultaneously [Schena et al., 1995]. Since then, the use of microarrays has become a powerful tool and a standard technique for analyzing the expression and regulation of thousands of genes in parallel. A DNA microarray consists of a glass slide containing thousands of DNA sequences fixed to the slide in a grid, each of which represents a portion of a gene. RNA samples under analysis are converted to cDNAs, labeled with fluorescent dyes, and hybridized to the array. Since the location and the sequence of the probe fixed in the array is known, the identity and relative abundance of sequences hybridized to the array can be determined [Sellheyer and Belbin, 2004]. There are two different types of DNA microarrays based on the types of probes used: cDNA microarrays and oligonucleotide microarrays. cDNA microarrays use probes generated by amplified cDNA (either from plasmids or amplified by RT-PCR). These probes are fixed on silane slides by covalently UV cross-linking the thymidine residues of the cDNA with the amine groups in the silane slide [Cheung et al., 1999]. Oligonucleotide microarrays use ~25mer oligonucleotides complementary to a selected gene or EST sequence [Lipshutz et al., 1999]. The arrays are designed in silico and placed in the slide. Array densities have increased rapid over the last few years, driven in part by rapid increases in genomic sequence and EST databases. The most recent human and mouse Affymetrix oligonucleotide arrays contain 47,000 and 37,000 gene sequences, respectively.
Applications of microarrays to analysis of nuclear receptor signaling
Microarrays have revolutionized the nuclear receptor field by providing us with snapshots of receptor signaling on genome-wide or near genome-wide scales, and the technique is by far the most heavily used in the field for large-scale studies of gene regulation. Microarrays have been used to dissect nuclear receptor functions both in normal and disease states, in tissues and in cell models. Examples include: numerous studies on nuclear receptor gene regulation for identification of downstream signaling pathways [Lee et al., 2003; Quinn et al., 2005; White, 2004]; comparative analyses of gene expression in nuclear receptor null versus wild type mice for several receptors, including VDR, PPAR, and ERR [Carrier et al., 2004; Lee et al., 2002; Li et al., 2003]; profiling of drug-induced gene expression through studies of CAR-mediated gene regulation in response to phenobarbital in the liver [Ueda et al., 2002]; identification of PXR-regulated genes encoding phase I, II and III xenobiotic metabolizing enzymes [Rosenfeld et al., 2003]; and comparison in PXR-regulated drug metabolism in intestine and liver to understand and predict drug-drug interactions [Hartley et al., 2004]. Analysis of PPAR activation in a high cholesterol context followed by microarray studies revealed potential target genes of triglyceride-lowering drugs [Frederiksen et al., 2004]. Microarray technology has also been used to understand the genome-wide effects of environmental estrogens [Moggs, 2005]. Nuclear receptor signaling has been analyzed in such diverse disease states as allograft rejection [Amuchastegui et al., 2005] and inflammatory bowel disease [Langmann et al., 2004]. Finally, gene expression patterns have been profiled by microarrays in several models of hormone-dependent cancer for examination of hormonal signal transduction and identification of biomarkers [Choi and Pinto, 2005; Hanekamp et al., 2003; Helms et al., 2005; Kristensen et al., 2005; Lee et al., 2005; Quinn et al., 2005], and classification of different cancer types and prediction of patient survival [Glinsky et al., 2004; Haqq et al., 2005; Helms et al., 2005; Yoshida et al., 2004].
While there are clearly a wide range of applications for microarrays in studies of nuclear receptor signaling, comparisons between different sets of experiments are not straightforward. Handling the extensive amount of information generated and sorting out the biologically meaningful data from the background noise has been a difficult challenge. MIAME (Minimum Information About a Microarray Experiment [Brazma et al., 2001] focuses on defining the content and structure of data presentation rather than standardization techniques for data analysis. Indeed, comparisons are complicated by differences in sample labeling, hybridization conditions, and imaging quantification techniques. Cut-offs of significant fold regulation used in filtering microarray data, which are often ‘noisy’ because of stochastic variations in gene expression vary from one study to another. Early experiments used microarrays with far fewer genes than more recent analyses. Moreover, there are no standards for determining statistically significant versus biologically relevant ways to analyze and interpret raw microarray data [van Bakel and Holstege, 2004].
Comparative analyses of microarray studies are further complicated by the remarkably limited overlaps in nuclear receptor target genes observed in different experimental models. For example, the vitamin D field has taken full advantage of microarray technology to identify a large number of target genes. Besides its classical physiological function in calcium homeostasis, the hormonal form of vitamin D, 1,25-dihydroxyvitamin D3 (1,25D3), regulates cellular proliferation, differentiation, apoptosis, survival, and is a modulator of immune responses. Several studies have shown that 1,25D3 arrests cell proliferation at G0/G1 in a variety of models [Akutsu et al., 2001; Krishnan et al., 2004; Lin et al., 2002; Lin and White, 2004; Swami et al., 2003]. With such a plethora of biological effects, differences in the types of genes regulated are to be expected between specialized cell types. Nonetheless, apart from the cyp24 gene, which encodes the enzyme regulating 1,25D3 catabolism, the lack of overlap of regulated genes identified in different studies is striking [Zhang et al., 2005]. Some differences are attributable to differential regulation of target genes in different cells. For example, the gene encoding the cyclin-dependent kinase inhibitor p21WAF1/CIP1 is strongly induced by 1,25D3 in myelomonocytic cells [Liu et al., 1996; Munker et al., 1996], but repressed in 1,25D3-sensitive squamous carcinoma cells [Hershberger et al., 1999; Prudencio et al., 2001]. However, while the specifics may vary, comparisons of microarrays studies have revealed that 1,25D3 signaling regulates the expression of genes controlling cell cycle arrest, DNA repair, markers of cellular differentiation, calcium and redox homeostasis, cell survival and immune responses [White, 2004]. The relative lack of overlap of specific gene targets between studies underlines the importance of carefully choosing the cell model that will best represent the physiological responses under investigation.
SAGE and related techniques
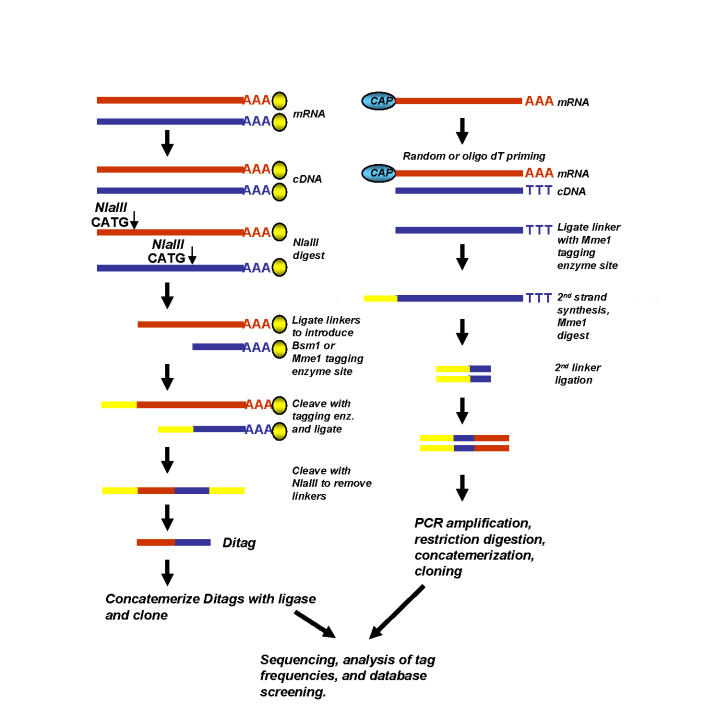
SAGE (serial analysis of gene expression), which was developed in the mid-1990’s [Velculescu et al., 1995], is a technique for gene expression profiling that entails the generation, concatemerization and sequencing of so-called short diagnostic sequence tags. Tags are generated in part by cleavage of cDNA sequences with class II restriction enzymes such as NlaIII, which recognizes frequently occurring CATG recognition sequences and leave 4bp overhangs (Figure 1). SAGE and its variant longSAGE [Saha et al., 2002] have been widely used to catalogue gene expression patterns in a number of tissues. Similar to microarray analyses, SAGE has been used to identify hormone-regulated genes in a variety of experimental models [Datson et al., 2001; Inadera et al., 2000; Robert-Nicoud et al., 2001; Seth et al., 2002], as well for comparative studies of gene expression profiles in hormone-dependent and -independent cancers [Abba et al., 2005; Enerback et al., 2002].
Figure 1. Generation of tag-based libraries by SAGE and CAGE.
Schematic representations of SAGE (left) and CAGE (right) techniques for generation of tagged expression libraries. In SAGE, mRNA sequences are captured on oligo(dT) magnetic beads (yellow spheres). In CAGE, 5’ ends of mRNAs are extracted from total or polyA+ RNA using the CAP-trapper approach. See [Harbers, 2005] and [Porter, 2006] for details.
Although technically quite distinct, the use of SAGE carries some of the same caveats associated with microarray studies. As with microarrays, comparisons between SAGE studies of hormone-regulated gene expression in different tissues or cell types will likely reveal relatively limited overlaps in gene expression profiles. Comparisons between different SAGE studies may be complicated by the number of different statistical methods used to analyze expression patterns in SAGE libraries [Ruijter et al., 2002], which, like microarrays, must deal with ‘noise’ associated with stochastic variations in gene expression. It should also be pointed out that use of SAGE is also technically considerably more demanding than microarrays, and unlike microarray studies which are readily performed in triplicate or more, comparative SAGE analyses are usually performed between single libraries.
Apart from its relative technical complexity SAGE also carries certain inherent drawbacks. Transcripts lacking class II restriction sites used to generate tags will not be detected. In addition, some tags may identify several genomic locations, and the technique can generate artifactual ‘orphan’ tags via a number of routes [Harbers and Carninci, 2005]. On the other hand, SAGE does have distinct advantages over microarrays [Porter et al., 2006]. Generation of SAGE libraries requires no a priori knowledge mRNA sequences of the organism under study. As indicated above, transcripts identified microarray analyses are limited by the number of sequences on the chip, which is in part limited by knowledge of the transcriptome under study. While this is becoming less limiting for human studies, it is more problematic for analyses of other model organisms with less well characterized transcriptomes. Moreover, multiple transcripts from the same gene can be identified by SAGE, something that is only possible with specialized microarrays bearing sequences of well-characterized genes [Porter et al., 2006].
Along with ‘classical’ SAGE, several other tag-based methods have been developed to profile gene expression [Harbers and Carninci, 2005]. These include CAGE (Figure 1), which generates tags from the 5’ ends of transcripts by trapping the mRNA CAPs of full length transcripts. In CAGE, tags are generated by cleavage with Mme1, which is introduced in a ligated linker sequence, thus circumventing the problem of elimination of messages that lack specific restriction enzyme recognition sequences.
Another offshoot of SAGE of interest to nuclear receptor researchers is the SACO (serial analysis of chromatin occupancy) technique [Impey et al., 2004]. This technique couples chromatin immunoprecipitation using an antibody against a transcription factor (in the case of Impey et al, against CREB) with longSAGE to generate so-called genome signature tags (GSTs). Many of the sequences identified by multiple GSTs in the Impey et al study lay adjacent to known transcription start sites or CpG islands. As SACO can be used to identify potential transcription factor binding sites in addition to target genes, it bears some relation to ChIP-on-chip studies, which is discussed below.
ChIP-on-chip analysis of nuclear receptor binding sites
Overview of the technique
ChIP-on-chip or genome-wide localization analysis was developed a few years ago [Ren et al., 2000] to identify transcription factor target genes on a chromosome- or genome-wide scale. The technique is a combination of chromatin immunoprecipitation followed by microarray analysis. Briefly, the transcriptional regulator is cross-linked in vivo to DNA (chromatin), and the DNA is sheared to ~300-500bp fragments. The transcription factor is immunoprecipitated, and protein complexes are de-crosslinked. The associated DNA is purified, converted into fluorescent-labeled probe and hybridized to a microarray. The advantage of ChIP-on-chip technique is clear; after immunoprecipitation the nuclear receptor can be unambiguously assigned to a specific chromosomal region. There are four types of arrays [Blais and Dynlacht, 2005] used for ChIP-on-chip analysis: proximal promoter arrays, where about ~1 kb PCR products encompassing transcription start sites are used as probes; arrays composed of CpG islands amplified by PCR; large promoter arrays, which consist of tiling oligonucleotides of promoter sequences extending up to several kb upstream of the transcription start site; and, finally, the genome tiling array, in which nonrepetive sequence from entire chromosomes is reconstituted from oligonucleotides. When chromosomal sequence is densely covered, very high resolution can be achieved with genome tiling microarrays [Blais and Dynlacht, 2005].
ChIP-on-chip analysis of signaling by estrogen receptor α
Two recent studies have used ChIP-on-chip approaches to identify ERα binding sites in estrogen-dependent MCF-7 breast cancer cells. Both papers made substantial contributions to understanding estrogen-dependent signaling. However, the two studies are noteworthy for both their similarities and their differences. Both analyzed ERα binding in MCF-7 cells treated for 45 min. with 100 nM estradiol. One study used an Affymetrix tiling array for chromosomes 21/22 [Carroll et al., 2005], and the other used a promoter-specific array composed of 1kb fragments ranging from -800 to +200 bp relative to known transcription start sites [Laganiere et al., 2005]. One advantage of chromosomal tiling arrays is their design, and the fact that the results obtained are not dependent on the reliability of previous mapping studies to identify promoter regions. Indeed, Carroll et al [Carroll et al., 2005] cloned by RACE sequences lying several dozen kb upstream of the known 5’ end of the gene encoding RIP140 after identifying far upstream EREs on a tiling array. Significantly, in addition to identifying numerous ERα binding sites in vivo, both studies provided evidence that the forkhead protein FoxA1 contributes to estrogen-dependent regulation of a number of ERα target genes. Carroll et al [Carroll et al., 2005] reached this conclusion after finding an enrichment of FoxA1 binding sites in the vicinity of EREs characterized on the array.
As ChIP-on-chip approaches use microarrays, they carry the same limitations as other microarray studies. Results will be cell-model and -context specific, with the likelihood of relatively limited overlap between studies performed with different experimental models. Moreover, the number and identity of binding sites will depend on those present on the array. Notably, with the exception of trefoil factor 1 (TFF-1) there was no overlap between the two analyses of ERα binding sites in the two studies cited above, in spite of the use of the same estradiol-treated cell model. Although the microarrays used were different (chromosome 21/22 tiling arrays vs. a short promoter-specific array), there were genes located on chromosomes 21 and 22 identified on the promoter-specific microarray that were not picked up on the tiling array [Carroll et al., 2005; Laganiere et al., 2005]. The study by Carroll et al [Carroll et al., 2005] is remarkable for having identified binding sites located >100kb from the 5’ ends of estrogen target genes. Function of a distal site in the gene encoding RIP140 as an enhancer was verified by chromatin conformation capture assay. Such sites could not have been identified by Laganiere et al [Laganiere et al., 2005] because of the use of proximal promoter sequences.
A high percentage of binding sites identified in both studies corresponded to consensus palindromic EREs [Carroll et al., 2005; Laganiere et al., 2005]. Although the palindromic ERE in the TFF-1 gene contains two non consensus substitutions, it represents a high affinity binding site [Bourdeau et al., 2004]. It is not clear whether the above studies identified only the most high affinity binding sites, or whether the majority of bona fide EREs in vivo are (near) consensus palindromic elements. Several other studies based largely on reporter gene assays of proximal promoter regions have shown that estrogen responsive promoter elements can be composed of ERE half-sites in association with binding sites for other transcription factors, or that ERα can associate with regulatory regions indirectly by interaction with other DNA-bound proteins [Sanchez et al., 2002]. Moreover, it is not clear whether the discrepancies in ERα binding sites on chromosomes 21 and 22 of Carroll et al [Carroll et al., 2005] and Laganiere et al [Laganiere et al., 2005] can be attributed to differences in filtering algorithms used to eliminate false positives or other differences in experimental protocols (such as different immunoprecipitating antibodies).
In silico hormone response element analyses
Overview of the technique
For the purpose of this review, we will refer to in silico binding site analysis as the computational techniques used to map consensus or near consensus hormone response elements on a genome wide scale with the aim of identifying potential target genes of the nuclear receptor of interest (as opposed to analysis of a particular promoter for different binding sites to identify regulatory factors). Such approaches have come to the fore with the availability of an increasing number of genome sequences. They have the advantage that they identify response elements at a genome-wide level, whereas microarray or ChIP-on-chip approaches are limited by the number of genes screened and are highly cell context specific. Algorithms are designed with specific “cut-offs” that limit sequences screened to defined distances from known 5’ ends of genes. These approaches are powerful and have identified numerous response elements in nuclear receptor target genes, as well as novel target genes (see below). However, the mapping of response elements is limited by the reliability of data used to identify the 5’ end(s) of a given gene. Transcription start sites will be thus better defined for genes whose mRNAs have been characterized using techniques to define 5’ ends (e.g. 5’ RACE, RNase protection). The approach is thus less reliable for genes whose existence is inferred from sequence analysis only.
In silico screening for hormone response elements
The nuclear receptor field has used this type of mapping approach to identify hormone response elements for ERs [Bourdeau et al., 2004; Jin et al., 2004; Jin et al., 2005; Kamalakaran et al., 2005; Tang et al., 2004], the AR [Horie-Inoue et al., 2004; Masuda et al., 2005], and the VDR [Wang et al., 2005]. One issue that must be addressed in setting up such a screen for response elements is how to define a response element consensus. The most rigorous definition of a consensus estrogen response element (ERE) is a palindromic sequence of PuGGTCA half-sites separated by 3 bp [Sanchez et al., 2002]. However, different in silico studies defined EREs slightly differently. Most used GGTCA as a consensus sequence half-site [Jin et al., 2004; Kamalakaran et al., 2005; Tang et al., 2004], one defined it as PuGGTC [Lin et al., 2004], and only one study used PuGGTCA [Bourdeau et al., 2004]. The study by Bourdeau et al [Bourdeau et al., 2004] screened the human and mouse genomes from -10 to +5kb of 5’ ends for consensus and near consensus EREs based on PuGGTCA motifs and identified conserved and non-conserved elements, often in multiple occurrences, in more than 230 estrogen-regulated human genes previously identified from microarray studies. Significantly, the screen identified distal consensus or near-consensus EREs in genes with previously characterized more degenerate promoter-proximal elements, suggesting that the latter may not be of primary importance in driving the hormonal response. The study showed that a number of distal EREs represented functional binding sites in vivo by ChIP assay. A similar screen for VDREs assigned response elements to several hundred 1,25D3 target genes identified from microarrays [Wang et al., 2005], as well as to responsive genes not picked up on arrays. For example, consensus VDREs were found in two genes encoding antimicrobial peptides, defB4 and camp, which led to the discovery that 1,25D3 is a direct regulator of antimicrobial innate immunity [Wang et al., 2004; Wang et al., 2005].
Another issue to be addressed in setting up in silico screens concerns which non-consensus nucleotide substitutions to include in response element sequences selected for in silico screening. Approaches based on nucleotide frequency matrices constructed via compilation of the relatively small number of known natural response elements can identify many variant sequences [Bajic et al., 2003; Podvinec et al., 2002]. For example, a number of putative VDREs containing multiple non-consensus nucleotides have been identified in vitamin D target genes [Toell et al., 2000]. Many of these sequences were identified in transient expression experiments with overexpressed VDR and reporter plasmids containing limited proximal promoter fragments. While matrix-based algorithms may appropriate for screening defined stretches of promoter regions, screening for numerous highly degenerate elements is impractical as it is difficult to define which elements will be functional. Systematic mutagenesis coupled to electrophoretic mobility shift assays with response element-containing oligonucleotides could be used to identify sequences that bind receptors at levels greater than an arbitrary cut-off (e.g. 25% of a consensus control element). These approaches are reliable for identifying high affinity binding sites in vitro. However, Wang et al [Wang et al., 2005] found that single nucleotide substitutions that disrupted binding in vitro were present in VDREs that functioned in vivo, as determined by positive results of ChIP assays in many instances, and response element function in reporter gene assays in another. It is likely that receptor affinity for a given response element sequence differs between naked and nucleosomal DNA. Alternatively, affinity of a receptor for a given binding site in vivo may be enhanced by association with other transcription factors bound to adjacent sites.
It should also be noted that, apart from the problems associated with identifying the sequences of all potential hormone response elements that are functional in vivo, in silico screens are limited by the fact that they focus on classical response element-mediated pathways of nuclear receptor regulated gene expression. However, it is now well established that nuclear receptors regulate gene expression by diverse mechanisms [Dostert and Heinzel, 2004; Sanchez et al., 2002], including recognition of non-canonical response elements in vivo, activation or repression of gene expression through interaction with other classes of transcription factors, and various non-genomic mechanisms.
Another limitation of in silico response element screens is the high rates of identification of false positives; i.e., nonregulated genes containing response element sequences. However, these are not easy to identify reliably. Lack of regulation of a gene in a given cell type is not sufficient to eliminate it as a potential target. Although many receptors are widely expressed, many of their target genes are regulated in a cell-specific manner. For example, promoters of the camp and defB4 antimicrobial peptide genes contain proximal, consensus VDREs [Wang et al., 2004]. Whereas expression of both defB4 and camp was induced by vitamin D in cells of epithelial origin, only camp expression was regulated in nonepithelial cells such as monocytes and neutrophils. An analysis of defB4 regulation in monocytes, would therefore have concluded that the gene represented a false positive. In addition, expression of several nuclear receptor target genes is only modestly affected by ligand treatment. Because expression of many genes is controlled by multiple signal transduction pathways, the effect of ligand may be amplified in the presence of other transducers. These effects could thus be missed if target gene expression is analyzed in the presence of ligand only.
Data generated from an in silico screen should be complemented by data from other types of screening techniques, bearing in mind the limitations of each technique. Experimental approaches to verify which sites are functional could include ChIP assays, RT/PCR, microarrays, ChIP-on-chip, with the caveat that these techniques will detect genes regulated in a given tissue/cell type, and should be interpreted in that relatively narrow context. The number of in silico hits, including consensus sequences, will generally far out number the number of elements detected in a ChIP-on-chip analysis, and together the two techniques would provide an indication of which regions of chromatin are in a transcriptionally ‘open’ state.
Support for the potential functionality of a response element identified in an in silico screen can be also provided by phylogenetic analysis and demonstration of conservation of element sequence (and location) broadly across species. Certainly, demonstration of broad conservation of a given element is strong evidence for function. However, lack of conservation is not necessarily evidence against function. For example, neither the regulation by 1,25D3, nor the consensus VDRE in the human camp gene are conserved in rodents, the element having been transposed into the gene on an Alu repeat sometime during primate evolution [Gombart et al., 2005; Wang et al., 2004], emphasizing the importance of performing phylogenetic screens as broadly as possible, and not just in species of experimental interest.
Concluding remarks
The rapid evolution of techniques for large-scale genomics studies, such as microarrays and tag-based expression screens, ChIP-on-chip, and in silico response element screens, has dramatically advanced our understanding of global gene regulation by nuclear receptors. We have moved from identification of target genes on a gene-by-gene basis to identification of several tens or hundreds of target genes in a single experiment. These approaches have uncovered new physiological actions of nuclear receptors and have revealed the pleiotropic nature of nuclear receptor signaling. However, each technique has its intrinsic strengths and limitations, and different studies are characterized by a remarkably limited overlap in experimental findings. Comparisons of results obtained for signaling by a given receptor using the same technique or across techniques are complicated by the use of different cell/tissue models, platforms, and/or different experimental or bioinformatic protocols. While standardization of protocols (both experimental and bioinformatic) for each technique will minimize to some degree the apparent discrepancies between experiments, the wide variability in experimental results underlines the importance of choosing the appropriate cell/tissue model for analysis of genomic responses underlying the physiological model under scrutiny. Ideally, any rigorous genome-wide analysis of hormone-regulated gene expression in a given experimental system would exploit multiple techniques to generate the most complete and extensively validated data set possible.
Acknowledgments
Genomics studies in the Mader and White laboratories were supported by operating grants from the Canadian Institutes of Health Research (CIHR) and Genome Canada. S.M. and J.H.W. are chercheurs-boursier of the Fonds de Recherche en Santé du Québec. L.T.M. was supported by funds from a CIHR training grant to the Montreal Centre for Experimental Therapeutics in Cancer (MCETC).
Abbreviations
- 1,25D3
1,25-dihydroxyvitamin D3
- AR
androgen receptor
- camp
cathelicidin antimicrobial peptide
- CAR
constitutive androstane receptor
- ChIP
chromatin immunoprecipitation
- cyp24
1,25-dihydroxyvitamin D3 24-hydroxylase
- defB4
defensin beta 2
- ERE
estrogen response element
- ERR
estrogen related receptor
- ERα
estrogen receptor α
- EST
expression sequence tag
- PPAR
peroxisomal proliferator-activated receptor
- PXR
pregnane X receptor
- RACE
rapid amplification of cDNA ends
- RIP140
receptor interacting protein 140
- SACO
serial analysis of chromatin occupancy
- SAGE
serial analysis of gene expression
- TFF-1
trefoil factor 1
- VDR
vitamin D receptor
- VDRE
vitamin D response element
References
- Abba M. C., Hu Y., Sun H., Drake J. A., Gaddis S., Baggerly K., Sahin A., Aldaz C. M. Gene expression signature of estrogen receptor α status in breast cancer. BMC Genomics. 2005;6 doi: 10.1186/1471-2164-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akutsu N., Lin R., Bastien Y., Bestawros A., Enepekides D. J., Black M. J., White J. H. Regulation of gene Expression by 1alpha,25-dihydroxyvitamin D3 and Its analog EB1089 under growth-inhibitory conditions in squamous carcinoma Cells. Mol Endocrinol. 2001;15:1127–39. doi: 10.1210/mend.15.7.0655. [DOI] [PubMed] [Google Scholar]
- Amuchastegui S., Daniel K. C., Adorini L. Inhibition of acute and chronic allograft rejection in mouse models by BXL-628, a nonhypercalcemic vitamin D receptor agonist. Transplantation. 2005;80:81–7. doi: 10.1097/01.tp.0000164619.49828.7a. [DOI] [PubMed] [Google Scholar]
- Bajic V. B., Tan S. L., Chong A., Tang S., Strom A., Gustafsson J. A., Lin C. Y., Liu E. T. Dragon ERE Finder version 2: A tool for accurate detection and analysis of estrogen response elements in vertebrate genomes. Nucleic Acids Res. 2003;31:3605–7. doi: 10.1093/nar/gkg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A., Dynlacht B. D. Devising transcriptional regulatory networks operating during the cell cycle and differentiation using ChIP-on-chip. Chromosome Res. 2005;13:275–88. doi: 10.1007/s10577-005-2167-y. [DOI] [PubMed] [Google Scholar]
- Bourdeau V., Deschenes J., Metivier R., Nagai Y., Nguyen D., Bretschneider N., Gannon F., White J. H., Mader S. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol. 2004;18:1411–27. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C. A., Causton H. C., Gaasterland T., Glenisson P., Holstege F. C., Kim I. F., Markowitz V., Matese J. C., Parkinson H., Robinson A., Sarkans U., Schulze-Kremer S., Stewart J., Taylor R., Vilo J., Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Carrier J. C., Deblois G., Champigny C., Levy E., Giguere V. Estrogen-related receptor α (ERRalpha) is a transcriptional regulator of apolipoprotein A-IV and controls lipid handling in the intestine. J Biol Chem. 2004;279:52052–8. doi: 10.1074/jbc.M410337200. [DOI] [PubMed] [Google Scholar]
- Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Cheung V. G., Morley M., Aguilar F., Massimi A., Kucherlapati R., Childs G. Making and reading microarrays. Nat Genet. 1999;21:15–9. doi: 10.1038/4439. [DOI] [PubMed] [Google Scholar]
- Choi Y., Pinto M. Estrogen receptor β in breast cancer: associations between ERbeta, hormonal receptors, and other prognostic biomarkers. Appl Immunohistochem Mol Morphol. 2005;13:19–24. doi: 10.1097/00129039-200503000-00004. [DOI] [PubMed] [Google Scholar]
- Datson N. A., van der Perk J., de Kloet E. R., Vreugdenhil E. Identification of corticosteroid-responsive genes in rat hippocampus using serial analysis of gene expression. Eur J Neurosci. 2001;14:675–89. doi: 10.1046/j.0953-816x.2001.01685.x. [DOI] [PubMed] [Google Scholar]
- Dostert A., Heinzel T. Negative glucocorticoid receptor response elements and their role in glucocorticoid action. Curr Pharm Des. 2004;10:2807–16. doi: 10.2174/1381612043383601. [DOI] [PubMed] [Google Scholar]
- Enerback C., Porter D. A., Seth P., Sgroi D., Gaudet J., Weremowicz S., Morton C. C., Schnitt S., Pitts R. L., Stampl J., Barnhart K., Polyak K. Psoriasin expression in mammary epithelial cells in vitro and in vivo. Cancer Res. 2002;62:43–7. [PubMed] [Google Scholar]
- Frederiksen K. S., Wulff E. M., Sauerberg P., Mogensen J. P., Jeppesen L., Fleckner J. Prediction of PPAR-α ligand-mediated physiological changes using gene expression profiles. J Lipid Res. 2004;45:592–601. doi: 10.1194/jlr.M300239-JLR200. [DOI] [PubMed] [Google Scholar]
- Glinsky G. V., Higashiyama T., Glinskii A. B. Classification of human breast cancer using gene expression profiling as a component of the survival predictor algorithm. Clin Cancer Res. 2004;10:2272–83. doi: 10.1158/1078-0432.ccr-03-0522. [DOI] [PubMed] [Google Scholar]
- Gombart A. F., Borregaard N., Koeffler H. P. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3. Faseb J. 2005;19:1067–77. doi: 10.1096/fj.04-3284com. [DOI] [PubMed] [Google Scholar]
- Hanekamp E. E., Gielen S. C., Smid-Koopman E., Kuhne L. C., de Ruiter P. E., Chadha-Ajwani S., Brinkmann A. O., Grootegoed J. A., Burger C. W., Huikeshoven F. J., Blok L. J. Consequences of loss of progesterone receptor expression in development of invasive endometrial cancer. Clin Cancer Res. 2003;9:4190–9. [PubMed] [Google Scholar]
- Haqq C., Li R., Khodabakhsh D., Frolov A., Ginzinger D., Thompson T., Wheeler T., Carroll P., Ayala G. Ethnic and racial differences in prostate stromal estrogen receptor α. Prostate. 2005;65:101–9. doi: 10.1002/pros.20272. [DOI] [PubMed] [Google Scholar]
- Harbers M., Carninci P. Tag-based approaches for transcriptome research and genome annotation. Nat Methods. 2005;2:495–502. doi: 10.1038/nmeth768. [DOI] [PubMed] [Google Scholar]
- Hartley D. P., Dai X., He Y. D., Carlini E. J., Wang B., Huskey S. E., Ulrich R. G., Rushmore T. H., Evers R., Evans D. C. Activators of the rat pregnane X receptor differentially modulate hepatic and intestinal gene expression. Mol Pharmacol. 2004;65:1159–71. doi: 10.1124/mol.65.5.1159. [DOI] [PubMed] [Google Scholar]
- Helms M. W., Packeisen J., August C., Schittek B., Boecker W., Brandt B. H., Buerger H. First evidence supporting a potential role for the BMP/SMAD pathway in the progression of oestrogen receptor-positive breast cancer. J Pathol. 2005;206:366–76. doi: 10.1002/path.1785. [DOI] [PubMed] [Google Scholar]
- Hershberger P. A., Modzelewski R. A., Shurin Z. R., Rueger R. M., Trump D. L., Johnson C. S. 1,25-Dihydroxycholecalciferol (1,25-D3) inhibits the growth of squamous cell carcinoma and down-modulates p21(Waf1/Cip1) in vitro and in vivo. Cancer Res. 1999;59:2644–9. [PubMed] [Google Scholar]
- Horie-Inoue K., Bono H., Okazaki Y., Inoue S. Identification and functional analysis of consensus androgen response elements in human prostate cancer cells. Biochem Biophys Res Commun. 2004;325:1312–7. doi: 10.1016/j.bbrc.2004.10.174. [DOI] [PubMed] [Google Scholar]
- Impey S., McCorkle S. R., Cha-Molstad H., Dwyer J. M., Yochum G. S., Boss J. M., McWeeney S., Dunn J. J., Mandel G., Goodman R. H. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–54. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Inadera H., Hashimoto S., Dong H. Y., Suzuki T., Nagai S., Yamashita T., Toyoda N., Matsushima K. WISP-2 as a novel estrogen-responsive gene in human breast cancer cells. Biochem Biophys Res Commun. 2000;275:108–14. doi: 10.1006/bbrc.2000.3276. [DOI] [PubMed] [Google Scholar]
- Jin V. X., Sun H., Pohar T. T., Liyanarachchi S., Palaniswamy S. K., Huang T. H., Davuluri R. V. ERTargetDB: an integral information resource of transcription regulation of estrogen receptor target genes. J Mol Endocrinol. 2005;35:225–30. doi: 10.1677/jme.1.01839. [DOI] [PubMed] [Google Scholar]
- Jin V. X., Leu Y. W., Liyanarachchi S., Sun H., Fan M., Nephew K. P., Huang T. H., Davuluri R. V. Identifying estrogen receptor α target genes using integrated computational genomics and chromatin immunoprecipitation microarray. Nucleic Acids Res. 2004;32:6627–35. doi: 10.1093/nar/gkh1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalakaran S., Radhakrishnan S. K., Beck W. T. Identification of estrogen-responsive genes using a genome-wide analysis of promoter elements for transcription factor binding sites. J Biol Chem. 2005;280:21491–7. doi: 10.1074/jbc.M409176200. [DOI] [PubMed] [Google Scholar]
- Krishnan A. V., Shinghal R., Raghavachari N., Brooks J. D., Peehl D. M., Feldman D. Analysis of vitamin D-regulated gene expression in LNCaP human prostate cancer cells using cDNA microarrays. Prostate. 2004;59:243–51. doi: 10.1002/pros.20006. [DOI] [PubMed] [Google Scholar]
- Kristensen V. N., Sorlie T., Geisler J., Langerod A., Yoshimura N., Karesen R., Harada N., Lonning P. E., Borresen-Dale A. L. Gene expression profiling of breast cancer in relation to estrogen receptor status and estrogen-metabolizing enzymes: clinical implications. Clin Cancer Res. 2005;11:878s–83s. [PubMed] [Google Scholar]
- Laganiere J., Deblois G., Lefebvre C., Bataille A. R., Robert F., Giguere V. From the Cover: Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A. 2005;102:11651–6. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmann T., Moehle C., Mauerer R., Scharl M., Liebisch G., Zahn A., Stremmel W., Schmitz G. Loss of detoxification in inflammatory bowel disease: dysregulation of pregnane X receptor target genes. Gastroenterology. 2004;127:26–40. doi: 10.1053/j.gastro.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Lee S. S., Tian L., Lee W. S., Cheung W. T. Application of fluorescent differential display and peroxisome proliferator-activated receptor (PPAR) α-null mice to analyze PPAR target genes. Methods Enzymol. 2002;357:214–40. doi: 10.1016/s0076-6879(02)57681-3. [DOI] [PubMed] [Google Scholar]
- Lee P., Rosen D. G., Zhu C., Silva E. G., Liu J. Expression of progesterone receptor is a favorable prognostic marker in ovarian cancer. Gynecol Oncol. 2005;96:671–7. doi: 10.1016/j.ygyno.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Olson P., Evans R. M. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144:2201–7. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- Li X., Zheng W., Li Y. C. Altered gene expression profile in the kidney of vitamin D receptor knockout mice. J Cell Biochem. 2003;89:709–19. doi: 10.1002/jcb.10547. [DOI] [PubMed] [Google Scholar]
- Lin C. Y., Strom A., Vega V. B., Kong S. L., Yeo A. L., Thomsen J. S., Chan W. C., Doray B., Bangarusamy D. K., Ramasamy A., Vergara L. A., Tang S., Chong A., Bajic V. B., Miller L. D., Gustafsson J. A., Liu E. T. Discovery of estrogen receptor α target genes and response elements in breast tumor cells. Genome Biol. 2004b;5 doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Nagai Y., Sladek R., Bastien Y., Ho J., Petrecca K., Sotiropoulou G., Diamandis E. P., Hudson T. J., White J. H. Expression profiling in squamous carcinoma cells reveals pleiotropic effects of vitamin D3 analog EB1089 signaling on cell proliferation, differentiation, and immune system regulation. Mol Endocrinol. 2002;16:1243–56. doi: 10.1210/mend.16.6.0874. [DOI] [PubMed] [Google Scholar]
- Lin R., White J. H. The pleiotropic actions of vitamin D. Bioessays. 2004a;26:21–8. doi: 10.1002/bies.10368. [DOI] [PubMed] [Google Scholar]
- Lipshutz R. J., Fodor S. P., Gingeras T. R., Lockhart D. J. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–4. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Liu M., Lee M. H., Cohen M., Bommakanti M., Freedman L. P. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes Dev. 1996;10:142–53. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- Masuda K., Werner T., Maheshwari S., Frisch M., Oh S., Petrovics G., May K., Srikantan V., Srivastava S., Dobi A. Androgen receptor binding sites identified by a GREF_GATA model. J Mol Biol. 2005;353:763–71. doi: 10.1016/j.jmb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Moggs J. G. Molecular responses to xenoestrogens: mechanistic insights from toxicogenomics. Toxicology. 2005;213:177–93. doi: 10.1016/j.tox.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Munker R., Kobayashi T., Elstner E., Norman A. W., Uskokovic M., Zhang W., Andreeff M., Koeffler H. P. A new series of vitamin D analogs is highly active for clonal inhibition, differentiation, and induction of WAF1 in myeloid leukemia. Blood. 1996;88:2201–9. [PubMed] [Google Scholar]
- Podvinec M., Kaufmann M. R., Handschin C., Meyer U. A. NUBIScan, an in silico approach for prediction of nuclear receptor response elements. Mol Endocrinol. 2002;16:1269–79. doi: 10.1210/mend.16.6.0851. [DOI] [PubMed] [Google Scholar]
- Porter D., Yao J., Polyak K. SAGE and related approaches for cancer target identification. Drug Discov Today. 2006;11:110–8. doi: 10.1016/S1359-6446(05)03694-9. [DOI] [PubMed] [Google Scholar]
- Prudencio J., Akutsu N., Benlimame N., Wang T., Bastien Y., Lin R., Black M. J., Alaoui-Jamali M. A., White J. H. Action of low calcemic 1alpha,25-dihydroxyvitamin D3 analogue EB1089 in head and neck squamous cell carcinoma. J Natl Cancer Inst. 2001;93:745–53. doi: 10.1093/jnci/93.10.745. [DOI] [PubMed] [Google Scholar]
- Quinn D. I., Henshall S. M., Sutherland R. L. Molecular markers of prostate cancer outcome. Eur J Cancer. 2005;41:858–87. doi: 10.1016/j.ejca.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Volkert T. L., Wilson C. J., Bell S. P., Young R. A. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Robert-Nicoud M., Flahaut M., Elalouf J. M., Nicod M., Salinas M., Bens M., Doucet A., Wincker P., Artiguenave F., Horisberger J. D., Vandewalle A., Rossier B. C., Firsov D. Transcriptome of a mouse kidney cortical collecting duct cell line: effects of aldosterone and vasopressin. Proc Natl Acad Sci U S A. 2001;98:2712–6. doi: 10.1073/pnas.051603198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld J. M., Vargas R., Jr., Xie W., Evans R. M. Genetic profiling defines the xenobiotic gene network controlled by the nuclear receptor pregnane X receptor. Mol Endocrinol. 2003;17:1268–82. doi: 10.1210/me.2002-0421. [DOI] [PubMed] [Google Scholar]
- Ruijter J. M., Van Kampen A. H., Baas F. Statistical evaluation of SAGE libraries: consequences for experimental design. Physiol Genomics. 2002;11:37–44. doi: 10.1152/physiolgenomics.00042.2002. [DOI] [PubMed] [Google Scholar]
- Saha S., Sparks A. B., Rago C., Akmaev V., Wang C. J., Vogelstein B., Kinzler K. W., Velculescu V. E. Using the transcriptome to annotate the genome. Nat Biotechnol. 2002;20:508–12. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- Sanchez R., Nguyen D., Rocha W., White J. H., Mader S. Diversity in the mechanisms of gene regulation by estrogen receptors. Bioessays. 2002;24:244–54. doi: 10.1002/bies.10066. [DOI] [PubMed] [Google Scholar]
- Schena M., Shalon D., Davis R. W., Brown P. O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–70. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Sellheyer K., Belbin T. J. DNA microarrays: from structural genomics to functional genomics. The applications of gene chips in dermatology and dermatopathology. J Am Acad Dermatol. 2004;51:681–92; quiz 693-6. doi: 10.1016/j.jaad.2004.03.038. [DOI] [PubMed] [Google Scholar]
- Seth P., Krop I., Porter D., Polyak K. Novel estrogen and tamoxifen induced genes identified by SAGE (Serial Analysis of Gene Expression) Oncogene. 2002;21:836–43. doi: 10.1038/sj.onc.1205113. [DOI] [PubMed] [Google Scholar]
- Swami S., Raghavachari N., Muller U. R., Bao Y. P., Feldman D. Vitamin D growth inhibition of breast cancer cells: gene expression patterns assessed by cDNA microarray. Breast Cancer Res Treat. 2003;80:49–62. doi: 10.1023/A:1024487118457. [DOI] [PubMed] [Google Scholar]
- Tang S., Tan S. L., Ramadoss S. K., Kumar A. P., Tang M. H., Bajic V. B. Computational method for discovery of estrogen responsive genes. Nucleic Acids Res. 2004;32:6212–7. doi: 10.1093/nar/gkh943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toell A., Polly P., Carlberg C. All natural DR3-type vitamin D response elements show a similar functionality in vitro. Biochem J. 2000;352 Pt 2:301–9. [PMC free article] [PubMed] [Google Scholar]
- Ueda A., Hamadeh H. K., Webb H. K., Yamamoto Y., Sueyoshi T., Afshari C. A., Lehmann J. M., Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- van Bakel H., Holstege F. C. In control: systematic assessment of microarray performance. EMBO Rep. 2004;5:964–9. doi: 10.1038/sj.embor.7400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu V. E., Zhang L., Vogelstein B., Kinzler K. W. Serial analysis of gene expression. Science. 1995;270:484–7. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Wang T. T., Nestel F. P., Bourdeau V., Nagai Y., Wang Q., Liao J., Tavera-Mendoza L., Lin R., Hanrahan J. W., Mader S., White J. H. Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J Immunol. 2004;173:2909–12. doi: 10.4049/jimmunol.173.5.2909. [DOI] [PubMed] [Google Scholar]
- Wang T. T., Tavera-Mendoza L. E., Laperriere D., Libby E., MacLeod N. B., Nagai Y., Bourdeau V., Konstorum A., Lallemant B., Zhang R., Mader S., White J. H. Large-scale in silico and microarray-based identification of direct 1,25-dihydroxyvitamin D3 target genes. Mol Endocrinol. 2005;19:2685–95. doi: 10.1210/me.2005-0106. [DOI] [PubMed] [Google Scholar]
- White J. H. Profiling 1,25-dihydroxyvitamin D3-regulated gene expression by microarray analysis. J Steroid Biochem Mol Biol. 2004;89-90:239–44. doi: 10.1016/j.jsbmb.2004.03.074. [DOI] [PubMed] [Google Scholar]
- Yoshida N., Omoto Y., Inoue A., Eguchi H., Kobayashi Y., Kurosumi M., Saji S., Suemasu K., Okazaki T., Nakachi K., Fujita T., Hayashi S. Prediction of prognosis of estrogen receptor-positive breast cancer with combination of selected estrogen-regulated genes. Cancer Sci. 2004;95:496–502. doi: 10.1111/j.1349-7006.2004.tb03239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li P., Bao J., Nicosia S. V., Wang H., Enkemann S. A., Bai W. Suppression of death receptor-mediated apoptosis by 1,25-dihydroxyvitamin D3 revealed by microarray analysis. J Biol Chem. 2005;280:35458–68. doi: 10.1074/jbc.M506648200. [DOI] [PMC free article] [PubMed] [Google Scholar]