Abstract
The use of transgenic plants to express orally immunogenic protein antigens is an emerging strategy for vaccine biomanufacturing and delivery. This concept has particular suitability for developing countries. One factor that has limited the development of this technology is the relatively modest levels of accumulation of some antigenic proteins in plant tissues. We used fusion protein design to improve expression of the hepatitis B surface antigen (HBsAg) by attempting to mimic the process of HBsAg targeting to the endoplasmic reticulum of human liver cells during hepatitis B virus infection. We created a gene encoding a recombinant HBsAg modified to contain a plant signal peptide fused to its amino terminus. The signal peptide from soybean vegetative storage protein vspA (VSPαS) directed endoplasmic reticulum targeting of HBsAg in plant cells, but was not cleaved and resulted in enhanced VSPαS-HBsAg fusion accumulation. This product was more stable and presented the protective “a” antigenic determinant to significantly higher levels than unmodified native HBsAg expressed in plant cells. It also showed a greater extent of intermolecular disulfide bond formation and formation of virus-like particles. Moreover, VSPαS-HBsAg stimulated higher levels of serum IgG than native HBsAg when injected into mice. We conclude that HBsAg tolerates a polypeptide fusion at the amino terminus and that VSPαS-HBsAg is an improved antigen for plant-based expression of a subunit vaccine for hepatitis B virus.
Keywords: transgenic plants‖tobacco NT-1 cell‖vaccine
Hepatitis B virus (HBV) infection is an important global health problem, and vaccination is a proven strategy to control HBV infection. Current vaccines use yeast-derived recombinant hepatitis B surface antigen (rHBsAg) delivered by intramuscular injection, requiring trained medical practitioners and refrigerated storage, and are thus expensive to use. We have evaluated a new strategy for biomanufacturing the antigen to create a plant-derived oral vaccine, which could provide lower capital costs of production, mucosal immunization, and needle-free delivery of the final product. These advantages are particularly important for immunization programs in developing countries. Plant-derived HBsAg assembles into virus-like particles (VLPs) (1, 2) as in human (3), Chinese hamster ovary (4), and yeast (5) cells. Potato-derived HBsAg is orally immunogenic in mice, but high doses are needed (2, 6), creating a need for enhanced accumulation of the antigen in plant tissues if acceptable amounts of the plant material are to be delivered orally.
HBsAg is a transmembrane protein with uncleaved internal signal sequences that facilitate cotranslational translocation and integration of HBsAg into the endoplasmic reticulum (ER) membrane (7). Subsequently, HBsAg dimers are rapidly formed via intermolecular disulfide bonds, catalyzed by protein disulfide isomerase (PDI). The protein–membrane complexes then assemble and extrude into the ER lumen (8) and are transported to the post-ER–pre-Golgi intermediate compartment, where further cross-linking to form higher-order oligomers occurs in the absence of PDI (9). We previously showed that a plant ER signal peptide from the soybean gene vspA (VSPαS) or an ER retention signal (KDEL) influenced HBsAg accumulation in transgenic potato (6). The vspA gene has a 35-residue N-terminal pro sequence (10), and the N-terminal 21 aa (VSPαS) contain a predicted ER signal peptide (11). The full 35-aa sequence (VSPαL) also contains a putative vacuolar targeting peptide at its C terminus (10). Proteins containing an ER retention signal (H/KDEL) at the C terminus are returned to the ER via KDEL receptor in cis-Golgi (12), and KDEL modification of the C terminus can enhance expression of the plant protein vicilin (13) in transgenic tobacco.
In this paper, we examine effects of plant ER-targeting motifs on HBsAg at the molecular level by using tobacco cells as a model. We show that the VSPαS signal peptide significantly enhances HBsAg expression, “a” determinant formation, and VLP formation in NT-1 cells, resulting in a fusion protein VSPαS-HBsAg that is more immunogenic in mice than the unmodified HBsAg.
Materials and Methods
Construction of Expression Plasmids.
The synthetic plant-optimized DNA sequence encoding HBsAg (sHBsAg) subtype adr (GenBank accession no. AF504292; unpublished work) was modified by N-terminal extension with either a 21-aa (VSPαS) or 35-aa (VSPαL) plant ER signal peptide (10), and/or C-terminal extension with the hexapeptide “SEKDEL” (6). The coding sequences were under the control of enhanced cauliflower mosaic virus (CaMV) 35S promoter with the 5′ UTR of the tobacco etch virus (TEV), and 3′ UTR and polyadenylation signal from the soybean vegetative storage protein gene (vspB) (14). The expression cassettes from HindIII to EcoRI were subcloned into pGPTV-KAN (15), a plant binary vector.
Plant Cell Transformation.
The binary vectors were electroporated into Agrobacterium LBA4404, which were used to transform Nicotiana tabacum cv. Bright Yellow 2 (NT-1) cells (16). Transformed calli were selected on NT-1 medium containing kanamycin and carbenicillin (Sigma) each at 300 mg/liter, and randomly picked lines from each construct were maintained on NT-1 selective medium. The five highest-expression lines of each construct were placed into liquid suspension culture in shaker flasks at 25°C and 120 rpm and passed every 7 days.
Protein Analysis.
Protein was extracted from NT-1 cells by a Fastprep FP120 (Bio 101) machine in HB-extracting buffer [50 mM NaPi, pH 6.6/100 mM NaCl/10 mM EDTA, pH 8.0/0.1% Triton X-100/50 mM sodium ascorbate/1 mM PMSF with Complete Protease Inhibitor (Roche, Indianapolis)]. Total HBsAg was assayed by polyclonal ELISA (17), using yeast recombinant HBsAg (Rhein Biotech, Düsseldorf, Germany) as reference standard, and HBsAg “a” determinant was assayed by Auszyme Monoclonal kit (Abbott), using human serum-derived HBsAg as reference standard. Total soluble protein was assayed by the Bradford dye-binding procedure (ref. 18; Bio-Rad), using BSA fraction V (Pierce) as a standard.
Pulse–Chase Analysis and Immunoprecipitation.
HBsAg stability experiments were performed as described (19) with some modifications. NT-1 cells (0.6 ml) at 7 days after passage were subcultured in 15 ml of NT-1 liquid medium. At day 5 after subculture, the cells were pulsed for 30 min with 133 μCi (1 Ci = 37 GBq) of 35S methionine/cysteine protein labeling mix (NEN) per milliliter of culture cells. After the radioactive pulse, cells were washed three times with NT-1 medium and chased with 1 mM each unlabeled methionine and cysteine in NT-1 medium. At time points, 0.1 g of cells were collected and homogenized by Fastprep in 1 ml of HBsAg extracting buffer plus 10× PBSTSD (4% Triton X-100/2.5% deoxycholate/0.5% SDS in 1× PBS). For immunoprecipitation (IP, Fitzgerald Industries, Concord, MA), mouse monoclonal anti-HBsAg (Biogenesis, Poole, U.K.) was incubated with cell extracts (1 μg/1 mg total soluble protein) for 3 h on a rotor at 4°C. Then, a Protein G Plus-Agarose (Calbiochem) suspension was added and incubated at 4°C for 2 h. The protein G–antibody complexes were washed four times with 1× PBSTSD and once with 10 mM Tris-acetate (pH 7.5), dried, and denatured with 6% SDS sample buffer containing 1 M DTT at 100°C for 10 min or 20 nM DTT at 37°C for 5 min. Immunoprecipitates were subjected to SDS/PAGE on 15% acrylamide gels, blotted to poly(vinylidene difluoride) (PVDF) membrane (Amersham Pharmacia), and probed with goat anti-HBsAg (Fitzgerald Industries, Concord, MA). The blot was imaged with a Storm chemifluorescence system (Molecular Dynamics). For 35S pulse–chase experiments, the gel was dried, detected with Storm system phosphor screen, and quantified with imagequant software.
Sucrose Gradient and Concentrated Partially Purified NT-1-Derived HBsAg.
Sucrose gradients of crude plant cell extracts were performed as described (1). Protein was extracted from NT-1 suspension culture cells by using 1 ml of extraction buffer (0.05% Triton X-100) per 0.1 g of cells (20). Gradient fractions were subjected to polyclonal ELISA and Auszyme monoclonal assay. For mouse immunization, gradient fractions containing HBsAg were pooled, concentrated with Centriprep (Amicon), and kept at 4°C. A nontransgenic NT-1 cell extract was prepared in the same way for negative control.
Mouse Immunization.
Antigens were adsorbed to Imject alum (Pierce) according to the manufacturer's instructions. BALB/c mice (five mice per group) received i.p. injections of 490 μg of alum-adsorbed particulate protein from nontransgenic NT-1 or 0.5 μg of rHBsAg from yeast (Rhein Biotech, Düsseldorf, Germany), NT-1 cell line HB117, or NT-1 cell line HB118 (transgenic lines generated with constructs pHB117 and pHB118) on days 0 and 14. Tail blood was collected before each injection and every 2 weeks after the last injection. Serum antibodies against HBsAg were determined by endpoint titer ELISA and AUSAB EIA assay (Abbott) as described (2). Geometric mean titer (GMT) was calculated using the reciprocal of the highest serum dilution that had an absorbance ≥0.1 OD unit above background. Anti-HBsAg IgG1 and IgG2a were determined by subisotyping with Mouse Typer Sub-Isotyping Kit (Bio-Rad) and 0.1 μg per well of yeast rHBsAg.
Statistical Analysis.
ANOVA for two-factor nested designs and Tukey–Kramer multiple comparison procedure at α = 0.05 were used to test least-squares means of HBsAg levels. Repeated measurement designs and Duncan's multiple range test were used in immune response analysis at α = 0.05.
Results
VSPαS Plant Signal Peptide Enhanced HBsAg and “a” Determinant Accumulation.
NT-1 cells were transformed with plasmids (Fig. 1) containing the plant-optimized sHBsAg coding sequence modified by N-terminal extension with vspA signal peptides and/or C-terminal extension with an ER retention signal. Sixty-four independent lines of each construct were randomly sampled and tested for total HBsAg by polyclonal antibody ELISA and for “a” determinant by Auszyme monoclonal assay (Table 1). The VSPαS signal peptide significantly enhanced total HBsAg accumulation in HB118 lines as compared with HB117, the unmodified antigen. Modifications of HBsAg with VSPαL or with the ER retention signal SEKDEL showed no significant enhancing effects on total HBsAg accumulation; HB123.1 (VSPαL plus SEKDEL) actually showed significantly lower antigen accumulation (Table 1).
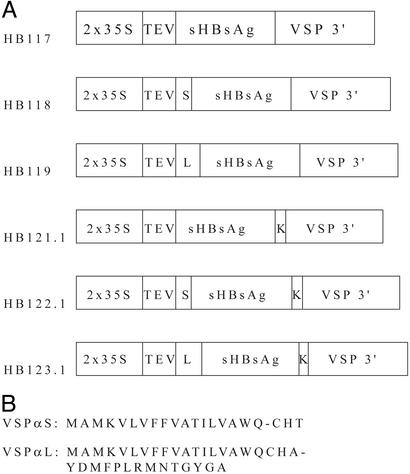
Figure 1.
(A) HBsAg constructs used in this paper. 2x35S, enhanced cauliflower mosaic virus (CaMV) 35S promoter; TEV, tobacco etch virus 5′ UTR; S, soybean vegetative storage protein vspA signal peptide (VSPαS); L, vspA signal peptide with putative vacuolar targeting signal (VSPαL); sHBsAg, synthetic plant optimized HBsAg gene; K, KDEL ER retention signal; VSP 3′, soybean vspB gene 3′ element. (B) VSPαS and VSPαL amino acid sequences with predicted cleavage sites between positions 18 and 19 (AWQ-CH) and positions 21 and 22 (CHA-YD), respectively.
Table 1.
Effect of plant signal peptides VSPαS and VSPαL and ER retention signal on HBsAg accumulation and HBsAg “a” determinant formation in transgenic NT-1 cells
| Construct | Modification | Least-squares means of HBsAg
|
|
|---|---|---|---|
| ng/mg TSP | % “a” determinant | ||
| HB117 | sHBsAg | 128bc | 18c |
| HB118 | αS-sHBsAg | 226a | 35ab |
| HB119 | αL-sHBsAg | 49cd | 26bc |
| HB121.1 | sHBsAg-KDEL | 61cd | 13c |
| HB122.1 | αS-sHBsAg-KDEL | 177ab | 46a |
| HB123.1 | αL-sHBsAg-KDEL | 17d | 20bc |
HBsAg levels (ng/mg TSP) were from duplicate assays in three independent experiments by polyclonal ELISA and % “a” determinant was calculated as (HBsAg by Auszyme assay)/(HBsAg by polyclonal ELISA) in two independent experiments. Tukey's studentized range test at α = 0.05 was used to test least-squares means of HBsAg and least-squares means of % “a” determinant. The superscript letters indicate the significant differences between constructs; data with the same letter indicated are not significantly different at α = 0.05.
The five best lines from each construct (based on polyclonal ELISA) were selected for further study. The relative efficiency of “a” determinant formation for each construct was calculated as the percentage of “a” determinant in the total HBsAg. The HBsAg modified with VSPαS in both HB118 and HB122.1 cell lines showed % “a” determinant at 35% and 46%, respectively, significantly higher than the unmodified form, HB117 (Table 1). The modification of HBsAg with either VSPαL or SEKDEL did not show any significant enhancing effect on “a” determinant formation.
We assayed “a” determinant in partially purified HBsAg stored at 4°C (later used for mouse immunization). The HB118 VSPαS-HBsAg contained 53% “a” determinant at day 1 and remained stable until day 18 (Table 2). However, HB117 HBsAg contained 12% “a” determinant at day 1 and progressively increased to 55% at day 27. Total HBsAg measured by polyclonal ELISA remained fairly stable over the storage period for both constructs.
Table 2.
HBsAg stability at 4°C storage of concentrated partially purified HBsAg
| Cell line | Day after concentrated
|
||
|---|---|---|---|
| 1 | 18 | 27 | |
| HB117 | |||
| ELISA, ng/μl | 5.8 | 4.8 | 4.5 |
| Auszyme, ng/μl | 0.7 | 2.2 | 2.5 |
| % “a” epitope | 12 | 45 | 55 |
| HB118 | |||
| ELISA, ng/μl | 2.1 | 2.1 | ND |
| Auszyme, ng/μl | 1.1 | 1.1 | ND |
| % “a” epitope | 53 | 53 | ND |
HBsAg was measure by either polyclonal ELISA or Auszyme monoclonal assay; % “a” epitope is calculated as in Table 1. ND, not determined.
VSPαS Signal Peptide Enhanced HBsAg Oligomerization and Stability.
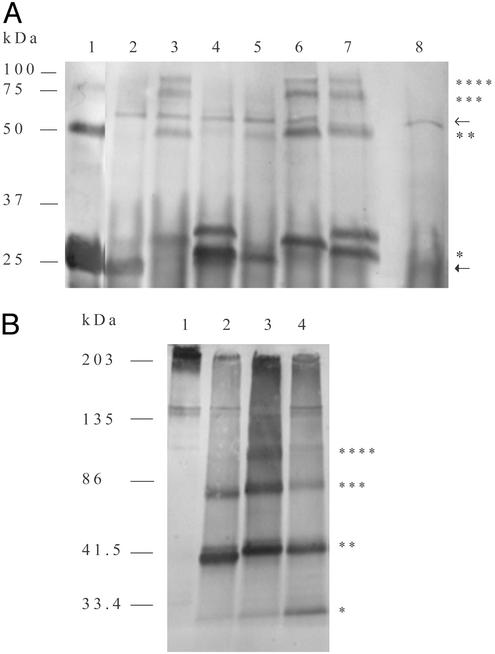
HBsAg was immunoprecipitated from NT-1 cell extracts with monoclonal antibody against HBsAg, reduced with 1 M or 20 nM DTT, and run in 15% acrylamide gels. Despite the stringent reducing condition, yeast rHBsAg (Fig. 2A, lane 1) showed a prominent dimer and weaker trimer band. HB117 (Fig. 2A, lane 2) showed a monomer at the same mobility as the yeast rHBsAg but no evidence of oligomers. The band at ≈55 kDa was the immunoprecipitating IgG heavy chain cross-reacting with the anti-goat horseradish peroxidase (HRP) conjugate, which also occurred in nontransgenic NT-1 (Fig. 2A, lane 8). HB118 showed distinct monomer, dimer, trimer, and tetramer bands (Fig. 2A, lane 3). Both VSPαS-modified HB118 and HB122.1 monomers (Fig. 2A, lanes 3 and 6, respectively) ran slower than yeast rHBsAg at a position consistent with the size of HBsAg extended by the uncleaved VSPαS signal peptide (adding ≈2.4 kDa). Both VSPαL-modified HB119 (Fig. 2A, lane 4) and HB123.1 (Fig. 2A, lane 7) showed partial processing to form the monomer comigrating with yeast rHBsAg (HB119) or slightly larger (HB123.1). Interestingly, HB119 showed only a faint dimer, whereas the SEKDEL-modified HB123.1 showed prominent dimer, trimer, and tetramer bands. Thus, HB118, HB122.1, and HB123.1 extracts showed more extensive disulfide-mediated oligomerization than HB117, HB119, and HB121.1. When samples were run at a slight reducing condition (20 nM DTT), the dimer was the most prominent form in HB117 unmodified and HB119 VSPαL-modified HBsAg (Fig. 2B, lanes 2 and 4, respectively), whereas the HB118 VSPαS-modified HBsAg showed strong dimer, trimer, tetramer, and higher-order oligomers that barely penetrated the resolving gel (Fig. 2B, lane 3). These data indicate that oligomer formation resulted from enhanced intermolecular disulfide bond formation in VSPαS-modified HBsAg.
Figure 2.
Effect of plant signal peptides VSPαS and VSPαL and ER retention signal on HBsAg oligomerization. (A) Western blots of immunoprecipitated HBsAg from yeast or transgenic NT-1 cells. Samples were denatured with 1 M DTT at 100°C for 20 min. Lane 1, yeast recombinant HBsAg, 50 ng; lane 2, HB117; lane 3, HB118; lane 4, HB119; lane 5, HB121.1; lane 6, HB122.1; lane 7, HB123.1; lane 8, nontransgenic NT-1; lower arrow, light chain of goat IgG against HBsAg; upper arrow, heavy chain of goat IgG against HBsAg; *, HBsAg monomers; **, HBsAg dimers; ***, HBsAg trimers; ****, HBsAg tetramers. (B) Western blot as in A at partially reducing condition (20 nM DTT) and fractionated in 4–20% polyacrylamide gel. Lanes 1–4 are as in A. *, HBsAg monomers; **, HBsAg dimers; ***, HBsAg trimers; ****, HBsAg tetramers.
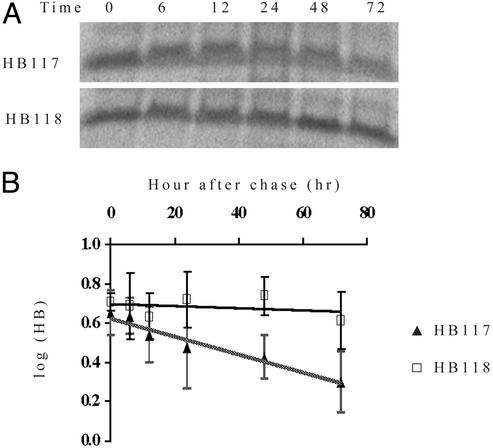
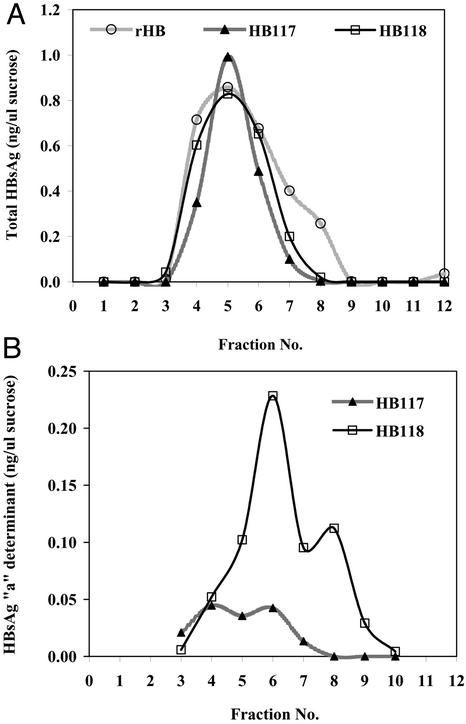
Because the signal peptide remained uncleaved, the VSPαS-modified HBsAg construct produced a fusion protein in plant cells. To determine whether the VSPαS-HBsAg fusion protein was more stable in plant cells, we studied HBsAg antigen degradation rates with 35S pulse–chase experiments. The HB117 unmodified HBsAg showed a half-life of ≈68 h (Fig. 3). In contrast, we could detect no degradation of the VSPαS-HBsAg fusion protein for up to 72 h of chase period. The slope of the HB117 degradation rate was significantly different from that of HB118 at P = 0.0002, using analysis of covariance. HBsAg expressed in potato assembles VLPs that accumulate in ER-derived vesicles (2). To determine the VLP nature of the VSPαS-HBsAg fusion protein, we performed sucrose gradient sedimentation experiments with HB117 and HB118 NT-1 cell extracts compared with yeast HBsAg. Both NT-1 lines showed polyclonal antibody-reactive antigen that cosedimented with the peak of the yeast rHBsAg (Fig. 4A). The HB118 VSPαS-HBsAg showed a somewhat broader peak than unmodified HB117 but did not display the faster-sedimenting shoulder produced by yeast rHBsAg. The HB118 profile of Auszyme monoclonal-reactive antigen (Fig. 4B) did show such a shoulder in fraction 8, and furthermore showed much greater reactivity in all particulate fractions than did the unmodified HB117 HBsAg.
Figure 3.
Pulse–chase analysis of HB117 and HB118 NT-1 lines. (A) Unmodified HBsAg (HB117) and VSPαS-modified HBsAg (HB118) were labeled with 35S and chased in unlabeled media. Samples were taken at indicated times, and extracts were immunoprecipitated with polyclonal antibody and run on a 15% acrylamide gel. (B) Stability of HBsAg and VSPαS-modified HBsAg. The x axis represents hours after labeling pulse, and the y axis represents log scale of percentage of HBsAg level present at time zero. Triangles, HB117; squares, HB118. Data are means ± SD from four independent experiments.
Figure 4.
Sucrose gradient analysis of NT-1 cell-derived HBsAg. The extracts contained 1.8 mg of total soluble protein. (A) Profiles of polyclonal antibody-reactive HBsAg for yeast recombinant HBsAg (circles) and NT-1 derived HBsAg of HB117 (triangles) and HB118 (squares). (B) Profiles of monoclonal antibody-reactive HBsAg (“a” determinant) for NT-1-derived HBsAg from HB117 (triangles) and HB118 (squares). Fraction 1 is top of gradient.
VSPαS-HBsAg Fusion Protein Is a More Potent Immunogen than Unmodified HBsAg.
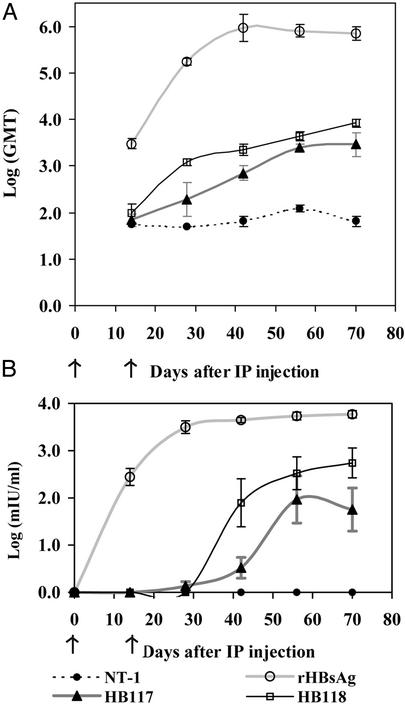
We tested the immunogenicity of partially purified extracts of NT-1 cell lines by i.p. injection in mice at days 0 and 14. Although our ultimate goal is oral delivery of a plant-derived vaccine, the aim of this experiment was to evaluate the immunogenic potential of the two antigen forms in a manner as precise as possible. Mice that received two 0.5-μg doses of alum-adsorbed yeast recombinant HBsAg showed very high titers for both AUSAB EIA assay [mean = 5,154 milli-international units (mIU)/ml at day 56] and endpoint titer (GMT = 704,636 at day 70). Mice that received HB117-unmodified HBsAg showed a GMT of 2,868 (five of five responding) at day 70 but only three of five showed an immune response higher than the protective limit (>10 mIU/ml) at day 56, with a mean of 12 mIU/ml (Fig. 5 A and B). The HB118 VSPαS-HBsAg fusion protein produced AUSAB responses in five of five mice (mean = 115 mIU/ml at day 56, GMT = 8,390 at day 70). The yeast rHBsAg caused antibody responses within 14 days after the first dose, with the anti-HBsAg reaching peak levels at 40 days and then leveling off (Fig. 5). The HB118 VSPαS-HBsAg mice reached the protective level of 10 mIU/ml in 42 days, but the HB117 HBsAg mice required 56 days to reach this level (Fig. 5B). The responses to HB117 HBsAg leveled off at day 56, but for HB118 both ELISA GMT and AUSAB titer responses continued to increase to day 70. Mice that received alum-adsorbed, partially purified total soluble protein from nontransgenic NT-1 cells showed no significant immune responses. The HB118 responses were significantly higher than those of HB117, as determined by Duncan's multiple range test at α = 0.05.
Figure 5.
Kinetics of serum IgG anti-HBsAg-specific responses of BALB/c mice after i.p. immunization. Each group of mice (n = 5) was injected with alum-adsorbed protein at days 0 and 14, as indicated by arrows: 490 μg of total soluble protein from nontransgenic NT-1 (filled circles), 0.5 μg of yeast rHBsAg (open circles), 0.5 μg of partially purified total HBsAg from HB117 (triangles), or 0.5 μg of partially purified total HBsAg from HB118 (squares). Data are means of log (GMT or mIU/ml) ± SEM. One mouse in the yeast rHBsAg group died at day 38; thus, the data for this group on days 42, 56, and 70 are derived from only four mice. (A) Serum IgG anti-HBsAg-specific responses measured by ELISA. Data are GMT ± SEM. (B) Serum IgG antiHBsAg-specific responses measured by AUSAB EIA assay. Data are means of [log(mIU/ml)] ± SEM.
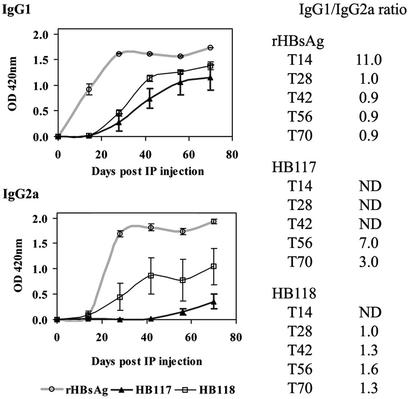
We examined the sera from immunized mice for IgG subtypes IgG1 and IgG2a (Fig. 6). The yeast rHBsAg induced more rapid and higher responses than either plant cell-derived antigen, but the HB118 VSPαS-HBsAg showed higher levels of both subtypes than HB117 unmodified HBsAg. Furthermore, the IgG1/IgG2a ratios obtained using HB118 antigen were much more similar to those obtained using the yeast rHBsAg.
Figure 6.
IgG1 and IgG2a levels in mice of Fig. 5 immunized with yeast or plant-cell HBsAg. The serum from each mouse was diluted 1:100 for reaction with yeast rHBsAg. The IgG1 and IgG2a were reported as a mean of OD420 from five mice of each group. The IgG1/IgG2a ratio was calculated by dividing the mean OD of IgG1 by that of IgG2a. ND, not determined when OD420 of either IgG1 or IgG2a was <0.05.
Discussion
VSPαS Plant Signal Peptide Enhanced HBsAg Accumulation in Transgenic NT-1 Cells.
HBsAg is a transmembrane protein that is synthesized, transported, and undergoes posttranslational modifications (disulfide bond formation and glycosylation), via the secretory pathway. To increase HBsAg accumulation in transgenic plant systems, we evaluated plant signal peptides and/or the ER retention signal SEKDEL for effects on expression and assembly of immunogenic HBsAg. The results show significantly enhanced expression of the VSPαS-HBsAg fusion protein (Table 1). If HBsAg signal I does not fully function in plant cells, some of the nascent chains may fold incorrectly in the reducing environment of the cytoplasm. Therefore, inefficient ER targeting could result in poor HBsAg accumulation in the plant system, as was observed with hordothionins expressed in transgenic tobacco (21). There is evidence in other systems that the host signal peptide functions more efficiently than a transgene's native signal peptide (22), findings consistent with our data on HBsAg expression. Conversely, VSPαL-modified HBsAg (HB119 and HB123.1) had a negative effect on HBsAg accumulation (Table 1). Because the VSPαL signal peptide contains the same N-terminal sequence as the VSPαS peptide, the putative vacuolar-targeting signal in the C terminus of VSPαL (10) might lead VSPαL-modified HBsAg to lytic vacuoles where protein degradation occurs (23), and thus limit accumulation of antigen.
The modification of HBsAg with ER retention signal, SEKDEL, showed no enhancing effects on HBsAg accumulation. However, an ER retention signal can retard protein transit through the ER yet be insufficient for protein retention in the ER (24). Further, HBsAg accumulates in post-ER–pre-Golgi intermediate compartments in Chinese hamster ovary cells (4), yeast cells (5), and plant cells (2). Thus, the recombinant antigen might never reach the KDEL receptor located in cis-Golgi (12), making the retention signal superfluous. This result is inconsistent with our previous findings in transgenic potato plants, where a marginal enhancement of HBsAg expression was found by using the native HBsAg gene extended with SEKDEL at the C terminus. Differences in the cellular sorting machinery in potato leaf versus NT-1 cells may account for this slight discrepancy.
VSPαS Plant Signal Peptide Enhanced Oligomerization, “a” Determinant Formation, and Stability of HBsAg.
The “a” determinant is a common antigenic determinant found in all hepatitis B virus (HBV) subtypes (25), thus providing cross protection. Disulfide bonding (intra- and intermolecular) plays an important role in HBsAg stability, correct antigenic determinant formation, and VLP assembly (26). We show here that extension of the HBsAg N terminus with an uncleaved VSPαS signal peptide enhanced intermolecular disulfide bond formation in plant cells, resulting in higher-order oligomer formation than with unmodified HBsAg (Fig. 2). The VSPαS-HBsAg fusion protein was also more stable in NT-1 cells (Fig. 3) and showed a significantly higher proportion of “a” determinant than unmodified form (Table 1). The VSPαS-HBsAg contained 53% of “a” determinant soon after extraction, whereas the unmodified HBsAg gradually increased to 55% of “a” determinant after 27 days in storage (Table 2). The observation that total HBsAg was stable during the storage period for both constructs suggests that the accumulating “a” determinant in HB117 material resulted from modification of the polypeptides in vitro, perhaps by oxidation to form correct disulfide bonds. This idea is supported by the observation that the reducing agent sodium ascorbate inhibits in vitro formation of “a” determinant and oligomerization of HBsAg in extracts of NT-1 cells (20).
These data suggest three possible mechanisms for the observed effects of the plant signal peptide. First, if the VSPαS peptide did enhance HBsAg targeting to the ER, the increased antigen concentration could enhance oligomerization and stability, as observed for influenza virus hemagglutinin (27). Second, VSPαS might directly affect folding of the HBsAg polypeptide, thus enhancing correct disulfide bond formation. The uncleaved VSPαS peptide is likely to form another hydrophobic transmembrane domain (28) that could attract the native transmembrane domains by hydrophobic interaction. Third, the cysteine residue in the VSPαS peptide, which is located outside the predicted helical transmembrane motif (28), might act as an internal chaperone to facilitate disulfide bond formation and protein folding. This mechanism was observed with a cysteine residue in the pro region of bovine pancreatic trypsin inhibitor that assists disulfide bond formation and folding (29).
The accumulation of VSPαS-HBsAg fusion in NT-1 cells indicates that amino acid fusions at the N terminus did not interfere with antigenic determinant formation, because VSPαS-HBsAg could be immunoprecipitated with monoclonal antibody specific to the “a” determinant and even increased the proportion of HBsAg detectable with the Auszyme monoclonal assay. The uncleaved signal did not interfere with VLP formation and actually enhanced “a” determinant in fast-sedimenting sucrose gradient fractions, compared with unmodified HBsAg (Fig. 4). The lack of cleavage of the VSPαS signal peptide might be due to poor presentation of the processing site to signal peptidase. The longer VSPαL signal peptide was partially processed (Fig. 2), perhaps because the linker between the two transmembrane domains allowed protrusion of the cleavage site from the ER membrane surface.
VSPαS-HBsAg Was More Immunogenic than Unmodified HBsAg.
We showed here that mice immunized with the HB118 VSPαS-HBsAg had stronger antibody responses and greater numbers of responders than mice that received the HB117 unmodified HBsAg (Fig. 5B). The yeast recombinant rHBsAg provoked an antibody response (mIU/ml) 106 times and 11 times stronger than tobacco cell-derived HB117 HBsAg and HB118 VSPαS-HBsAg, respectively (Fig. 5). Although we injected all mice with equal amounts of total HBsAg (as determined by polyclonal ELISA), the relative proportion of correctly folded antigen (Table 2, % “a” epitope) and the level of subunit oligomerization (Fig. 2) were lower in the plant material, especially in unmodified HBsAg from HB117 cells. Furthermore, the plant extracts were only partially purified, and thus contained contaminants that might interfere with antigen processing or presentation in mice.
In mice, Th1 lymphocytes stimulate production of complement-fixing IgG subtypes (e.g., IgG2a) and cell-mediated immune responses, whereas Th2 cells stimulate noncomplement fixing IgG subtype (e.g., IgG1) production and promote humoral immune responses (30, 31). Therefore, the mouse IgG1/IgG2a profile is usually used as an indicator of the Th1- versus Th2-biased immune response. Both the yeast rHBsAg and VSPαS-modified HBsAg induced relatively balanced immune responses (ratio near 1.0), whereas the unmodified HBsAg induced a more Th2-biased response. Alum is an adjuvant that can induce a strong Th2 response (32) and probably contributed to this effect. The early IgG1 responses were followed by increasing levels of IgG2a; however, HB117 cell antigen caused a lower IgG2a response than did either the yeast rHBsAg or HB118 VSPαS-HBsAg.
In conclusion, our results indicate that the VSPαS-HBsAg was a more efficient immunogen than the unmodified HBsAg expressed in plant cells. The enhanced immunogenicity of the VSPαS-HBsAg fusion probably results from the observed increase in oligomerization, antigen stability, and “a” determinant presentation in plant cells.
Acknowledgments
We thank Dr. Monchai Duangjinda, Dr. Russell Lloyd, and Karen Grace-Martin for assistance with statistical analysis. This work was supported by National Institutes of Health Grant AI42836 and a grant from the Park Foundation.
Abbreviations
- ER
endoplasmic reticulum
- GMT
geometric mean titer
- HBsAg
hepatitis B surface antigen
- mIU
milli-international units
- rHBsAg
recombinant HBsAg
- VLP
virus-like particle
- VSPαS
vegetative storage protein A signal peptide
- VSPαL
VSPαS with N-terminal propeptide
- VSPαS-HBsAg
VSPαS fused to HBsAg
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF504292).
References
- 1.Mason H S, Lam D M, Arntzen C J. Proc Natl Acad Sci USA. 1992;89:11745–11749. doi: 10.1073/pnas.89.24.11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kong Q, Richter L, Yang Y F, Arntzen C J, Mason H S, Thanavala Y. Proc Natl Acad Sci USA. 2001;98:11539–11544. doi: 10.1073/pnas.191617598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamimura T, Yoshikawa A, Ichida F, Sasaki H. Hepatology. 1981;1:392–397. doi: 10.1002/hep.1840010504. [DOI] [PubMed] [Google Scholar]
- 4.Patzer E J, Nakamura G R, Simonsen C C, Levinson A D, Brands R. J Virol. 1986;58:884–892. doi: 10.1128/jvi.58.3.884-892.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biemans R, Thines D, Petre-Parent B, De Wilde M, Rutgers T, Cabezon T. DNA Cell Biol. 1992;11:621–626. doi: 10.1089/dna.1992.11.621. [DOI] [PubMed] [Google Scholar]
- 6.Richter L J, Thanavala Y, Arntzen C J, Mason H S. Nat Biotechnol. 2000;18:1167–1171. doi: 10.1038/81153. [DOI] [PubMed] [Google Scholar]
- 7.Eble B E, MacRae D R, Lingappa V R, Ganem D. Mol Cell Biol. 1987;7:3591–3601. doi: 10.1128/mcb.7.10.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon K, Lingappa V R, Ganem D. J Cell Biol. 1988;107:2163–2168. doi: 10.1083/jcb.107.6.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huovila A P, Eder A M, Fuller S D. J Cell Biol. 1992;118:1305–1320. doi: 10.1083/jcb.118.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mason H S, Guerrero F D, Boyer J S, Mullet J E. Plant Mol Biol. 1988;11:845–856. doi: 10.1007/BF00019524. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 12.Semenza J C, Hardwick K G, Dean N, Pelham H R. Cell. 1990;61:1349–1357. doi: 10.1016/0092-8674(90)90698-e. [DOI] [PubMed] [Google Scholar]
- 13.Wandelt C I, Khan M R, Craig S, Schroeder H E, Spencer D, Higgins T J. Plant J. 1992;2:181–192. doi: 10.1046/j.1365-313x.1992.t01-41-00999.x. [DOI] [PubMed] [Google Scholar]
- 14.Mason H S, Haq T A, Clements J D, Arntzen C J. Vaccine. 1998;16:1336–1343. doi: 10.1016/s0264-410x(98)80020-0. [DOI] [PubMed] [Google Scholar]
- 15.Becker D, Kemper E, Schell J, Masterson R. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- 16.An G. Plant Physiol. 1985;79:568–570. doi: 10.1104/pp.79.2.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dogan B, Mason H S, Richter L, Hunter J B, Shuler M L. Biotechnol Progr. 2000;16:435–441. doi: 10.1021/bp0000454. [DOI] [PubMed] [Google Scholar]
- 18.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 19.Dombrowski J E, Schroeder M R, Bednarek S Y, Raikhel N V. Plant Cell. 1993;5:587–596. doi: 10.1105/tpc.5.5.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith M L, Keegan M E, Mason H S, Shuler M L. Biotechnol Progr. 2002;18:538–550. doi: 10.1021/bp010169w. [DOI] [PubMed] [Google Scholar]
- 21.Florack D E, Dirkse W G, Visser B, Heidekamp F, Stiekema W J. Plant Mol Biol. 1994;24:83–96. doi: 10.1007/BF00040576. [DOI] [PubMed] [Google Scholar]
- 22.Tessier D C, Thomas D Y, Khouri H E, Laliberte F, Vernet T. Gene. 1991;98:177–183. doi: 10.1016/0378-1119(91)90171-7. [DOI] [PubMed] [Google Scholar]
- 23.Vitale A, Raikhel N V. Trends Plant Sci. 1999;4:149–155. doi: 10.1016/s1360-1385(99)01389-8. [DOI] [PubMed] [Google Scholar]
- 24.Pueyo J J, Chrispeels M J, Herman E M. Planta. 1995;196:586–596. doi: 10.1007/BF00203660. [DOI] [PubMed] [Google Scholar]
- 25.Wallace L A, Carman W F. Viral Hepatitis. 1997;3:5–16. [Google Scholar]
- 26.Mangold C M, Unckell F, Werr M, Streeck R E. Arch Virol. 1997;142:2257–2267. doi: 10.1007/s007050050240. [DOI] [PubMed] [Google Scholar]
- 27.Ceriotti A, Colman A. J Cell Biol. 1990;111:409–420. doi: 10.1083/jcb.111.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claros M G, von Heijne G. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 29.Weissman J S, Kim P S. Cell. 1992;71:841–851. doi: 10.1016/0092-8674(92)90559-u. [DOI] [PubMed] [Google Scholar]
- 30.Abbas A K, Murphy K M, Sher A. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 31.Mosmann T R, Sad S. Immunol Today. 1996;17:138–146. doi: 10.1016/0167-5699(96)80606-2. [DOI] [PubMed] [Google Scholar]
- 32.Cox J C, Coulter A R. Vaccine. 1997;15:248–256. doi: 10.1016/s0264-410x(96)00183-1. [DOI] [PubMed] [Google Scholar]