Abstract
Zeatin is the most active and ubiquitous of the naturally occurring cytokinins. The O-glucoside of zeatin, found in all plants examined, is considered to be important in cytokinin transport, storage, and protection against cytokinin oxidases. The enzyme UDPglucose:zeatin O-glucosyltransferase (EC 2.4.1.203) was previously isolated from Phaseolus lunatus seeds. Immunoscreening of an expression library with monospecific antibody resulted in the isolation of a cDNA encoding the enzyme. The recombinant protein efficiently converts labeled zeatin to O-glucosylzeatin and has properties similar to the native enzyme. The cDNA of 1.5 kb contains an ORF encoding a 51.4-kDa polypeptide of 459 amino acids. The sequence is unique based on a blast search of data bases. The genomic sequence, isolated with PCR using specific primers based on the cDNA sequence, does not contain introns. The cloning of this gene provides the tools for further study of the regulation of cytokinin metabolism and analysis of the precise role of O-glucosylzeatin in plant development.
Cytokinins are plant hormones mediating cell division and differentiation (1). Naturally occurring cytokinins are adenine derivatives with a substituted N6-isoprenoid side chain (2). Zeatin is a highly active and ubiquitous cytokinin first identified in maize (3). Based on studies of the Agrobacterium gene ipt (4), the de novo synthesis of cytokinins in microbial systems involves the formation of Δ2-isopentenyladenosine monophosphate from Δ2-isopentenylpyrophosphate and AMP, catalyzed by an isopentenyltransferase. Subsequent hydroxylation of the N6-side chain leads to the formation of zeatin-related cytokinins. It is possible that a similar pathway exists in plants (5, 6). Conversion of zeatin to its riboside and ribonucleotides probably uses enzymes common to adenine metabolic pathways (7). The modifications of the zeatin side chain include reduction to dihydrozeatin, glycosylation to O-glucosyl- and O-xylosyl-zeatin, isomerization to and from cis-zeatin, and cleavage to form adenine (8, 9). The corresponding enzymes zeatin reductase, O-glucosyltransferase, O-xylosyltransferase, isomerase, and oxidases have been identified and appear to be highly specific for cytokinin substrates (7).
Zeatin O-glucoside (10) occurs in all plants examined (8) and is thought to be important in transport (11), storage (11), and protection against zeatin oxidases (12, 13). Detection of O-glucosylzeatin in stem exudates (11), the ready conversion back to zeatin by β-glucosidases (14–16), and the inability of cytokinin oxidases to degrade the O-glucosylated side chain (13) lend support to the suggested roles of O-glucosylzeatin. We have isolated and characterized the UDPglucose:zeatin O-β-d-glucosyltransferase (EC 2.4.1.203) from immature seeds of Phaseolus lunatus (17). The enzyme can also use UDPxylose as the sugar donor to form O-xylosylzeatin but at a much lower affinity. Thus far, this is the only cytokinin O-glucosyltransferase characterized. A mAb recognizing the enzyme was generated (18). In this article, we report the isolation of a cDNA encoding the enzyme by use of the antibody to screen a cDNA expression library. Based on the nucleotide information of the cDNA, the genomic sequence was isolated by using PCR and inverse PCR. One of the interesting features of the gene is the absence of introns. Genetic manipulation leading to controlled expression of the enzyme should allow a more precise assessment of the function of O-glucosylated cytokinins in regulating growth and development.
MATERIALS AND METHODS
Plant Materials.
Immature seeds of P. lunatus cv. Kingston were the source of mRNA for the construction of cDNA libraries. DNA and RNA were extracted from other plant parts for Northern and Southern blot analyses as specified.
Construction of cDNA Expression Library and Screening with mAbs.
Total RNA was isolated from immature seeds of P. lunatus with Tri Reagent (Molecular Research Center, Cincinnati) by following the manufacturer’s protocols with modifications for samples with a high content of polysaccharides. mRNA was further enriched with the PolyATtract mRNA isolation system according to the manufacturer’s instructions (Promega). An expression library was constructed by using the SuperScript λ system for cDNA synthesis and λ ZipLox cloning (BRL). First-strand synthesis was primed with a NotI-oligonucleotide–d(T) primer-adapter. After second-strand synthesis, SalI adapters were ligated to the cDNA, which was subsequently digested with NotI for directional cloning of cDNA into the λ ZipLox expression vector (BRL). The constructed λ cDNA library was packaged by following the manufacturer’s instructions (BRL λ packaging system). The λ ZipLox cDNA library was plated with Escherichia coli Y1090(ZL) on top agar and incubated at 37°C for 3–4 h. The plates were then overlaid with nitrocellulose filters, which had been presoaked with 10 mM isopropyl β-d-thiogalactoside and dried, and incubated at 37°C overnight. Filters were marked for orientation, removed from the plates, and washed briefly in TBS. The membranes were blocked in TBS containing 2% nonfat dry milk for 1–2 h at room temperature with constant agitation. Filters were then incubated with blocking solution containing primary antibody for 2 h at 30°C. Filters were washed for three 5-min periods in blocking solution. Secondary antibody (rabbit anti-mouse immunoglobulin conjugated to an alkaline phosphatase, Jackson ImmunoResearch) in blocking solution was applied to the filters for 2 h at 30°C. Filters were washed and the positive clones identified with an alkaline phosphatase substrate kit (Vector Laboratories). Immunopositive plaques were removed, replated, and rescreened until pure plaques were obtained.
Subcloning and Sequencing.
The plasmid pZL1 containing the insert was excised in vivo by infecting DH10B(ZIP) E. coli. pZL1 clones were purified with a Qiagen plasmid midi kit. Clones were sequenced by the Central Services Laboratory (Center for Gene Research and Biotechnology, Oregon State Univ.) using an Applied Biosystems model 370A DNA sequence analyzer. Primers synthesized by the Central Service Lab on an Applied Biosystems DNA Synthesizer were used for sequencing the inner portions of the clones.
Isolation of Recombinant Proteins.
To obtain recombinant proteins, purified plasmids were transformed into Epicurian Coli BL21(DE3) pLysS-competent cells (Stratagene) by the recommended protocol. Colonies were selected on Amp-LB plates. An individual colony was grown overnight in SOC-Amp medium and 0.5 ml of the culture was used to inoculate 50 ml of SOC-Amp medium. After cells were allowed to grow for 3–4 h (final OD595 = 0.8–1.0), they were induced with isopropyl β-d-thiogalactoside (0.5 ml of a 0.5 M stock solution) for 4 h. Cells were collected, frozen at −80°C overnight, and lysed the next day by thawing and resuspending in 0.5 ml of 0.2 M Tris⋅HCl (pH 7.5) containing lysozyme (1 mg/ml). DNase I (1 μg/10 μl of sample) was added after a 10-min cycle of freezing (−80°C) and thawing. Soluble proteins were collected after centrifugation. The recombinant protein was further purified on a Blue Sepharose 6B affinity column (17), followed by DEAE-anion-exchange chromatography (Bio-Rad Biologic System). Sample was applied to the DEAE column in buffer A (0.02 M Tris⋅HCl, pH 7.5/0.5 mM EDTA) and the enzyme was eluted with a linear gradient (0–50% over 20 min) of buffer B (buffer A/1.0 M NaCl) at 1 ml/min.
Enzyme Assays and Analysis of Reaction Products.
Enzyme activity was determined as reported (17). Briefly, 14C-labeled cytokinins (specific activity of 24 mCi/mmol; 1 Ci = 37 GBq), glycosyl donor (3 mM UDPglucose or UDPxylose ), MgCl2 (0.05 M) and ATP (0.5 mM) in 100 mM Tris⋅HCl (pH 8.0) were incubated with the recombinant protein. Reaction products were separated by HPLC (17).
Liquid Chromatography–MS.
After an enzyme assay with zeatin as the substrate and separation of products by HPLC, fractions corresponding to the elution of O-glucosylzeatin were subjected to liquid chromatography–MS analyses. The chromatography system consisted of a Perkin-Elmer Applied Biosystems model 140B syringe pump, a Rheodyne model 8125 injector with 5-μm sample loop, and a 5-μm particle Spherisorb octadecylsilane (ODS) column (2.1 mm × 100 mm). Compounds were separated at a linear gradient of 5–60% acetonitrile in 0.1% trifluoroacetic acid in 25 min at a flow rate of 200 μl/min. A Perkin-Elmer Sciex API III+ triple quadrupole Ionspray Mass Spectrometer was used, with an atmospheric pressure chemical ionization source.
Isolation of Genomic Sequence of Zeatin O-Glucosyltransferase from P. lunatus.
The genomic sequence was isolated by standard and inverse PCR (19). DNA was isolated from P. lunatus cv. Kingston by using a modified CTAB (hexadecyl trimethylammonium bromide) method (20). To obtain the genomic sequence of the expressed region (cDNA), standard PCR reactions were performed with pairs of primers based on the sequence of the cDNA. To obtain the genomic sequence upstream of the cDNA by using inverse PCR, the genomic DNA was digested with HindIII. After digestion, the restriction enzyme was inactivated by heating the sample to 75°C for 10 min. After dilution, T4 DNA ligase (Promega) was added to allow intramolecular ligation (circularization) at 15°C for 24 h. The DNA was precipitated and PCR was performed with primers designed to amplify regions outside the cDNA. The products obtained from PCR and inverse PCR reactions were analyzed on a 1% Sea-Plaque gel. Bands of interest were excised and DNA was purified with a Qiaex II gel extraction kit (Qiagen, Chatsworth, CA). The products were ligated into a pGem-T vector (Promega) for sequencing.
Northern Blot Analysis.
Poly(A)+ RNA was isolated from various tissues as described for cDNA library construction. mRNA (4 μg) was separated on a 1.2% formaldehyde gel (21). Northern blotting (RNA capillary transfer) to the Zeta-Probe GT membrane (Bio-Rad) was performed according to the manufacturer’s instructions. The cDNA [minus the poly(A) portion] was used to synthesize an [α-32P]dCTP-labeled probe with Ready-To-Go DNA labeling beads (Pharmacia). Membranes were prehybridized in a solution of 0.25 M Na2HPO4, pH 7.2/7% SDS/1 mM EDTA at 65°C for 30 min. The prehybridization solution was replaced with fresh solution plus the labeled probe. After 24 h at 65°C, membranes were washed twice in 1× SSC at room temperature for 10 min, followed by washing for three 30-min periods in 1× SSC with 0.1% SDS at 65°C. The membranes were then exposed to x-ray films at −80°C.
Southern Blot Analysis.
DNA was extracted from young leaves of P. lunatus by the modified CTAB procedure (20). DNA (30 μg) was digested with restriction enzymes, separated on a 1.1% gel, and transferred to a Zeta-Probe GT membrane. Hybridization was performed as for the Northern blotting.
RESULTS
Isolation and Authentication of cDNA Clones.
Approximately 106 plaques were screened with the mAb and nine immunopositive phage plaques were selected. The biological activity and the antigenicity of the recombinant proteins were assessed by enzyme assays and Western blot analysis, respectively.
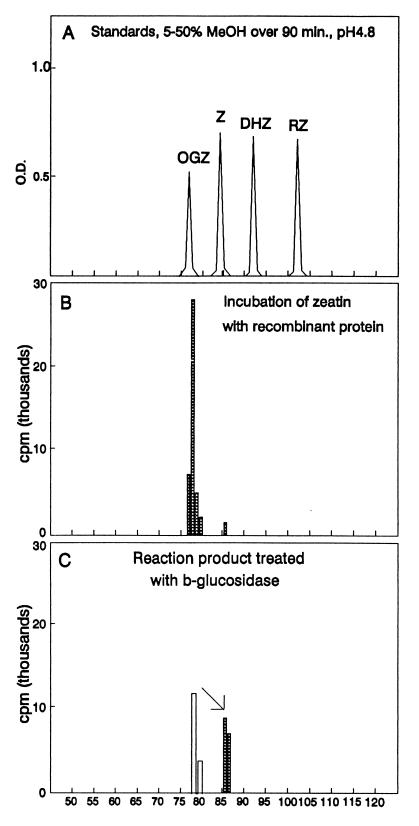
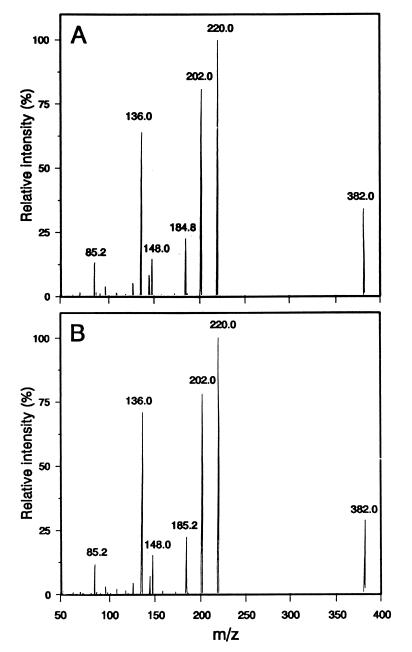
Recombinant protein from two clones, 21G and 27G, efficiently converted [14C]zeatin to O-[14C]glucosylzeatin (Fig. 1B) when UDPglucose was added as the glucosyl donor. The authenticity of the labeled product as O-glucosylzeatin was confirmed by treatment with β-glucosidase, resulting in conversion back to zeatin (Fig. 1C), and by an identical mass spectrum to authentic O-β-d-glucosylzeatin standard (Fig. 2).
Figure 1.
Separation of cytokinin standards and products of enzymatic reactions by HPLC as reported (17). (A) Cytokinin standards. (B) Products of enzymatic reaction after incubating soluble recombinant protein from 10 ml of cell culture with [14C]zeatin (0.8 nmol; 45,000 cpm), UDPglucose (3 mM), ATP (0.5 mM), and MgCl2 (0.05 M) for 2 h. (C) Reaction product (open bar) obtained from B treated with β-glucosidase for 2 h as reported (22), resulting in zeatin (hatched bar).
Figure 2.
Mass spectra of standard of O-β-d-glucosylzeatin (A) and product obtained from incubating zeatin with recombinant protein (B).
The substrate specificity of the recombinant protein was tested using both the crude preparation and the more purified form (Table 1). Similar to the native zeatin O-glucosyltransferase of P. lunatus, the recombinant proteins encoded by clones 21G and 27G were able to use UDPxylose as the donor to form O-xylosylzeatin but at a much lower affinity than UDPglucose (17). Interestingly, the recombinant protein could also use dihydrozeatin as substrate, resulting in the formation of O-glucosyldihydrozeatin, but the relative affinity to dihydrozeatin was much lower than to zeatin. Only when a large quantity of the recombinant protein was used were detectable amounts of O-glucosyldihydrozeatin formed. As expected based on the properties of the native enzyme isolated from P. lunatus seeds, ribosylzeatin and cis-zeatin were not substrates to the recombinant protein. These results indicate that the selected cDNAs encode the zeatin O-glucosyltransferase.
Table 1.
Relative affinity of recombinant protein to cytokinin and glycosyl substrates
| Exp. | Substrates | Product, cpm (%) | Substrate left, cpm (%) |
|---|---|---|---|
| A | Z + UDPG | OGZ | Z |
| 39,715 (100) | 0 (0) | ||
| Z + UDPX | OXZ | Z | |
| 4,956 (18) | 23,078 (82) | ||
| B | Z + UDPG | OGZ | Z |
| 3,731 (14) | 23,115 (86) | ||
| DZ + UDPG | OGDHZ | DHZ | |
| 329 (1) | 24,178 (99) |
DHZ, dihydrozeatin; OGZ, O-glucosylzeatin; OGDHZ, O-glucosyldihydrozeatin; OXZ, O-xylosylzeatin; Z, trans-zeatin; UDPG, UDPglucose; UDPX, UDPxylose. For experiment A, 200 μl of supernatant from lysed cells containing the recombinant protein was incubated with approximately 1 nmol of 14C-labeled zeatin for 2 h, and the reaction products were separated by HPLC. UDPglucose as the glycosyl donor resulted in zeatin depletion, and UDPxylose serving as the glycosyl donor resulted in much lower formation of the corresponding product, OXZ. For experiment B, DEAE-purified proteins (4 μg) were incubated with approximately 0.5 nmol of [14C]cytokinins for 2 h. Only a trace of O-glucosyldihydrozeatin was formed.
Sequence of the cDNA.
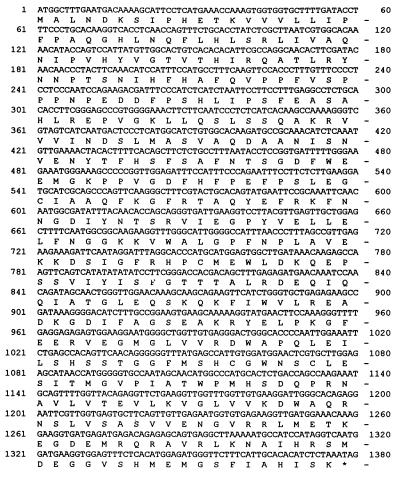
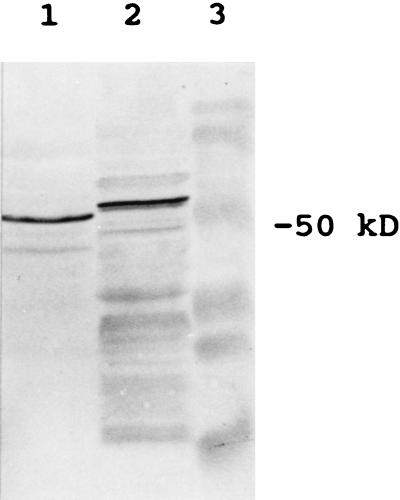
The nucleotide sequences of clones 21G and 27G were determined. The two clones were found to be identical. The cDNA is 1.5 kb long containing an ORF encoding a polypeptide of 459 amino acids (Fig. 3) with the molecular mass of 51.4 kDa. The cDNA includes a 5′ untranslated region of 32 bp and, with the α-peptide of the cloning vector, is expected to produce a fusion protein of 56 kDa. The antigenic protein of the selected clones was of the expected size (Fig. 4). A blast search of GenBank and EMBL data bases indicates that the sequence is unique, although short stretches of homology to UDPglucose:flavanoid and UDPglucose:indole acetate glucosyltransferases are present.
Figure 3.
Nucleotide and deduced amino acid sequence of ORF of zeatin O-glucosyltransferase cDNA isolated from P. lunatus seeds.
Figure 4.
Immunoblots after SDS/PAGE of native enzyme and recombinant proteins. Lanes: 1, native enzyme; 2, recombinant fusion protein.
Genomic Sequence of Zeatin O-Glucosyltransferase of P. lunatus.
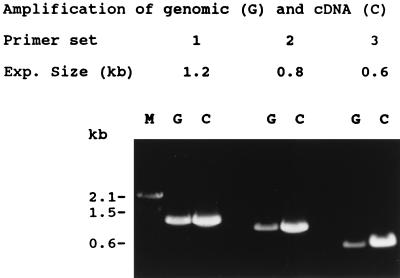
Genomic DNA of P. lunatus was isolated and amplified with three pairs of primers based on the sequence of the cDNA. PCR products obtained from genomic DNA were identical in length to those when cDNA was used as the template (Fig. 5). The genomic PCR products were cloned into pGEM-T vector and sequenced. The nucleotide sequences of genomic products were identical to those of the corresponding regions of the cDNA, indicating this gene has no introns. We have designated the gene ZOG1, for zeatin O-glucosyltransferase.
Figure 5.
PCR products, using three sets of primers, obtained from genomic (G) and cDNA (C) templates. M, size marker.
To obtain the 5′ upstream region of the gene, inverse PCR was used. Genomic DNA was digested with HindIII, recircularized, and amplified with primers based on the cDNA sequence. The PCR product was cloned into pGem-T vector and the sequence was determined. A region of approximately 200 bp upstream from the cDNA clone containing a TATA-like region was isolated.
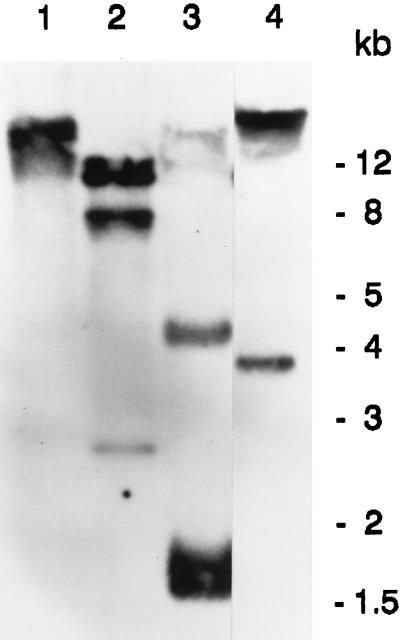
In Southern blot analyses, digestion with the enzymes KpnI, XbaI, HindIII, and BamHI, which do not cut the cDNA probe, revealed two or three fragments (Fig. 6). The results indicate that there are at least two homologous genes in P. lunatus.
Figure 6.
Southern blot analysis of P. lunatus DNA digested with KpnI (lane 1), XbaI (lane 2), HindIII (lane 3), and BamHI (lane 4), and hybridized with the labeled cDNA 21G.
Northern Blot Analysis.
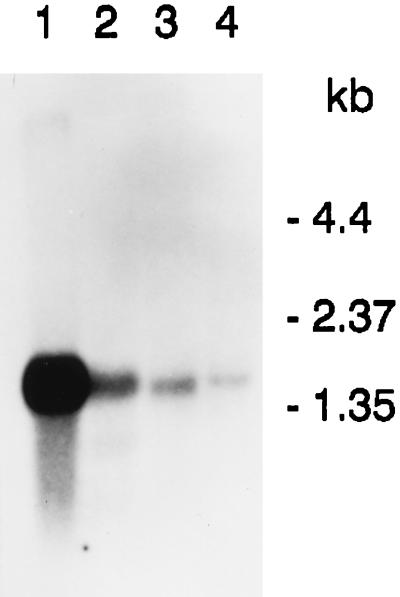
To determine the expression of the zeatin O-glucosyltransferase, mRNA was isolated from roots, leaves, and seeds of P. lunatus cv. Kingston and probed with radiolabeled 21G. The level of mRNA in young seeds smaller than 5 mm (Fig. 7, lane 1) was very high. Larger seeds, 10 mm long, had much reduced mRNA (lane 2). In vegetative tissues, roots, and leaves, the gene was expressed at a low level (lanes 3 and 4).
Figure 7.
Northern blot analysis of mRNA from P. lunatus cv. Kingston tissues hybridized with labeled cDNA 21G. Lanes: 1, 5-mm seeds; 2, 10-mm seeds; 3, roots; 4, leaves.
DISCUSSION
The recombinant protein produced by the selected cDNA has properties very similar to the native enzyme isolated from P. lunatus seeds (17). Like the native enzyme, it converts zeatin to O-glucosylzeatin when UDPglucose is supplied and forms O-xylosylzeatin in the presence of UDPxylose but with lower efficiency. Both the recombinant and the native protein do not recognize ribosylzeatin and cis-zeatin as substrates. With the availability of large quantity of the recombinant protein, however, we were able to determine that dihydrozeatin can be converted to its O-glucoside, but the affinity of the enzyme to dihydrozeatin is much lower than to zeatin (Table 1). Previously, because of limiting amounts of native enzyme available for assays, we were unable to detect the formation of O-glucosyldihydrozeatin from dihydrozeatin. Thus the substrate recognition by the enzyme is highly specific. O-Glucosyl derivatives of zeatin, dihydrozeatin, and their ribosides have been found in plant tissues (14). Because the zeatin O-glucosyltransferase reported herein does not catalyze glucosylation of zeatin riboside, either ribosylation occurs after O-glucosylation or perhaps other enzymes recognizing cytokinin ribosides exist.
Based on the sequence of the cDNA, primers were designed and PCR was used to isolate the genomic sequence. The products of the genomic PCR indicate that at least one of the genes encoding zeatin O-glucosyltransferase does not contain any intron. This is probably the gene that is highly expressed in seeds, because the sequence is identical to the seed-derived cDNA. The sequences of the other gene(s) remain to be determined. The significance of the absence of introns is not known.
Compared with reproductive organs, enzyme activity was low in vegetative tissues, based on incubation studies with labeled zeatin and equal amounts of plant material (unpublished result). The Northern blot analyses confirmed that the ZOG gene is indeed expressed only at relatively low levels in roots and leaves. Nevertheless, gene expression in roots lends support to the assumption that O-glucosylzeatin found in xylem sap is derived from roots (11). Gene expression is high in young seeds as predicted from Western blot analyses of proteins from seeds of different sizes. Because only the cDNA obtained from seed was used, it is not possible at present to distinguish gene-specific messages.
Very few genes specific to cytokinins have been isolated although numerous cytokinin-related mutants have been reported (23, 24). The ipt (4) gene from Agrobacterium is the most studied gene, relating to cytokinin biosynthesis in microbial systems. However, no ipt-homologous sequence has been found in plants. Recently, several Arabidopsis genes most likely involved in cytokinin signaling pathways have been identified. The gene CKI1 shows similarities to histidine kinase genes and appears to have cytokinin receptor properties, because constitutive production of the protein leads to cytokinin autonomy in regeneration experiments (25). The IBC6 gene (26) is rapidly induced by cytokinin and may function in combination with CKI1. The CGR1 gene (27) encodes a transmembrane protein with some similarity to G proteins and, when disrupted, affects cytokinin perception. We have centered our efforts to isolate genes involved in zeatin metabolism on Phaseolus (7), and this article reports the cloning of a gene encoding a zeatin metabolic enzyme. On the basis of the gene sequence of zeatin O-glucosyltransferase, we have recently isolated, by PCR and inverse PCR, a genomic sequence from P. vulgaris seeds exhibiting O-xylosyltransferase activity. The nucleotide sequence is 93% identical to the O-glucosyltransferase gene and the deduced amino acid sequence shows 90% similarity (details to be published elsewhere). The activity of these enzymes may be affected by other genes, because the product of a P. vulgaris cDNA constitutively expressed in transgenic tobacco stimulates low level of O-xylosyltransferase activity (28). Although genes encoding β-glucosidases able to degrade O-glucosylzeatin have been reported (15, 29), they do not exhibit the high specificity of the zeatin O-glucosyltransferase.
Zeatin is the most active and ubiquitous cytokinin. The metabolism of zeatin, along with its biosynthesis, is likely to affect the level of active cytokinins in plant tissues. Unlike other metabolic steps such as N-glucosylation, the formation of O-glucosylzeatin is reversible by β-glucosidase (14–16). This reversibility is important because it indicates that the metabolite has a possible regulatory function. In addition, O-glucosylzeatin is resistant to cytokinin oxidases, which selectively degrade cytokinins with an unsaturated isoprenoid side chain (12, 13). More importantly, O-glucosylzeatin is found in all plant species examined, and an understanding of how its formation is regulated is likely to have broad implications. The isolation of the cDNA and the gene encoding zeatin O-glucosyltransferase will allow a more precise assessment of the role of O-glucosylzeatin in growth and development, through manipulation of expression of this gene. In cereals, grain development was suggested to be related to elevated zeatin (30–32). Because the zeatin O-glucosyltransferase level in seeds is stage-specific, the enzyme likely regulates active vs. storage forms of cytokinins and, therefore, could have an impact on seed growth. This and other possible functions of O-glucosylation of zeatin are being examined via genetic manipulations.
Acknowledgments
Research was supported by grants from the National Science Foundation (IBN-9116490) and the U.S. Department of Agriculture–National Research Initiative Competitive Grants Program (9801398), and by the Oregon Agricultural Experiment Station (paper 11430).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF101972).
References
- 1.Miller C O, Skoog F, Okumura F S, Von Saltza M H, Strong F M. J Am Chem Soc. 1956;78:1375–1380. [Google Scholar]
- 2.Shaw G. In: Cytokinins: Chemistry, Activity and Function. Mok D W S, Mok M C, editors. Boca Raton, FL: CRC Press; 1994. pp. 15–34. [Google Scholar]
- 3.Letham D S. Life Sci. 1963;8:569–573. doi: 10.1016/0024-3205(63)90108-5. [DOI] [PubMed] [Google Scholar]
- 4.Barry G F, Rogers S R, Fraley R T, Brand L. Proc Natl Acad Sci USA. 1984;81:4776–4780. doi: 10.1073/pnas.81.15.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C M, Melitz D K. FEBS Lett. 1979;107:15–20. doi: 10.1016/0014-5793(79)80452-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen C M, Ertl J R. In: Cytokinins: Chemistry, Activity and Function. Mok D W S, Mok M C, editors. Boca Raton, FL: CRC Press; 1994. pp. 81–85. [Google Scholar]
- 7.Mok D W S, Martin R C. In: Cytokinins: Chemistry, Activity and Function. Mok D W S, Mok M C, editors. Boca Raton, FL: CRC Press; 1994. pp. 129–137. [Google Scholar]
- 8.Jameson P E. In: Cytokinins: Chemistry, Activity and Function. Mok D W S, Mok M C, editors. Boca Raton, FL: CRC Press; 1994. pp. 113–128. [Google Scholar]
- 9.Chen C M. Physiol Plant. 1997;101:665–673. [Google Scholar]
- 10.Letham D S, Parker C W, Duke C C, Summons R E, MacLeod J K. Ann Bot. 1976;40:261–263. [Google Scholar]
- 11.Letham D S. In: Cytokinins: Chemistry, Activity and Function. Mok D W S, Mok M C, editors. Boca Raton, FL: CRC Press; 1994. pp. 57–80. [Google Scholar]
- 12.Whitty C D, Hall R H. Can J Biochem. 1974;52:781–799. doi: 10.1139/o74-112. [DOI] [PubMed] [Google Scholar]
- 13.Armstrong D J. In: Cytokinins: Chemistry, Activity and Function. Mok D W S, Mok M C, editors. Boca Raton, FL: CRC Press; 1994. pp. 139–154. [Google Scholar]
- 14.Letham D S, Palni M S. Annu Rev Plant Physiol. 1983;34:162–197. [Google Scholar]
- 15.Brzobohaty B, Moore I, Kristoffersen P, Bako L, Campos N, Schell J, Palme K. Science. 1993;262:1051–1054. doi: 10.1126/science.8235622. [DOI] [PubMed] [Google Scholar]
- 16.Faiss M, Strnad M, Redig P, Dolezal K, Hanus J, Van Onckelen H, Schmülling T. Plant J. 1996;10:33–46. [Google Scholar]
- 17.Dixon S C, Martin R C, Mok M C, Mok D W S. Plant Physiol. 1989;90:1316–1321. doi: 10.1104/pp.90.4.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin R C, Martin R R, Mok M C, Mok D W S. Plant Physiol. 1990;94:1290–1294. doi: 10.1104/pp.94.3.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochman H, Medhora M M, Garza D, Hartl D L. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 219–227. [Google Scholar]
- 20.Doyle J J, Doyle J L. Focus. 1990;12:13–15. [Google Scholar]
- 21.Davis L G, Dibner M D, Battey J F. Basic Methods in Molecular Biology. New York: Elsevier; 1986. [Google Scholar]
- 22.Lee Y H, Mok M C, Mok D W S, Griffin D A, Shaw G. Plant Physiol. 1985;77:635–641. doi: 10.1104/pp.77.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deikman J. Plant Growth Regul. 1998;23:33–40. [Google Scholar]
- 24.Wang T L. In: Cytokinins: Chemistry, Activity and Function. Mok D W S, Mok M C, editors. Boca Raton, FL: CRC Press; 1994. pp. 255–268. [Google Scholar]
- 25.Kakimoto T. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- 26.Brandstatter J, Kieber J J. Plant Cell. 1998;10:1009–1019. doi: 10.1105/tpc.10.6.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plakidou-Dymock S, Dymock D, Hooley R. Curr Biol. 1998;8:315–324. doi: 10.1016/s0960-9822(98)70131-9. [DOI] [PubMed] [Google Scholar]
- 28.Martin R C, Mok M C, Mok D W S. Plant J. 1997;12:306–312. doi: 10.1046/j.1365-313x.1997.12020305.x. [DOI] [PubMed] [Google Scholar]
- 29.Falk A, Rask L. Plant Physiol. 1995;108:1369–1377. doi: 10.1104/pp.108.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheikh N, Jones R J. Plant Physiol. 1994;106:45–51. doi: 10.1104/pp.106.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dietrich J T, Kaminek M, Blevins D G, Reinbott T M, Morris R O. Plant Physiol Biochem. 1995;33:327–336. [Google Scholar]
- 32.Jameson P E, McWha J A, Wright G J. Z Pflanzenphysiol. 1982;106:27–36. [Google Scholar]