Mitochondrial oxidative phosphorylation (OXPHOS) has been observed to decline with age in association with the accumulation of somatic mtDNA rearrangement (1) and base substitution (2–4) mutations. These observations have lead to the theory that the mtDNA is the aging clock. To this aging story was added a new twist in a recent issue of PNAS, an mtDNA mutation that promotes longevity (5).
MtDNA rearrangement mutations accumulate exponentially with age in postmitotic tissues, with the inflection point generally occurring about age 45. Although any one mtDNA rearrangement is rare in a particular tissue, surveys of the array of mtDNA rearrangement mutations using long extension-PCR have revealed that a broad spectrum of different mtDNA rearrangements accumulate in each tissue with age (1).
Certain control region (CR) mtDNA mutants have also been reported to accumulate with age in specific tissues (4), and evidence of heteroplasmic (mixtures of mutant and normal mtDNAs) CR mutants has been reported in a variety of contexts including human brain tissue (6), different hairs of single individuals (7), tissues of the Grand Duke of the Romanov family (8), and cancer cells (9, 10).
A surprising aspect of these age-related CR mtDNA mutants is their tissue specificity. One somatic CR mtDNA mutation, a T to G transversion occurring at nucleotide position (np) 414 (T414G), is generally found in skin fibroblast cultures from individuals over 60 years of age with the level of mutant mtDNAs exceeding 60% in some cases (Table 1). Two additional mtDNA CR mutations, A189G and T408A, have been reported to accumulate with age in skeletal muscle (3, 5), but unlike T414G these mutations have also been reported to be germ-line-transmitted polymorphisms associated with specific mtDNA haplogroups (groups of related mtDNA haplotypes) (Table 1). Thus, it appears that each tissue may accumulate its own unique somatic mtDNA CR mutations with age, but some of these same variants might also be inherited.
Table 1.
Age-related heteroplasmic mtDNA CR variant reported as somatic mutations and germ-line variations in various haplogroups
| Sequence variation | Germ line
|
Somatic tissue specificity | |
|---|---|---|---|
| Clusters | No. of cases (total 444) | ||
| C150T | Lo (26, 27) | 2 | Leukocyte, fibroblast (5) |
| L2 (24, 26–28) | 9 | ||
| L3 (26, †) | 8 | ||
| G (26) | 1 | ||
| M* (†) | 2 | ||
| D (29, 30, †) | 3 | ||
| N no R (26, 30, 31) | 4 | ||
| N1b (24) | 1 | ||
| U* (†) | 1 | ||
| U2 (28) | 1 | ||
| U3 (28) | 2 | ||
| U5 (26, 28, 32, †) | 11 | ||
| J (24, 32–34, †) | 9 | ||
| T (26, 28, †) | 3 | ||
| HV* (28) | 1 | ||
| H (26, †) | 2 | ||
| A189G | Lo (24, 26–28, 32, 35) | 7 | Skeletal muscle (2) |
| U5 (†) | 1 | ||
| L3 (26) | 1 | ||
| W (24, 28, 32) | 39 | ||
| B (36) | 2 | ||
| J (32) | 2 | ||
| V (32) | 1 | ||
| T408A | L3 (26) | 1 | Skeletal muscle (3) |
| M* (29) | 1 | ||
| T414G | — | — | Fibroblast (4), skeletal muscle (2) |
References are given in parentheses.
Unpublished data.
This story is further complicated in a recent issue of PNAS (5) by the report of a C150T variant that accumulates with age in skin fibroblasts, but is also present in the blood cell lymphocytes of centenarians and twins (Table 1). The association of the C150T mutation with centenarians and twins suggested to the authors that the C150T imparts resistance to stress and thus promotes longevity. Yet, this mutation has also been reported to be an inherited polymorphism in some instances (Table 1).
The unusual behavior of the CR mtDNA mutations raises a myriad of questions. Why is it that mtDNA rearrangement mutations are ubiquitous in aging postmitotic tissues, but each individual rearrangement is rare; whereas the age-related CR mtDNA base substitution mutations are tissue-specific, but each mutation can be present at high levels throughout a tissue? How can most somatic CR mtDNA mutations be deleterious and contribute to aging in their tissue of origin, whereas the C150T mutation may be advantageous when occurring in lymphocytes? Why is it that certain CR somatic mutations are thought to contribute to aging, and yet the same variants have been reported to be inherited in certain mtDNA haplogroups? Finally, how can one distinguish between germ-line and somatic mutations in forensic specimens, and what does this ambiguity imply about the reliability of CR sequences in individual identification?
Cell-Specific Amplification of Rearrangement Mutations.
The low level of each mtDNA rearrangement in a tissue has been interpreted as each individual mutation arising de novo within a single cell or a region of skeletal muscle fiber. Once the mutation appears, it is selectively amplified until it comes to be the predominate mtDNA in the cell or fiber domain of origin. This idea has been substantiate by in situ hybridization to mtDNAs within individual muscle fibers from the elderly which revealed the clonal amplification of different mtDNA rearrangements in different regions of the same muscle fiber (11), and by studying cultured cardiomyocytes which revealed that each cardiomyocyte harbored only one mtDNA rearrangement that was clonally expanded within that cell (12, 13). Thus, each somatic mtDNA rearrangement reaches high levels of heteroplasmy within its cell of origin, however, when a tissue is homogenized, all of the rearranged mtDNA are mixed together diluting each of the molecules to low average levels.
But why would the deleterious mtDNA rearrangement mutations be selectively amplified? The human mtDNA encodes 13 polypeptides that are essential for the mitochondrial energy generating system, OXPHOS, plus the 12S and 16S rRNA genes and 22 tRNA genes necessary for their expression. Thus, any mutation in the mtDNA coding region will alter mitochondrial energy production. The mitochondria use OXPHOS to generate most of the cellular ATP, but as a toxic by-product they produce most of the endogenous reactive oxygen species (ROS). The ROS, in turn, can damage the OXPHOS enzymes and the mtDNA, eroding mitochondrial function. When mitochondrial energy production gets too low and/or mitochondrial ROS damage becomes too high, the mitochondrial permeability transition pore (mtPTP) is activated and the cell with its damaged mitochondria is removed by apoptosis (14).
So why might somatic mtDNA rearrangements be selectively amplified? Apparently, the only way that a cell can cope with chronic energy deficiency is to make more mitochondria; but only those cells or muscle regions that have mutant mtDNAs have energy deficiencies. Hence, only the nuclei associated with the defective mitochondria are induced to make more mitochondria. Thus, the mutant mtDNAs are selectively amplified.
But what tells the nuclei to make more mitochondria? One possibility is that the nuclei monitor the redox state of the cell and thus the mitochondria. The mitochondria oxidize NADH derived from carbohydrates and fats with O2 to generate H2O and trap the energy released in ATP. Thus, a defect in OXPHOS will reduce ATP production and increase the NADH/NAD+ ratio making the cell more reduced. The more reduced environment of the cell could be monitored by a ubiquitous transcription factor, such as the REBOX-binding factor. This factor only binds to its 5′ cis-elements in nDNA bioenergetic genes when in a reducing environment (15). Thus, when the defective mitochondria inadequately oxidize the NADH the increasingly reduced environment would activate the REBOX-binding factor and this in turn would induce the transcription of nuclear DNA (nDNA) proteins involved in the bioenergetics and biogenesis of mitochondria. Only the nuclei associated with mutant mtDNA would be activated and hence the mutant mtDNAs would be preferentially amplified.
So why don't rearranged mtDNAs come to predominate in elderly tissues? Because once the rearranged mtDNA level becomes too high within a cell, the reduced energy output and increased oxidative stress activates the mtPTP, and the cell and its mutant mtDNAs are eliminated (14).
Tissue-Specific Amplification of CR Mutations.
Unlike individual mtDNA rearrangements, specific CR mtDNA variants can come to represent the majority of the mtDNAs in a tissue. This means that most of the cells must contain high levels of the same mutant. How could this be? One possibility is that the CR base substitution mutation could arise by chance at various times during development. The earlier the mutation, the greater the percentage of cells in the tissue that would inherit the mutant mtDNA. If this particular CR mutant imparted a replicative advantage in a particular tissue, then the mutant mtDNAs would be amplified in all of the cells of that tissue. Thus, the more cells that inherited the mutant mtDNA, the greater the percentage of the mtDNAs in the tissue that would ultimately be mutant.
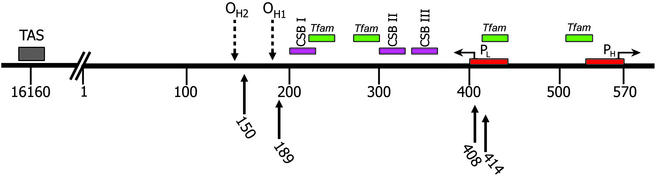
But why might specific CR mutants be amplified in specific cells? The CR is known to contain regulatory elements. Its 1,122 nps encompasses the H- and L-strand promoters (PL, PH), the origin of H-strand replication (OH), three conserved sequence blocks (CSB I, II, III), and the termination associated sequences (TAS). Yet these elements represents only for a small proportion of the CR sequence in a genome notorious for its coding sequence economy (Fig. 1).
Figure 1.
Schematic diagram of the mtDNA-CR. The functional sequences are indicated with boxes. Dashed arrows point to H-strand origin of replication 1 and 2 (OH1 and OH2). The arrows pointing left and right show transcription orientation of H- and L-strand promoters (PH and PL). Arrows below the line indicate the heteroplasmic nucleotide positions associated with aging. TAS, termination association sequence; CSB I, II, III, conserved sequence block I, II, and III; Tfam, Tfam binding sites.
What then is the function of the rest of the CR sequence? This might be deduced from two observations. First, both PL and PH are associated with binding sites for the nDNA coded transcription factor Tfam. The T414G mutation occurs in close proximity to the Tfam binding site of the PL and thus could perturb both L-strand transcription and, because the L-strand acts as its primer, H-strand replication (4). Thus, CR sequences may bind nDNA-encoded regulatory factors. Second, two novel cis-regulatory elements, the OXBOX and the REBOX, are found 5′ to multiple nDNA energy genes (15, 16). Moreover, homologous cis-elements are also observed in the CR of the mtDNA, and these mtDNA sequences can gel-shift the nDNA-encoded transcription factors. Whereas the REBOX-binding factor is ubiquitous, the OXBOX-binding factor is heart and muscle specific (15). Such trans-acting factors might coordinate nDNA and mtDNA transcription and replication. If so, there might be many such factors, some of which are tissue-specific. In this case, a chance CR mtDNA mutation in the cis-binding site of a tissue-specific regulatory factor would remove the tissue-specific regulation. If this factor imposed a negative control on mtDNA replication, then the mtDNA harboring the CR mutant would be selectively amplified in every cell of a tissue expressing that trans-factor.
This same logic could explain the excess of mtDNA CR mutants recently reported in cancer cells (9, 10). Loss of cis-binding sites that negatively regulated mtDNA replication could impart a significant growth advantage to a cancer cell.
CR mtDNA mutations do have the potential of being functionally important. Recently, a polymorphism in the TAS region at np 16,189 has been reported to correlate with an increased risk for type II diabetes (17).
So why don't the amplified CR mtDNA mutations induce apoptosis and remove the cells with the mutant mtDNAs? The answer probably lies with the fact that the CR mutants would not generate defective mtDNA gene products. Rather, they would alter the ratio of the mtDNA and nDNA gene products. This might erode the efficiency of a tissue without actually inhibiting OXPHOS.
Advantages of the C150T Mutation
How then could the C150T mutation have both a replicative advantage in skin fibroblasts and be protective to centenarians and twins in lymphocytes? The authors suggest that the C150T mutation alters the location of H-strand replication origin and this imparts a replicative advantage to the mtDNA.
But why might C150T be advantageous? One possibility is that it enhances the immune system in some way, possibly by slowing the turnover of memory T-cells. Alternatively, because most of the centenarians with the C150T mutation appear to have inherited it through the germ line, the C150T mutation might simply have been linked to other variants in the mtDNA that promote longevity.
Somatic Versus Germ-Line CR mtDNA Mutations
Previously, the authors of the C150T paper reported that in this same group of centenarians there was enrichment for the European haplogroup J (18). Since their report, haplogroup J has been associated with longevity in three other European studies (19–21). Furthermore, a subset of haplogroup M mtDNAs has also been associated with longevity in a Japanese study (22).
We have surveyed the mtDNA sequence of over 400 mtDNAs for the presence of the C150T mutation, and found that it was present in certain African-specific haplogroups (L0, L2, L3), European-specific haplogroups (NnoR, N1b, U*, U2, U3, U5, J, T, HV*, and H), and Asian-specific haplogroups (M* and D). The presence of C150T in European haplogroup J is particularly interesting. Possibly the centenarians with haplogroup J mtDNAs are also the ones with the germ-line C150T variant (Table 1).
How might C150T and/or haplogroup J contribute to longevity? Human mtDNAs radiated out of Africa and around the world through successive transmission from mother to daughter, only changing by the accumulation of sequential mutations along radiating maternal linage (23). Africans encompass the African-specific mtDNA haplogroups L0-L3, and the M and N lineages derived from L3. However, only M and N left Africa to colonize Eurasia. In Europe, lineage N radiated to give the plethora of European mtDNA haplogroups including J. In Asia, both M and N radiated into numerous mtDNA lineages, but only four of the southern Asian lineages (A, C, D, and G) came to predominate in northern Siberia. Only three of these (A, C, and D) crossed the Bering land bridge to populate the Americas.
The striking transitions in mtDNA types between Africa and Eurasia and between southern Asia and Siberia correlate with latitude and indicate that mtDNA diversity has been subjected to climatic selection (24). The mitochondria burn calories to make ATP to do work, but also to generate heat to maintain our body temperature. The balance between these two mutually exclusive processes is determined by the OXPHOS efficiency, which is regulated in large measure by the mtDNA ATP6 gene protein. Highly efficient OXPHOS generates primarily ATP with little waste heat. Less efficient OXPHOS generates more heat for the same amount of ATP. The former is preferable in the tropics, whereas the later would be critical for survival in the arctic. Interestingly, the amino acid sequence of the human mtDNA ATP6 protein is hypervariable in the arctic, implying that mutations in this mtDNA gene have been important for human adaptation to extreme cold. A similar argument can be made for variation in the mtDNA cytochrome gene among the European mtDNA lineages (24).
A reduction in OXPHOS efficiency would also burn more calories. As a result, fewer reducing equivalents (electrons) from the diet would be available to make ROS. A life-long reduction in mitochondrial ROS stress would, in turn, decrease apoptosis and increase longevity. Hence, the C150T variant or a linked polymorphism in haplogroup J might have changed OXPHOS efficiency and thus ROS production, reducing oxidation stress and increasing longevity.
CR mtDNA Variants and Forensics
It is clear that the same CR mutations can occur either as tissue-specific somatic mutations or systemic germ-line mutations (Table 1). At the same time the mtDNA CR sequence has become increasingly popular as a forensic tool for individual identification, primarily because of its hypervariability. However, this application is predicated on the assumption that the mtDNA CR sequence variants of a person will be the same in all tissues of his or her body. But it is now clear that the CR sequences from two different tissues of the same person could harbor different somatic mutations, erroneously excluding a match. For example, the C150T mutation has already been observed in the heteroplasmic state in the blood samples of one Spanish subject (25). Clearly, more research is still required on tissue-specific somatic mtDNA mutations before CR sequences can be fully reliably for individual identification in forensics.
Footnotes
See companion article on page 1116 in issue 3 of volume 100.
References
- 1.Wallace D C. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 2.Murdock D G, Christacos N C, Wallace D C. Nucleic Acids Res. 2000;28:4350–4355. doi: 10.1093/nar/28.21.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Michikawa Y, Mallidis C, Bai Y, Woodhouse L, Yarasheski K E, Miller C A, Askanas V, Engel W K, Bhasin S, Attardi G. Proc Natl Acad Sci USA. 2001;98:4022–4027. doi: 10.1073/pnas.061013598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Asin-Cayuela J, Fish J, Michikawa Y, Bonafé M, Olivieri F, Passarino G, De Benedictis G, Franceschi C, Attardi G. Proc Natl Acad Sci USA. 2003;100:1116–1121. doi: 10.1073/pnas.242719399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jazin E E, Cavelier L, Eriksson I, Oreland L, Gyllensten U. Proc Natl Acad Sci USA. 1996;93:12382–12387. doi: 10.1073/pnas.93.22.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendall K E, Macaulay V A, Sykes B C. Am J Hum Genet. 1997;61:1303–1308. doi: 10.1086/301636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanov P L, Wadhams M J, Roby R K, Holland M M, Weedn V W, Parsons T J. Nat Genet. 1996;12:417–420. doi: 10.1038/ng0496-417. [DOI] [PubMed] [Google Scholar]
- 9.Polyak K, Li Y, Zhu H, Lengauer C, Willson J K, Markowitz S D, Trush M A, Kinzler K W, Vogelstein B. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 10.Fliss M S, Usadel H, Caballero O L, Wu L, Buta M R, Eleff S M, Jen J, Sidransky D. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 11.Muller-Hocker J, Schneiderbanger K, Stefani F H, Kadenbach B. Mutat Res. 1992;275:115–124. doi: 10.1016/0921-8734(92)90016-i. [DOI] [PubMed] [Google Scholar]
- 12.Muller-Hocker J. Brain Pathol. 1992;2:149–158. doi: 10.1111/j.1750-3639.1992.tb00683.x. [DOI] [PubMed] [Google Scholar]
- 13.Muller-Hocker J, Seibel P, Schneiderbanger K, Zietz C, Obermaier-Kusser B, Gerbitz K D, Kadenbach B. Hum Pathol. 1992;23:1431–1437. doi: 10.1016/0046-8177(92)90065-b. [DOI] [PubMed] [Google Scholar]
- 14.Kokoszka J E, Coskun P, Esposito L A, Wallace D C. Proc Natl Acad Sci USA. 2001;98:2278–2283. doi: 10.1073/pnas.051627098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haraguchi Y, Chung A B, Neill S, Wallace D C. J Biol Chem. 1994;269:9330–9334. [PubMed] [Google Scholar]
- 16.Chung A B, Stepien G, Haraguchi Y, Li K, Wallace D C. J Biol Chem. 1992;267:21154–21161. [PubMed] [Google Scholar]
- 17.Poulton J, Luan J, Macaulay V, Hennings S, Mitchell J, Wareham N J. Hum Mol Genet. 2002;11:1581–1583. doi: 10.1093/hmg/11.13.1581. [DOI] [PubMed] [Google Scholar]
- 18.De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafe M, Monti D, Baggio G, Bertolini S, et al. FASEB J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 19.Niemi A K, Hervonen A, Hurme M, Karhunen P J, Jylha M, Majamaa K. Hum Genet. 2003;112:29–33. doi: 10.1007/s00439-002-0843-y. [DOI] [PubMed] [Google Scholar]
- 20.Rose G, Passarino G, Carrieri G, Altomare K, Greco V, Bertolini S, Bonafe M, Franceschi C, De Benedictis G. Eur J Hum Genet. 2001;9:701–707. doi: 10.1038/sj.ejhg.5200703. [DOI] [PubMed] [Google Scholar]
- 21.Ross O A, McCormack R, Curran M D, Duguid R A, Barnett Y A, Rea I M, Middleton D. Exp Gerontol. 2001;36:1161–1178. doi: 10.1016/s0531-5565(01)00094-8. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka M, Gong J S, Zhang J, Yoneda M, Yagi K. Lancet. 1998;351:185–186. doi: 10.1016/S0140-6736(05)78211-8. [DOI] [PubMed] [Google Scholar]
- 23.Wallace D C, Brown M D, Lott M T. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 24.Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark A G, Hosseini S, Brandon M, Easley K, Chen E, Brown M D, et al. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crespillo M, Luque J A, Paredes M, Fernandez R, Ramirez E, Valverde J L. Int J Legal Med. 2000;114:130–132. doi: 10.1007/s004140000158. [DOI] [PubMed] [Google Scholar]
- 26.Ingman M, Kaessmann H, Paabo S, Gyllensten U. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 27.Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, Calderon F L, Simionati B, Valle G, Richards M, Macaulay V, Scozzari R. Am J Hum Genet. 2001;69:1348–1356. doi: 10.1086/324511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maca-Meyer N, Gonzalez A M, Larruga J M, Flores C, Cabrera V M. BMC Genet. 2001;2:13. doi: 10.1186/1471-2156-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kivisild T, Tolk H V, Parik J, Wang Y, Papiha S S, Bandelt H J, Villems R. Mol Biol Evol. 2002;19:1737–1751. doi: 10.1093/oxfordjournals.molbev.a003996. [DOI] [PubMed] [Google Scholar]
- 30.Ozawa T, Tanaka M, Ino H, Ohno K, Sano T, Wada Y, Yoneda M, Tanno Y, Miyatake T, Tanaka T, et al. Biochem Biophys Res Commun. 1991;176:938–946. doi: 10.1016/s0006-291x(05)80276-1. [DOI] [PubMed] [Google Scholar]
- 31.Ozawa T. Exp Gerontol. 1995;30:269–290. doi: 10.1016/0531-5565(94)00057-a. [DOI] [PubMed] [Google Scholar]
- 32.Finnila S, Lehtonen M S, Majamaa K. Am J Hum Genet. 2001;68:1475–1484. doi: 10.1086/320591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown M D, Zhadanov S, Allen J C, Hosseini S, Newman N J, Atamonov V V, Mikhailovskaya I E, Sukernik R I, Wallace D C. Hum Genet. 2001;109:33–39. doi: 10.1007/s004390100538. [DOI] [PubMed] [Google Scholar]
- 34.Brown M D, Starikovskaya E, Derbeneva O, Hosseini S, Allen J C, Mikhailovskaya I E, Sukernik R I, Wallace D C. Hum Genet. 2002;110:130–138. doi: 10.1007/s00439-001-0660-8. [DOI] [PubMed] [Google Scholar]
- 35.Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N. Proc Natl Acad Sci USA. 1995;92:532–536. doi: 10.1073/pnas.92.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamasoba T, Goto Y, Oka Y, Nishino I, Tsukuda K, Nonaka I. Neuromuscul Disord. 2002;12:506–512. doi: 10.1016/s0960-8966(01)00329-7. [DOI] [PubMed] [Google Scholar]