Abstract
Background
In vitro systems have inherent limitations in their ability to model whole organism gene responses, which must be identified and appropriately considered when developing predictive biomarkers of in vivo toxicity. Systematic comparison of in vitro and in vivo temporal gene expression profiles were conducted to assess the ability of Hepa1c1c7 mouse hepatoma cells to model hepatic responses in C57BL/6 mice following treatment with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD).
Results
Gene expression analysis and functional gene annotation indicate that Hepa1c1c7 cells appropriately modeled the induction of xenobiotic metabolism genes in vivo. However, responses associated with cell cycle progression and proliferation were unique to Hepa1c1c7 cells, consistent with the cell cycle arrest effects of TCDD on rapidly dividing cells. In contrast, lipid metabolism and immune responses, representative of whole organism effects in vivo, were not replicated in Hepa1c1c7 cells.
Conclusion
These results identified inherent differences in TCDD-mediated gene expression responses between these models and highlighted the limitations of in vitro systems in modeling whole organism responses, and additionally identified potential predictive biomarkers of toxicity.
Background
Advances in microarray and related technologies continue to revolutionize biomedical research and are being incorporated into toxicology and risk assessment. These technologies not only facilitate a more comprehensive elucidation of the mechanisms of toxicity, but also support mechanistically-based quantitative risk assessment [1-5]. In addition, these technologies are being used to develop predictive toxicity screening assays to screen drug candidates with adverse characteristics earlier in the development pipeline in order to prioritize resources and maximize successes in clinical trials [6-8]. Comparable screening strategies are also being proposed to rank and prioritize commercial chemicals, natural products, and environmental contaminants that warrant further toxicological investigation. Traditionally, rodent models or surrogates for ecologically-relevant species are typically used in regulatory testing. However, public and regulatory pressure, especially in Europe, seek to minimize the use of animals in testing [9]. Similar policies in the US, such as the ICCVAM Authorization Act of 2000, provide guidelines to facilitate the regulatory acceptance of alternative testing methods. These initiatives combined with the need to assess an expanding list of drug candidates and commercial chemicals for toxicity, have increased demand for the development and implementation of high-throughput in vitro screening assays that are predictive of toxicity in humans and ecologically-relevant species.
Various in vitro hepatic models including the isolated perfused liver, precision cut liver slices, isolated primary liver cells and a number of immortalized liver cell lines, have been used as animal alternatives [10]. In addition to providing a renewable model, in vitro systems are a cost-effective alternative and are amenable to high-throughput screening. These models, particularly immortalized cell lines, also allow for more in-depth biochemical and molecular investigations, such as over-expression, knock-down, activation or inhibition strategies, thus further elucidating mechanisms of action. However, inherent limitations in the ability of cell cultures to model whole organism responses must also be considered when identifying putative biomarkers for high-throughput toxicity screening assays, and elucidating relevant mechanisms of toxicity that support quantitative risk assessment. Despite several in vitro toxicogenomic reports [11-13], few have systematically examined the ability of in vitro systems to predict in vivo gene expression profiles in response to chemical treatment [10,14].
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is a widespread environmental contaminant that elicits a number of adverse effects including tumor promotion, teratogenesis, hepatotoxicity, and immunotoxicity as well as the induction of several metabolizing enzymes [15]. Many, if not all of these effects, are due to alterations in gene expression mediated by the aryl hydrocarbon receptor (AhR), a basic-helix-loop-helix-PAS (bHLH-PAS) transcription factor [15,16]. Ligand binding to the cytoplasmic AhR complex triggers the dissociation of interacting proteins and results in the translocation of the ligand-bound AhR to the nucleus where it heterodimerizes with the aryl hydrocarbon receptor nuclear translocator (ARNT), another member of the bHLH-PAS family. The heterodimer then binds specific DNA elements, termed dioxin response elements (DREs), within the regulatory regions of target genes leading to changes in expression that ultimately result in the observed responses [17]. Although the role of AhR is well established, the gene regulatory pathways responsible for toxicity are poorly understood and warrant further investigation to assess the potential risks to humans and ecologically relevant species.
Hepa1c1c7 cells and C57BL/6 mice are well-established models routinely used to examine the mechanisms of action of TCDD and related compounds. In this study, TCDD-elicited temporal gene expression effects were systematically compared in order to assess the ability of Hepa1c1c7 cells to replicate C57BL/6 hepatic tissue responses. Our results indicate that several phase I and II metabolizing enzyme responses are aptly reproduced. However, many responses were model-specific and reflect inherent in vitro and in vivo differences that must be considered in mechanistic studies and during the selection of biomarkers for developing toxicity screening assays.
Results
In vitro microarray data analysis
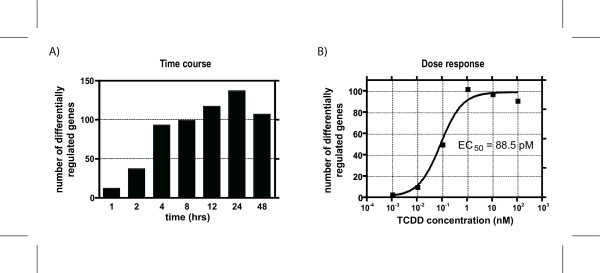
Temporal gene expression profiles were assessed in Hepa1c1c7 wild type cells following treatment with 10 nM TCDD using cDNA microarrays with 13,362 spotted features. Empirical Bayes analysis of the in vitro time course data identified 331 features representing 285 unique genes with a P1(t) value greater than 0.9999 at one or more time points, and differential expression greater than ± 1.5 fold relative to time-matched vehicle controls. The number of differentially regulated genes gradually increased from 1 to 24 hrs, followed by a slight decrease at 48 hrs (Figure 1A). In vitro dose-response data performed at 12 hrs with TCDD covering 6 different concentrations (0.001, 0.01, 0.1, 1.0, 10 and 100 nM), identified 181 features representing 155 unique genes (P1(t) > 0.9999 and an absolute fold change > 1.5 at one or more doses; Figure 1B). Complete in vitro time course and dose-response data are available in Additional file 1 and 2, respectively.
Figure 1.
Number of genes differentially regulated (P 1(t) > 0.9999 and Ifold changel > 1.5-fold) as measured by microarray analysis for the (A) time course and (B) and dose-response studies in mouse hepatoma Hepa1c1c7 cells. For the time course study, cells were treated with 10 nM TCDD and harvested at 1, 2, 4, 8, 12, 24 or 48 hrs after treatment. Cells for the 12 hr dose-response study were treated with 0.001, 0.01, 0.1, 1.0, 10 and 100 nM of TCDD
As a control, the gene expression effects elicited by 10 nM TCDD in ARNT-deficient c4 Hepa1c1c7 mutants [18] were examined at 1 and 24 hrs (data not shown). Only ATPase, H+ transporting, V1 subunit E-like 2 isoform 2 (Atp6v1e2) and SUMO/sentrin specific peptidase 6 (Senp6) exhibited a significant change in expression using the same criteria (P1(t) > 0.9999 and an absolute fold change > 1.5). Neither Atp6v1e2 nor Senp6 were among the active genes in wild-type Hepa1c1c7 cells or in C57BL/6 liver samples [19]. These results provide further evidence that the AhR/ARNT signaling pathway mediates TCDD-elicited gene expression responses, which are consistent with in vivo microarray results with AhR knockout mice [20].
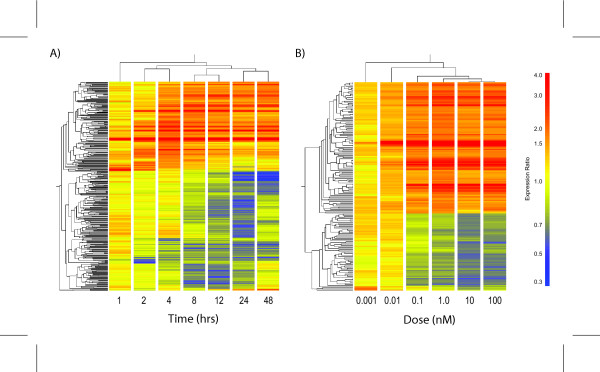
Hierarchical clustering of the genes expressed in Hepa1c1c7 time course indicate that 2 and 4 hrs were most similar, as were 8 and 12 hrs, and 24 and 48 hrs, while the 1 hr time point was segregated (Figure 2A). A strong dose-response relationship was also evident with clusters sequentially branching out with increasing concentration (Figure 2B). At 12 hrs, 117 genes were differentially expressed with 112 exhibiting a dose-dependent response. Moreover, the fold changes measured in both the time course and dose-response studies using 10 nM TCDD were comparable. For example, xanthine dehydrogenase (Xdh) and NAD(P)H dehydrogenase, quinone 1 (Nqo1) were induced 2.39- and 4.89-fold respectively in the time course and 2.93- and 4.71-fold in the dose-response study. There is a strong correlation (R = 0.97) between the differentially expressed genes at 12 hrs in the time course with the differentially regulated genes in the dose-response study at 10 nM, demonstrating the reproducibility between independent studies and providing further evidence that these genes are regulated by TCDD.
Figure 2.
Hierarchical clustering of the differentially regulated gene lists for A) temporal and B) dose-response microarray studies in mouse hepatoma Hepa1c1c7 cells. The results illustrate time and dose-dependent clustering patterns. From the A) temporal results, the early (2 hr and 4 hr), intermediate (8 hr and 12 hr) and late (24 hr and 48 hr) time points cluster separately while the 1 hr time point clusters alone. Results from the B) dose-response show that highest doses clustered together, while the remaining doses branched out in a dose-dependent manner
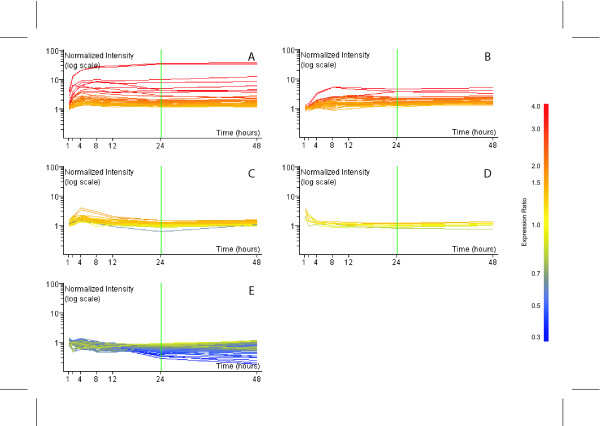
The list of temporally regulated genes was subjected to k-means clustering using the standard correlation distance metrics. Five k-means clusters best characterized the dataset and identified clusters representing A) up-regulated early and sustained, B) up-regulated intermediate and sustained, C) up-regulated intermediate, D) up-regulated immediate and E) down-regulated late (Figure 3). These were comparable to the k-means clusters identified in hepatic tissue of C57BL/6 mice following treatment with 30 μg/kg TCDD [19]. Although, no discernable functional category is over-represented in any one cluster, the sustained up-regulation of early (Cluster A) and intermediate (Cluster B) responding genes include classic TCDD-responsive genes such as cytochrome P450, family 1, subfamily a, polypeptide 1 (Cyp1a1), Xdh and Nqo1. Many down-regulated late genes were associated with cell cycle regulation such as myelocytomatosis oncogene (Myc). Additionally, targets of Myc, including cyclin D1 and ornithine decarboxylase (Odc1), were also down-regulated suggesting a mechanism for cell cycle arrest [21-23], a common in vitro response to TCDD.
Figure 3.
K-means clustering of temporally differentially regulated genes in vitro. Five k-mean clusters corresponding to (A) up-early and sustained, (B) up-intermediate and sustained, (C) up-regulated intermediate, (D) up-regulated immediate and (E) down-regulated late. Time and expression ratio are indicated on the x- and y-axis respectively. The color of individual gene expression profiles reflects the expression ratio observed at 24 hrs
Classification of gene expression responses for common regulated genes
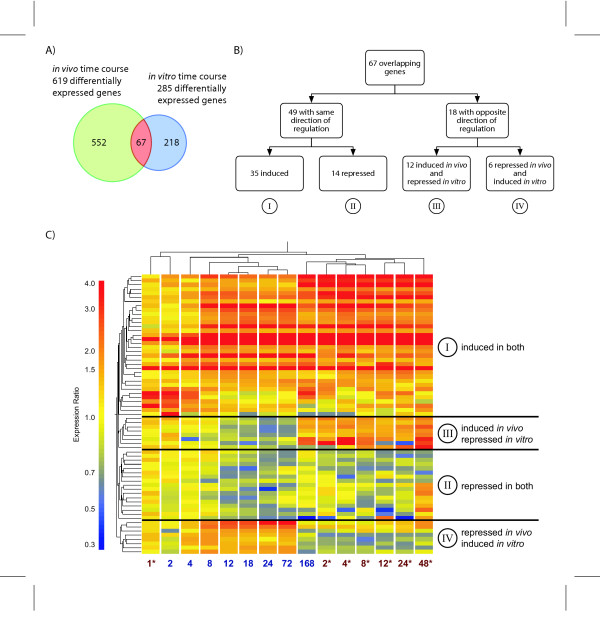
Using the same filtering criteria (P1(t) > 0.9999 and an absolute fold change > 1.5), 678 features representing 619 unique genes were differentially expressed as previously reported in a time course study conducted in hepatic tissue from C57BL/6 mice orally gavaged with 30 μg/kg TCDD [19]. The number of responsive in vivo genes and their temporal expression patterns closely paralleled the results from this in vitro study. The fewest number of active genes was observed at 2 hrs, followed by a large increase at 4 hrs, which was sustained to 72 hrs. However, the substantial increase in expressed in vivo genes at 168 hrs was attributed to triglyceride accumulation and immune cell infiltration, which was not observed in Hepa1c1c7 cells. This list of 619 of in vivo genes served as the basis for subsequent comparisons against TCDD-elicited in vitro responses.
Comparison of in vitro and in vivo differentially expressed gene lists identified common and model specific responses (Figure 4A). TCDD treatment resulted in a total of 838 regulated genes in either model and with 67 common to both. TCDD elicited 218 gene expression changes unique to Hepa1c1c7 cells while 552 genes were specific to C57BL/6 hepatic samples. Although 67 genes were regulated in both models, not all possessed similar temporal patterns of expression. Contingency analysis using a 2 × 2 table and the χ2 test resulted in a p-value < 0.001 (α = 0.05) illustrate a statistically significant association between the lists of differentially regulated genes in vitro and in vivo. Further stratification revealed genes that were either induced in both models (class I), repressed in both models (class II), induced in vivo while repressed in vitro (class III), or repressed in vivo while induced in vitro (class IV; Figure 4B). Genes regulated in a similar fashion in both models (classes I and II) accounted for 49 of the 67 common active genes, while the remaining genes exhibited divergent expression profiles (classes III and IV). Hierarchical clustering of the temporal expression values for the 67 overlapping genes identified the same four classes (Figure 4C). The pattern across model and time illustrates that the earliest time points (i.e. 1 hr in vitro and 2 hr in vivo time points) cluster together while the remaining clusters branch into in vitro or in vivo clusters according to time. These results suggest that potential biomarkers of acute TCDD-mediated responses may best be predicted by the immediate-early in vitro gene responses.
Figure 4.
Comparison of common significant in vitro and in vivo TCDD elicited time-dependent gene expression changes. A) 285 differentially regulated in vitro genes and 619 differentially regulated in vivo genes were identified, with 67 genes common to both studies. B) The temporal gene expression profiles from both studies were categorized into (I) induced in both, (II) repressed in both, (III) induced in vivo and repressed in vitro, and (IV) repressed in vitro and induced in vivo. C) Hierarchical clustering identified similar classification groups. Clustering across both time and model, separated samples from in vitro and in vivo, with the exception of the early time points from both studies (1 hr in vitro and 2 hr in vivo), which clustered together. * identifies in vitro time points
In vitro and in vivo induced genes (class I) include xenobiotic and oxidoreductase enzymes such as abhydrolase domain containing 6 (Abhd6), Cyp1a1, dehydrogenase/reductase (SDR family) member 3 (Dhrs3), Nqo1, prostaglandin-endoperoxide synthase 1 (Ptgs1), UDP-glucose dehydrogenase (Ugdh) and Xdh (Table 1). These genes have previously been reported to be TCDD-responsive [19,24], with Cyp1a1 and Nqo1 being members of the "AhR gene battery" [25]. Glutathione S-transferase, alpha 4 (Gsta4) was also induced in vitro and in vivo, 1.7- and 2.0-fold respectively, consistent with TCDD-mediated induction of phase I and II metabolizing enzymes. Of the 35 genes responding similarly in both models, approximately 71% of were similarly up-regulated (class I) while the remaining genes were repressed across both models (class II). Repressed class II genes include minichromosome maintenance deficient 6 (Mcm6), glycerol kinase (Gyk) and ficolin A (Fcna) (repressed 1.6-, 1.6- and 1.7-fold in vitro, respectively). Overall, repressed genes did not share any common discernable biological function.
Table 1.
Classification of common differentially regulated temporal gene expression responses to TCDD in both in vitro and in vivo models
| Accession | Gene name | Gene symbol | Entrez Gene ID | In vivo | In vitro | ||||
| Fold changea | Time pointsb | EC50c,d (μg/kg) | Fold changea | Time pointsb | EC50c,d (pM) | ||||
| I) Induced both in vivo and in vitroe | |||||||||
| BE689910 | RIKEN cDNA 2310001H12 gene | 2310001H12Rik | 69504 | 2.7 | 2f, 168 | 48.02 | 3.9 | 1f | ND |
| BF226070 | RIKEN cDNA 2600005C20 gene | 2600005C20Rik | 72462 | 2.1 | 4, 12, 18, 24f, 72, 168 | 2.18 | 2.3 | 4, 8, 12f, 24, 48 | 265.50 |
| AI043124 | RIKEN cDNA 2810003C17 gene | 2810003C17Rik | 108897 | 1.6 | 12f | 37.02 | 1.7 | 4f | ND |
| AW537038 | expressed sequence AA959742 | AA959742 | 98238 | 7.2 | 4, 8, 12f, 18, 24, 72, 168 | 1.71 | 5.2 | 4, 8f, 12, 24, 48 | 67.79 |
| W34507 | abhydrolase domain containing 6 | Abhd6 | 66082 | 1.7 | 4, 8f, 12, 18, 24, 72, 168 | 154.30 | 1.5 | 48f | 138.50 |
| NM_026410 | cell division cycle associated 5 | Cdca5 | 67849 | 8.8 | 4, 8, 12, 18, 24, 72f, 168 | ND | 1.7 | 4f | ND |
| BG063743 | craniofacial development protein 1 | Cfdp1 | 23837 | 3.6 | 4, 8, 12f, 18, 24, 72, 168 | 14.27 | 2.3 | 4f, 8, 12, 24, 48 | 42.64 |
| AA073604 | procollagen, type I, alpha 1 | Col1a1 | 12842 | 1.7 | 18, 24, 72f | 0.65 | 1.6 | 4, 8, 12f | 17.25 |
| NM_009992 | cytochrome P450, family 1, subfamily a, polypeptide 1 | Cyp1a1 | 13076 | 38.4 | 2, 4, 8, 12, 18, 24f, 72, 168 | 0.05 | 37.7 | 1, 2, 4, 8, 12, 24, 48f | 14.06 |
| BE457542 | Dehydrogenase/reductase (SDR family) member 3 | Dhrs3 | 20148 | 2.0 | 4, 8, 12f, 18, 72, 168 | 0.67 | 1.5 | 8f | 2.43 |
| AW552715 | DnaJ (Hsp40) homolog, subfamily B, member 11 | Dnajb11 | 67838 | 1.7 | 12, 18, 24f,168 | 3.95 | 1.6 | 8, 12f | 9.85 |
| AK015223 | dermatan sulphate proteoglycan 3 | Dspg3 | 13516 | 6.2 | 4, 8, 12, 18, 24f, 72, 168 | 0.13 | 8.4 | 2, 4, 8, 12, 24, 48f | 16.34 |
| NM_008655 | growth arrest and DNA-damage-inducible 45 beta | Gadd45b | 17873 | 4.6 | 2f, 4, 72 | 133.30 | 3.7 | 1f, 2 | 1440.00 |
| W54349 | glutathione S-transferase, alpha 4 | Gsta4 | 14860 | 2.0 | 18, 24, 72f | 0.48 | 1.7 | 8f, 12 | 56.38 |
| BG067127 | interferon regulatory factor 1 | Irf1 | 16362 | 1.5 | 168f | ND | 1.7 | 2f, 4 | ND |
| AA015278 | integrin beta 1 (fibronectin receptor beta) | Itgb1 | 16412 | 1.6 | 4, 18, 24, 168f | 97.23 | 4.2 | 4, 8, 12, 24, 48f | 72.92 |
| AA041752 | Jun proto-oncogene related gene d1 | Jund1 | 16478 | 2.0 | 12f, 18, 24 | 0.99 | 2.1 | 4, 8, 12, 24, 48f | 50.34 |
| BF538945 | lectin, mannose-binding, 1 | Lman1 | 70361 | 1.9 | 12, 72, 168f | 13.49 | 2.0 | 4f, 8, 24, 48 | 40.72 |
| BG066626 | lipin 2 | Lpin2 | 64898 | 3.0 | 4, 12, 24f, 72 | 3.13 | 2.3 | 2, 4f, 8, 12, 24, 48 | 23.83 |
| BI440950 | leucine rich repeat containing 39 | Lrrc39 | 109245 | 2.9 | 2f, 4 | 49.71 | 3.1 | 1f, 2 | 68.57 |
| AW413953 | mitochondrial ribosomal protein L37 | Mrpl37 | 56280 | 8.3 | 2, 4, 8f, 12, 18, 24, 72, 168 | 8.77 | 2.7 | 2, 4f, 8, 12, 24, 48 | 49.59 |
| BE623489 | NAD(P)H dehydrogenase, quinone 1 | Nqo1 | 18104 | 4.6 | 4, 8, 12f, 18, 24, 72, 168 | 1.00 | 5.2 | 4, 8f, 12, 24, 48 | 33.74 |
| NM_026550 | PAK1 interacting protein 1 | Pak1ip1 | 68083 | 3.8 | 4, 8, 12, 18, 24, 72f, 168 | 0.26 | 2.2 | 4f, 8, 12, 24, 48 | 7.00 |
| AA152754 | prostaglandin-endoperoxide synthase 1 | Ptgs1 | 19224 | 1.6 | 168f | 1.11 | 2.3 | 4, 8f, 12, 24, 48 | 37.96 |
| BG063583 | solute carrier family 20, member 1 | Slc20a1 | 20515 | 2.2 | 2, 4f, 8 | ND | 1.8 | 2f, 4 | ND |
| AJ223958 | solute carrier family 27 (fatty acid transporter), member 2 | Slc27a2 | 26458 | 1.9 | 12f, 18, 24, 72, 168 | 2.88 | 2.1 | 8, 12f, 24, 48 | 17.42 |
| BG066820 | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | Slc6a6 | 21366 | 1.8 | 4f, 12 | 2.48 | 1.7 | 48f | 3.06 |
| AI592773 | suppression of tumorigenicity 5 | St5 | 76954 | 1.6 | 8, 12f | 28.85 | 1.7 | 4f, 8, 12 | 14.69 |
| BG067168 | TCDD-inducible poly(ADP-ribose) Polymerase | Tiparp | 99929 | 10.3 | 2, 4f, 12, 18, 24, 72, 168 | 36.49 | 6.4 | 1, 2f, 4, 8, 12, 24, 48 | 18.03 |
| BG065761 | tumor necrosis factor, alpha-induced protein 2 | Tnfaip2 | 21928 | 5.5 | 2, 4f, 12, 18, 72 | 36.41 | 6.3 | 2, 4f, 8, 12, 24, 48 | 41.15 |
| AA067191 | UDP-glucose dehydrogenase | Ugdh | 22235 | 3.1 | 4, 8, 12 f, 18, 24, 72, 168 | 0.79 | 1.5 | 2, 4, 8, 12f, 48 | 4.33 |
| NM_011709 | whey acidic protein | Wap | 22373 | 5.9 | 2, 4, 8, 12, 18, 24, 72, 168f | 0.12 | 4.2 | 2, 4, 8, 12, 24, 48f | 17.44 |
| BG075778 | Xanthine dehydrogenase | Xdh | 22436 | 2.7 | 4, 8, 12f, 18, 24, 72, 168 | 1.24 | 2.6 | 4, 8, 12, 24, 48f | 34.92 |
| BG073881 | zinc finger protein 36, C3H type-like 1 | Zfp36l1 | 12192 | 2.2 | 2f | ND | 1.7 | 1, 2f | 2427.00 |
| AA031146 | zinc finger protein 672 | Zfp672 | 319475 | 1.6 | 4f | 3.09 | 1.5 | 2f | ND |
| II) Repressed both in vivo and in vitroe | |||||||||
| BG146493 | RIKEN cDNA 6330406L22 gene | 6330406L22Rik | 70719 | -1.5 | 18f | 0.51 | -1.8 | 8, 12f | 25.67 |
| AA140059 | DNA methyltransferase (cytosine-5) 1 | Dnmt1 | 13433 | -1.9 | 168f | ND | -1.6 | 8f, 12 | ND |
| AI327022 | ficolin A | Fcna | 14133 | -1.6 | 18, 24f | ND | -1.7 | 12, 24f | ND |
| AA288963 | fibrinogen-like protein 1 | Fgl1 | 234199 | -1.9 | 24f | ND | -1.5 | 24f, 48 | ND |
| BE626913 | GTP binding protein 6 (putative) | Gtpbp6 | 107999 | -3.4 | 24, 72f | ND | -1.7 | 24, 48f | 116.40 |
| AA275564 | glycerol kinase | Gyk | 14933 | -1.5 | 12f | 10.2 | -1.6 | 24f | ND |
| BG070106 | lipocalin 2 | Lcn2 | 16819 | -2.8 | 24f | ND | -1.5 | 24, 48f | ND |
| AW049427 | leucine zipper domain protein | Lzf | 66049 | -1.6 | 24f | ND | -1.6 | 48f | 78.29 |
| AA016759 | minichromosome maintenance deficient 6 | Mcm6 | 17219 | -1.6 | 18f | 3.34 | -1.6 | 8f | 58.04 |
| BF011268 | mitochondrial methionyl-tRNA formyltransferase | Mtfmt | 69606 | -1.8 | 24, 72f, 168 | ND | -1.6 | 24, 48f | ND |
| AA683699 | RNA (guanine-7-) methyltransferase | Rnmt | 67897 | -2.0 | 12f | ND | -1.6 | 8f | ND |
| syntrophin, gamma 1 | Sntg1 | 71096 | -1.6 | 24f | 15.27 | -1.7 | 4f | 66.61 | |
| AA199550 | syntaxin 12 | Stx12 | 100226 | -1.5 | 18f | ND | -1.6 | 48f | ND |
| AA047942 | thymidine kinase 1 | Tk1 | 21877 | -1.7 | 18f, 24, 72 | 0.34 | -2.0 | 8, 12f | 153.90 |
| III) Induced in vivo and repressed in vitroe | |||||||||
| AA122925 | carbonic anhydrase 2 | Car2 | 12349 | 2.4 | 12, 72, 168f | 2.00 | -1.8 | 24f, 48 | 55.96 |
| AI327078 | coactosin-like 1 | Cotl1 | 72042 | 1.6 | 168f | ND | -1.7 | 24, 48f | 25.19 |
| NM_007935 | enhancer of polycomb homolog 1 | Epc1 | 13831 | 1.6 | 168f | 1.16 | -2.7 | 12, 24f | 75.21 |
| BC002008 | fatty acid binding protein 5, epidermal | Fabp5 | 16592 | 3.9 | 8, 12f | 2.43 | -1.9 | 8, 12f, 24 | 54.14 |
| NM_026320 | growth arrest and DNA-damage- inducible, gamma interacting protein 1 | Gadd45gip1 | 102060 | 1.8 | 168f | 4.67 | -1.5 | 8f | 40.49 |
| W11419 | inhibitor of DNA binding 3 | Id3 | 15903 | 1.8 | 168f | 0.34 | -1.5 | 24, 48f | 88.83 |
| AA009268 | myelocytomatosis oncogene | Myc | 17869 | 3.7 | 4, 12f, 168 | 5.59 | -2.2 | 2f | 148.40 |
| NM_011033 | poly A binding protein, cytoplasmic 2 | Pabpc2 | 18459 | 7.0 | 2f | ND | -1.6 | 12f | ND |
| REST corepressor 1 | Rcor1 | 217864 | 1.9 | 4, 8, 18, 72, 168 | 3.70 | -1.6 | 24f | 116.50 | |
| BE980584 | secretory granule neuroendocrine protein 1, 7B2 protein | Sgne1 | 20394 | 3.3 | 168f | 0.74 | -1.5 | 48f | 175.00 |
| AA462951 | transcription factor 4 | Tcf4 | 21413 | 1.6 | 12f, 168 | 5.77 | -1.5 | 24f | 74.44 |
| AA003942 | tenascin C | Tnc | 21923 | 1.6 | 168f | 0.37 | -1.8 | 24f, 48 | 59.34 |
| IV) Repressed in vivo and induced in vitroe | |||||||||
| W36712 | B-cell translocation gene 2, anti- proliferative | Btg2 | 12227 | -1.8 | 18f, 24 | ND | 1.5 | 4f | ND |
| AA174215 | cathepsin L | Ctsl | 13039 | -1.6 | 24f, 72, 168 | ND | 1.6 | 8, 48f | ND |
| AA419858 | cysteine rich protein 61 | Cyr61 | 16007 | -1.6 | 2f | 0.07 | 1.6 | 8, 48f | ND |
| AW488956 | polo-like kinase 3 | Plk3 | 12795 | -1.6 | 4f | ND | 1.6 | 4, 48f | ND |
| BG068288 | solute carrier organic anion transporter family, member 1b2 | Slco1b2 | 28253 | -1.7 | 8f, 12, 18, 24, 72, 168 | ND | 1.6 | 4f | 1.18 |
| NM_011470 | small proline-rich protein 2D | Sprr2d | 20758 | -1.6 | 18f, 72 | 1.97 | 1.6 | 4f | ND |
aMaximum absolute fold change determined by microarray analysis
bTime point where genes are differentially regulated with P1(t) > 0.9999 and |fold change| > 1.5
cEC50 valued determined from microarray results
dND = not determined from microarray results
eClassification groups as defined in Figure 4B
fTime point representing the maximum |fold change|
Forty-two of the 67 common differentially expressed genes were dose responsive at 12 and 24 hrs in vitro and in vivo, respectively, further suggesting the role of the AhR in mediating these responses. Microarray-based EC50 values spanned at least 3 orders of magnitude ranging from 0.05 μg/kg to >150 μg/kg in vivo, and 0.00118 nM to 2.4 nM in vitro (Table 1). Cyp1a1, the prototypical marker of TCDD exposure, had EC50 values of 0.05 μg/kg and 0.014 nM, in vivo and in vitro respectively, and was induced 38-fold in both time course studies. Complete data sets for the in vivo time course and dose-responses experiments are available in Additional file 3 and 4.
Of the 67 overlapping genes, 18 exhibited divergent temporal profiles (classes III and IV). Class III contains 12 genes induced in vivo but repressed in vitro, while 6 were repressed in vivo and induced in vitro (class IV). Example genes include Myc (class III) and B-cell translocation gene 2 (Btg2, class IV) which are both involved in regulating cell cycle progression [23,26-30]. Myc was induced 3.7-fold in vivo and repressed 2.2-fold in vitro, while Btg2 was repressed 1.8-fold in vivo and induced 1.5-fold in vitro.
In addition to the regulated genes common to both models, 218 in vitro- and 559 in vivo-specific genes were identified. Many of the unique in vitro responses are involved in cell cycle regulation, including cyclins D1 and B2 (Table 2). Cyclin D1, which complexes with cyclin-dependent kinase 4 (Cdk4) to regulate the progression from G1 to S phase [31,32], was down-regulated early and repressed 1.7-fold to 48 hrs. Furthermore, cyclin B2 and cell division cycle 2 homolog A (Cdc2a) which interact to form an active kinase required for G2 promotion, were down-regulated, 1.8-fold and 1.5-fold, respectively. In addition to cell cycle related genes, UDP glucuronosyltransferase 1 family, polypeptide A2 (Ugt1a2), a phase II metabolizing enzyme, was induced 2.8-fold in vitro, but not significantly regulated in vivo.
Table 2.
Examples of TCDD-elicited gene expression responses unique to Hepa1c1c7 cells
| Accession | Gene name | Gene Symbol | Entrez Gene ID | Fold changea | Time pointsb (hrs) |
| AA111722 | cyclin D1 | Ccnd1 | 12443 | -1.7 | 4, 8, 12, 24c, 48 |
| AA914666 | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | Cdkn2b | 12579 | 2.4 | 4c, 8, 48 |
| BC008247 | cyclin B2 | Ccnb2 | 12442 | -1.8 | 24c |
| BG064846 | cell division cycle 2 homolog A (S. pombe) | Cdc2a | 12534 | -1.5 | 12, 24c |
| AA011839 | minichromosome maintenance deficient 2 mitotin (S. cerevisiae) | Mcm2 | 17216 | -1.8 | 8c, 12 |
| BG074721 | minichromosome maintenance deficient 7 (S. cerevisiae) | Mcm7 | 17220 | -1.7 | 8, 12c, 24 |
| AA003042 | myeloblastosis oncogene-like 2 | Mybl2 | 17865 | -2.2 | 8c, 12, 24 |
| L27122 | UDP glucuronosyltransferase 1 family, polypeptide A2 | Ugt1a2 | 22236 | 2.8 | 4, 8, 12c, 24, 48 |
aMaximum absolute fold change determined by microarray analysis
bDifferentially regulated genes with P1(t) > 0.9999 and |fold change| > 1.5
cTime point representing the maximum |fold change|
Analysis of the C57BL/6 hepatic time course identified 552 unique genes that were solely regulated in vivo. This included TCDD induced transcripts for microsomal epoxide hydrolase 1 (Ephx1) and carbonyl reductase 3 (Cbr3) which both function as xenobiotic metabolizing enzymes. Notch gene homolog 1 (Notch1) and growth arrest specific 1 (Gas1) which are both associated with development and differentiation but serve undetermined roles in the liver, were also induced by TCDD (Table 3). Genes related to immune cell accumulation were also specific to the in vivo study, coincident with immune cell accumulation at 168 hr as determined by histopathological examination [19].
Table 3.
Examples of TCDD-elicited gene expression responses unique to C57BL/6 hepatic tissue
| Accession | Gene name | Gene Symbol | Entrez Gene ID | Fold changea | Time pointsb (hrs) |
| AA170585 | carbonic anhydrase 3 | Car3 | 12350 | -3.5 | 12c, 18, 24, 168 |
| AK003232 | carbonyl reductase 3 | Cbr3 | 109857 | 2.2 | 12, 18c |
| AA571998 | CD3 antigen, delta polypeptide | Cd3d | 12500 | -2.4 | 12, 18c, 24, 72, 168 |
| BG072496 | ELOVL family member 5, elongation of long chain fatty acids | Elovl5 | 68801 | 2.0 | 8, 12c, 18, 24, 72, 168 |
| BG072453 | epoxide hydrolase 1, microsomal | Ephx1 | 13222 | 1.9 | 8, 12 18, 24c |
| W84211 | growth arrest specific 1 | Gas1 | 14451 | -1.9 | 4, 8, 18, 24, 72, 168c |
| W41175 | glycerol phosphate dehydrogenase 2, mitochondrial | Gpd2 | 14571 | -2.3 | 8, 12, 18, 24, 72c, 168 |
| W29265 | glutathione S-transferase, alpha 2 (Yc2) | Gsta2 | 14858 | 7.2 | 12, 18, 24, 72c, 168 |
| AA145865 | lymphocyte antigen 6 complex, locus A | Ly6a | 110454 | 2.5 | 72, 168c |
| W98998 | Notch gene homolog 1 (Drosophila) | Notch1 | 18128 | 3.3 | 2, 4c, 8, 12, 18, 24, 72, 168 |
aMaximum absolute fold change determined by microarray analysis
bDifferentially regulated genes with P1(t) > 0.9999 and |fold change| > 1.5
cTime point representing the maximum |fold change|
Comparison of basal gene expression levels in Hepa1c1c7 cells and hepatic tissue
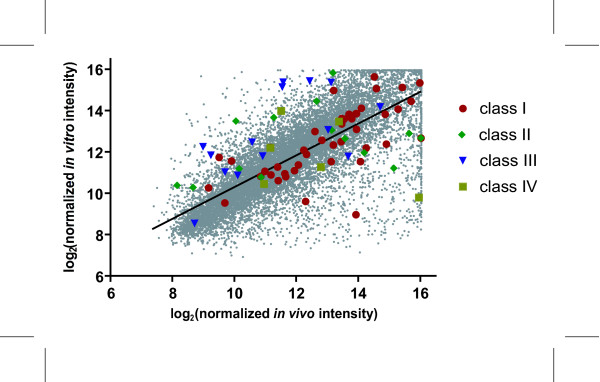
In order to further investigate differences in gene expression levels, Hepa1c1c7 cells and C57BL/6 liver samples were directly compared by competitive hybridization on the same array, to identify basal gene expression level differences. Subsequent linear regression analysis of the mean normalized signal intensities from the untreated samples resulted in a correlation value of R = 0.75 (Figure 5), which is consistent with basal gene expression comparisons of various in vitro rat hepatic systems against whole livers, where correlation values decreased between liver slices (R = 0.97), primary cells (R = 0.85), BRL3A (R = 0.3) and NRL clone 9 (R = 0.32) rat liver cell lines [10]. Overall, the correlation illustrates reasonable concordance in basal gene expression levels between the two models. However, data points which deviate from the fitted line indicate differences in the basal expression of individual genes between the Hepa1c1c7 cells and hepatic tissue from C57BL/6 mice. Although there are differences, they may be negligible if the TCDD-elicited responses are conserved in vitro and in vivo. Complete microarray data for the untreated comparisons are available in Additional file 5.
Figure 5.
Comparison of Hepalclc7 cell and C57BL/6 hepatic tissue basal gene expression. Untreated samples from Hepalclc7 cells and hepatic tissue from immature ovariectomized C57BL/6 mice taken at 0 hrs were competitively hybridized to the 13,362 feature cDNA microarray. Log2 normalized signal intensities were plotted for in vitro versus in vivo data to generate the correlation coefficient. The linear correlation coefficient R was 0.75 between in vitro and in vivo models
The relative basal expression of the 67 common active features was further investigated (Figure 5). In general, class I (i.e. induced in both models) genes fell close to the regression line, indicating that the basal expression of induced genes were comparable as were their in vitro and in vivo responses to TCDD. In contrast, basal expression levels of class III genes (i.e. induced in vivo while repressed in vitro) were generally higher in the Hepa1c1c7 cells, while levels in class II and IV (i.e. repressed in both models and repressed in vivo while induced in vitro, respectively) genes were scattered around the fitted linear line in Figure 5.
Quantitative real-time PCR verification of microarray responses
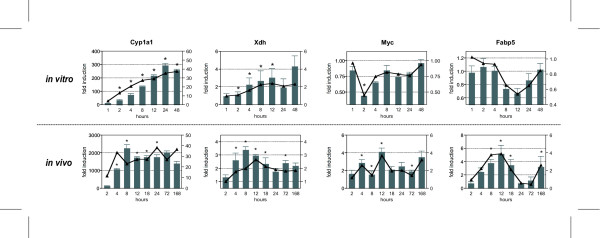
In total, 14 in vitro and 24 in vivo responsive genes representing common and model-specific genes were verified by quantitative real-time PCR (QRTPCR) (see Additional file 6). Of the selected genes regulated in both models, all displayed temporal patterns comparable to the microarray data (Figure 6). For example, Xdh, Myc and fatty acid binding protein (Fabp5) exhibited good agreement in fold change and temporal expression pattern when comparing microarray and QRTPCR data. However, significant data compression was evident when comparing in vitro and in vivo Cyp1a1 induction by QRTPCR, although in vitro and in vivo microarray induction levels were comparable. Previous studies suggest this is likely due to the limited fluorescence intensity range (0 – 65,535) of microarrays resulting in signal saturation and compression of the true magnitude of induction of transcript levels [33,34]. Cross hybridization of homologous probes to a given target sequence on the microarray may also be a contributing factor, especially in comparison to other, more gene-specific measurement techniques [35].
Figure 6.
Quantitative real-time PCR verification of in vitro and in vivo microarray results. The same RNA used for cDNA microarray analysis was examined by QRTPCR. All fold changes were calculated relative to time-matched vehicle controls. Bars (left axis) and line (right axis) represent data obtained by QRTPCR and cDNA microarrays, respectively. Genes are indicated by official gene symbols, and results are the average of four biological replicates. Classes refer to the respective classification categories as illustrated in Figure 4B. Error bars represent the standard error of measurement for the average fold change. *p < 0.05 for QRTPCR
Discussion
Microarrays have become an invaluable tool in toxicogenomics for comprehensively characterizing gene expression responses following treatment with an environmental contaminant, commercial chemical, natural product or drug as well as for investigating complex mixtures relevant to human and wildlife exposures. An emerging consensus suggests that toxicogenomics will accelerate drug development and significantly improve quantitative risk assessments [36,37]. In addition, toxicogenomics supports the development and refinement of predictive in vitro high-throughput toxicity screening assays that can be used as alternatives to traditional in vivo testing. Ideally, in vitro high-throughput toxicity screens can be used to rank and prioritize drug candidates, environmental contaminants, and commercial chemicals, which warrant further development or testing. Although in vitro responses are assumed to reflect a subset of comparable in vivo responses, few studies have completed a comprehensive and systematic comparison. This study closely examined two well-established models, and comprehensively compared the TCDD-elicited gene expression to assess the predictive value of in vitro systems.
Comparative analysis of Hepa1c1c7 cell and hepatic C57BL/6 microarray data identified 67 differentially expressed genes co-regulated by TCDD. Four classes based on their temporal expression patterns were identified (Figure 4B and 4C), with 42 of the 67 common regulated genes exhibiting dose-response characteristics in both models. In vitro EC50 values ranged from 0.001182 nM to 2.4 nM, while in vivo the values ranged from 0.05 μg/kg to >150 μg/kg. The wide range of EC50 values illustrate the varying sensitivity of regulated genes to TCDD in both models.
Hepa1c1c7 cells and hepatic tissue from C57BL/6 mice are the prototypical models used to investigate the mechanisms of action of TCDD and other related compounds and both exhibited the classic induction of phase I and II metabolizing enzymes including Cyp1a1 and Nqo1 [38,39]. Gsta4 and Xdh were also up-regulated in both models further demonstrating Hepa1c1c7 cells as a suitable model for investigating TCDD-regulated induction of xenobiotic metabolizing genes. In addition to these genes, the responses of Nqo1, Ugdh and Tnfaip2 were also conserved across models and were categorized as class I genes (similarly induced in both models; Figure 4B and 4C). However, Gsta2 was induced in vivo while no significant effect was detected in Hepa1c1c7 cells, and Ugt1a2 was induced in vitro but not differentially expressed in C57BL/6 hepatic tissue. Although many phase I and II metabolizing enzyme responses were conserved, differences exist that may limit Hepa1c1c7 cells from accurately modeling the full spectrum of in vivo hepatic responses elicited by TCDD.
A direct comparison of untreated Hepa1c1c7 cells and C57BL/6 hepatic tissue was performed to further investigate innate differences between the two models. Comparison of the normalized signal intensities revealed a good correlation (R = 0.75) between in vitro and in vivo basal expression levels (Figure 5). This illustrates that many genes are basally expressed to similar levels in both models as illustrated by the cluster of class I (similarly induced genes) closely surrounding the fitted line. Although a correlation exists, there are still differences in basal expression which may be associated with the origins of the models (i.e. normal hepatic tissue versus hepatoma derived Hepa1c1c7 cells), as well as the inability of in vitro systems to effectively model complex interactions between different cell types (e.g. Kupffer and stellate cells). For example, Myc, a G1 to S phase cell cycle regulator [23,26-29], was repressed in vitro while being induced in vivo and the model-specific responses may be related to difference in basal expression levels between the two models (Table 1). The levels of Myc transcripts in untreated Hepa1c1c7 cells were higher relative to untreated C57BL/6 hepatic tissue, consistent with the proliferative state of the in vitro system (data not shown). Examination of other class III genes suggests that they are more highly expressed in vitro when compared to in vivo (Figure 5). Consequently, differences in basal expression may be a factor contributing to divergent in vitro – in vivo responses. Another possible source for the model-specific responses may be related to DNA methylation status of the promoter region of TCDD-responsive genes in either model. DNA methylation results in gene silencing [40,41] and a previous study with Hepa1c1c7 has shown that TCDD-elicited gene expression responses are influenced by DNA methylation status [42]. The differing methylation states between the in vitro and in vivo systems may further contribute to the model-specific gene expression responses.
Many in vitro specific gene expression responses elicited by TCDD were associated with cell cycle progression and cell cycle arrest. Myc and its downstream target, cyclin D1, which forms a kinase complex with Cdk4 [43,44] were both repressed by TCDD. In contrast, Cdkn1a, an inhibitor of cyclin-dependent kinase 2 (Cdk2)-cyclin E complex kinase activity [43], was induced. Inactivation of the Cdk2-cyclin E complex prevents the phosphorylation of pRb resulting in cell cycle arrest during G1. Additionally, the in vitro induction of Btg2 suggests an alternative mechanism for cell cycle arrest during the G2 phase. Constitutively active BTG2 in human leukemia U937 cells, induces G2/M cell cycle arrest by inhibiting the formation of the cyclin B1 and Cdc2 complex, thereby inhibiting the active kinase function of the complex [30]. Collectively, these results corroborate and extend previous in vitro TCDD-mediated cell cycle arrest studies [45-48].
TCDD treatment resulted in a number of divergent gene responses across both models as represented by classes III and IV (Figures 4B and 4C). Genes related to immune cell accumulation, including major histocompatibility complex (MHC) molecules were only observed in vivo, and are likely a response to hepatic damage mediated by ROS or fatty accumulation and therefore independent of direct AhR action [19]. This is characteristic of the complex interaction between different cell types responding to liver injury that cannot be modeled in homogenous cultures of cells.
Pharmacokinetics may also contribute to response differences between the two models. Hepa1c1c7 cells were directly treated, whereas in vivo, TCDD must first be delivered to the liver and targeted cells prior to eliciting its effects. Additionally, C57BL/6 studies were able to be carried out to 168 hrs following TCDD treatment, while in vitro studies were limited to 48 hrs to minimize potentially confounding effects due to cell confluency. However, early responses associated with classes I and II (induced or repressed in both models; Figure 4B and 4C) are well conserved and exhibit comparable levels of induction or repression in both models. Hierarchical clustering of the common active genes (Figure 4C) illustrates gene induction occurs early while gene repression occurs later in both models. Clustering across both time and model revealed that gene expression profiles at 1 hr in vitro and 2 hr in vivo were most similar. This clustering pattern implies that early in vitro responses may accurately model early in vivo gene expression effects.
Conclusion
Comparative analysis of global gene expression from Hepa1c1c7 cells and hepatic tissue from C57BL/6 mice identified several model-specific responses to TCDD that should be considered when extrapolating in vitro results to potential in vivo effects. Despite these differences, immortalized cells as well as other emerging in vitro systems (e.g., primary cells, stem cells and 3-D culture systems) provide valuable mechanistic information that supports the further development of high-throughput toxicity screening assays. However, the relevance of in vitro responses requires complementary in vivo verification. Furthermore, comparative studies exploiting other in vitro and in vivo systems, different structurally diverse ligands and other relevant model species will not only corroborate the relevance of the mechanisms, but will also support more appropriate extrapolations between rodent studies and potential effects in humans and ecologically-relevant species.
Methods
Culture and treatment of cell lines
Hepa1c1c7 wild-type and c4 ARNT-deficient cell lines (gifts from O. Hankinson, University of California, Los Angeles, CA) were maintained in phenol-red free DMEM/F12 media (Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum (FBS) (Hyclone, Logan, UT), 2.5 μg/mL amphotericin B (Invitrogen), 2.5 μg/mL amphotericin B (Invitrogen), 50 μg/mL gentamycin (Invitrogen), 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen). 1 × 106 cells were seeded into T175 culture flasks (Sarstedt, Newton, NC) and incubated under standard conditions (5% CO2, 37°C). Time course studies were performed with wild-type and c4 mutant cells where both were dosed with either 10 nM TCDD (provided by S. Safe, Texas A&M University, College Station, TX) or DMSO (Sigma, St. Louis, MO) vehicle and harvested at 1, 2, 4, 8, 12, 24 or 48 hrs. Additional untreated control cells were harvested at the time of dosing (i.e. 0 hrs). For the dose-response study, wild-type cells were treated with DMSO vehicle or 0.001, 0.01, 0.1, 1.0, 10 or 100 nM TCDD and harvested at 12 hrs. The treatment and harvesting regimen for cell culture studies are illustrated in Additional file 7.
Animal treatment
The handling and treatment of female C57BL/6 mice has been previously described [19]. Briefly, immature ovariectomized mice were orally gavaged with 30 μg/kg TCDD for the time course study and sacrificed at 2, 4, 8, 12, 18, 24 72 or 168 hrs after treatment. For the dose-response study, mice were treated with 0.001, 0.01, 0.1, 1, 10, 100 or 300 μg/kg TCDD and sacrificed 24 hrs after dosing. Animals were sacrificed by cervical dislocation and tissue samples were removed, weighed, flash frozen in liquid nitrogen and stored at -80°C until further use.
RNA isolation
Cells were harvested by scraping in 2.0 mL of Trizol Reagent (Invitrogen). Frozen liver samples (approximately 70 mg) were transferred to 1.0 mL of Trizol Reagent and homogenized in a Mixer Mill 300 tissue homogenizer (Retsch, Germany). Total RNA from each study was isolated according to the manufacturer's protocol with an additional acid phenol:chloroform extraction. Isolated RNA was resuspended in The RNA Storage Solution (Ambion Inc., Austin, TX), quantified (A260), and assessed for purity by determining the A260/A280 ratio and by visual inspection of 1.0 μg on a denaturing gel.
Microarray experimental design
Changes in gene expression were assessed using customized cDNA microarrays containing 13,362 features representing 8,284 unique genes. For the time course study, TCDD-treated samples were compared to time-matched vehicle controls using an independent reference design [49]. In this design, treated Hepa1c1c7 cell or hepatic tissue samples were compared to the corresponding time-matched vehicle control with two independent labelings (dye swaps; Additional file 8). Four replicates of this design were performed, each using independent cell culture samples or different animals. Dose-response changes in gene expression were analyzed using a common reference design in which samples from TCDD-treated cells or mice were co-hybridized with a common vehicle reference (i.e. independent DMSO treated Hepa1c1c7 cell samples, hepatic samples from independent sesame oil treated C57BL/6 mice) using two independent labelings (Additional file 8). Four replicates with two independent labelings were performed for both in vitro and in vivo samples. Co-hybridizations of untreated Hepa1c1c7 cells and hepatic tissue from C57BL/6 mice were performed to investigate differences in basal gene expression levels between models (Additional file 8). Four replicates were performed with two independent labelings per sample (dye swap).
More detailed protocols regarding the microarray assay, including microarray preparation, labeling of the cDNA probe, sample hybridization and washing can be obtained from the dbZach website [50]. Briefly, polymerase chain reaction (PCR) amplified cDNAs were robotically arrayed onto epoxy-coated glass slides (Schott-Nexterion, Duryea, PA) using an Omnigrid arrayer (GeneMachines, San Carlos, CA) equipped with 48 (4 × 12) Chipmaker 2 pins (Telechem) at Michigan State University's Research Technology Support Facility [51]. Total RNA (30 μg) was reverse transcribed in the presence of Cy3- or Cy5-deoxyuridine triphosphate (dUTP) to create fluorescence-labeled cDNA, which was purified using a Qiagen PCR kit (Qiagen, Valencia, CA). Cy3 and Cy5 samples were mixed, vacuum dried and resuspended in 48 μL of hybridization buffer (40% formamide, 4× SSC, 1% sodium dodecyl sulfate [SDS]) with 20 μg polydA and 20 μg of mouse COT-1 DNA (Invitrogen) as competitor. This probe mixture was heated at 95°C for 3 min and hybridized on the array under a 22 × 60 mm LifterSlip (Erie Scientific Company, Portsmouth, NH) in a light-protected and humidified hybridization chamber (Corning Inc., Corning, NY) for 18–24 hrs in a 42°C water bath. Slides were then washed, dried by centrifugation and scanned at 635 nm (Cy5) and 532 nm (Cy3) on an Affymetrix 428 Array Scanner (Santa Clara, CA). Images were analyzed for feature and background intensities using GenePix Pro 5.0 (Molecular Devices, Union City, CA).
Microarray data quality assurance, normalization and analysis
Microarray data were first passed through a quality assurance protocol prior to further analysis to ensure consistently high quality data throughout the dose-response and time course studies prior to normalization and further analysis [52]. All the collected data were then normalized using a semi-parametric approach [53]. Empirical Bayes analysis was used to calculate posterior probabilities (P1(t) value) of activity on a per gene and time point or dose group basis using the model-based t-value [54]. The data were filtered using a P1(t) cutoff of 0.9999 and ± 1.5 fold change to identify the most robust changes in gene expression and to obtain an initial subset of differentially regulated genes for further investigation and data interpretation. Subsequent analysis included agglomerative hierarchical and k-means clustering using the standard correlation distance metric implemented in GeneSpring 6.0 (Silicon Genetics, Redwood City, CA). Functional categorization of differentially regulated genes were mined and statistically analyzed from Gene Ontology [55] using GOMiner [56].
Quantitative real-time PCR analysis
For each sample, 1.0 μg of total RNA was reverse transcribed by Superscript II using an anchored oligo-dT primer as described by the manufacturer (Invitrogen). The cDNA (1.0 μL) was used as a template in a 30 μL PCR reaction containing 0.1 μM of forward and reverse gene-specific primers designed using Primer 3 [57], 3 mM MgCl2, 1.0 mM dNTPs, 0.025 IU AmpliTaq Gold, and 1× SYBR Green PCR buffer (Applied Biosystems, Foster City, CA). PCR amplification was conducted in MicroAmp Optical 96-well reaction plates (Applied Biosystems) on an Applied Biosystems PRISM 7000 Sequence Detection System under the following conditions: initial denaturation and enzyme activation for 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. A dissociation protocol was performed to assess the specificity of the primers and the uniformity of the PCR-generated products. Each plate contained duplicate standards of purified PCR products of known template concentration covering 7 orders of magnitude to interpolate relative template concentrations of the samples from the standard curves of log copy number versus threshold cycle (Ct). No template controls (NTC) were also included on each plate. Samples with a Ct value within 2 standard deviations of the mean Ct values for the NTCs were considered below the limits of detection. The copy number of each unknown sample for each gene was standardized to the geometric mean of three house-keeping genes (β-actin, Gapd and Hprt) to control for differences in RNA loading, quality, and cDNA synthesis. For graphing purposes, the relative expression levels were scaled such that the expression level of the time-matched control group was equal to 1. Statistical analysis was performed with SAS 8.02 (SAS Institute, Cary, NC). Data were analyzed by analysis of variance (ANOVA) followed by Tukey's post hoc test. Differences between treatment groups were considered significant when p < 0.05. Official gene names and symbols, RefSeq and Entrez Gene IDs, forward and reverse primer sequences, and amplicon sizes are listed in Table 4.
Table 4.
Gene names and primer sequences for QRTPCR
| RefSeq | Gene name | Gene Symbol | Entrez Gene ID | Forward Primer | Reverse Primer | Product Size (bp) |
| NM_007393 | actin, beta, cytoplasmic | Actb | 11461 | GCTACAGCTTCACCACCACA | TCTCCAGGGAGGAAGAGGAT | 123 |
| NM_009992 | cytochrome P450, family 1, subfamily a, polypeptide 1 | Cyp1a1 | 13076 | AAGTGCAGATGCGGTCTTCT | AAAGTAGGAGGCAGGCACAA | 140 |
| NM_010634 | fatty acid binding protein 5, epidermal | Fabp5 | 16592 | TGTCATGAACAATGCCACCT | CTGGCAGCTAACTCCTGTCC | 87 |
| NM_008084 | glyceraldehyde-3- phosphate dehydrogenase | Gapd | 2597 | GTGGACCTCATGGCCTACAT | TGTGAGGGAGATGCTCAGTG | 125 |
| NM_013556 | hypoxanthine phosphoribosyl transferase | Hprt | 24465 | AAGCCTAAGATGAGCGCAAG | TTACTAGGCAGATGGCCACA | 104 |
| NM_010849 | myelocytomatosis oncogene | Myc | 17869 | CTGTGGAGAAGAGGCAAACC | TTGTGCTGGTGAGTGGAGAC | 127 |
| NM_011723 | xanthine dehydrogenase | Xdh | 22436 | GTCGAGGAGATCGAGAATGC | GGTTGTTTCCACTTCCTCCA | 124 |
Authors' contributions
In vitro work and associated microarrays and QRTPCR were conducted by ED, similarly, in vivo studies and associated microarrays and QRTPCR were performed by DRB. LDB provided the normalization, statistical analysis and database support of microarray data. Comparison of in vitro and in vivo data was primarily carried out by ED with support by DRB. ED produced the initial draft of the manuscript. TRZ was responsible for the design and oversaw the completion of the study.
Supplementary Material
Hepalclc7 TCDD time course microarray data. Ratios represent expression relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene and time-point basis using the model-based t-value.
Hepalclc7 TCDD dose-response microarray data. Ratios represent expression relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene and dose basis using the model-based t-value.
C57BL/6 mice hepatic tissue TCDD time course microarray data. Ratios represent expression relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene and time-point basis using the model-based t-value.
C57BL/6 mice hepatic tissue TCDD dose-response microarray data. Ratios represent expression relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene and dose basis using the model-based t-value.
Untreated Hepalclc7 and C57BL/6 sample microarray data. Ratios represent basal expression of Hepalclc7 cells relative to hepatic tissue from C5VBL/6 mice. P1(t)-values represent posterior probabilities of activity on a per gene and dose basis using the model-based t-value.
Gene names and primer sequences (5'-3') for transcripts verified by QRTPCR. Primer pair sequences used to verify in vitro and in vivo microarray results using QRTPCR.
Hepa1c1c7 TCDD treatment and harvesting regimen. For the time course study, wild-type and ARNT-deficient c4 mutant cells were treated with 10 nM TCDD or 0.1% DMSO vehicle and harvested at 1, 2, 4, 8, 12, 24, or 48 hrs post-treatment. Untreated controls were harvested at 0 hrs (as indicated by *). The dose-response study was done performed with Hepa1c1c7 wild-type cells and treated with 0.001, 0.01, 0.1, 1.0, 10, 100 nM TCDD or 0.1% DMSO vehicle and harvested 12 hrs post-treatment (as indicated by ‡).
Microarray experimental designs for A) temporal, B) dose-response and C) basal expression studies. A) Temporal gene expression patterns were analyzed by an independent reference design in which cells treated with TCDD (T) were co-hybridized to time-matched vehicle controls (V). This design involves two independent labelings per sample for a total of 14 arrays per replicate. Four biological replicates were conducted for a total of 56 microarrays. Numbers indicate time points for comparison. B) Dose-dependent changes in gene expression were analyzed 12 hrs after treatment using a common reference design in which cells treated with TCDD were co-hybridized with a common vehicle control. This design involves two independent labelings per sample for a total of 12 arrays per replicate. Four biological replicates were conducted for a total of 48 microarrays. Numbers indicate TCDD concentration in nM units. C) Comparative basal gene expression levels between untreated in vitro and in vivo samples were analyzed by an independent reference design. Four biological replicates of untreated Hepa1c1c7 cells and hepatic tissue from C57BL/6 mice harvested at 0 hrs were co-hybridized and two independent labelings were performed per sample for a total of 8 arrays. Double-headed arrows indicate dye swaps (each sample labeled with Cy3 and Cy5 on different microarrays).
Acknowledgments
Acknowledgements
Special thanks to Joshua Kwekel for helpful discussions and critical review of this manuscript. DRB is supported by a fellowship from the Michigan Agricultural Experimental Station. TRZ is partially supported by the Michigan Agricultural Experimental Station. This work was supported by funds from NIH Grant R01 ES 12245.
Contributor Information
Edward Dere, Email: dereedwa@msu.edu.
Darrell R Boverhof, Email: boverho5@msu.edu.
Lyle D Burgoon, Email: burgoonl@msu.edu.
Timothy R Zacharewski, Email: tzachare@msu.edu.
References
- Battershill JM. Toxicogenomics: regulatory perspective on current position. Hum Exp Toxicol. 2005;24:35–40. doi: 10.1191/0960327105ht495oa. [DOI] [PubMed] [Google Scholar]
- Bishop WE, Clarke DP, Travis CC. The genomic revolution: what does it mean for risk assessment? Risk Anal. 2001;21:983–987. doi: 10.1111/0272-4332.216167. [DOI] [PubMed] [Google Scholar]
- Olden K, Wilson S. Environmental health and genomics: visions and implications. Nat Rev Genet. 2000;1:149–153. doi: 10.1038/35038586. [DOI] [PubMed] [Google Scholar]
- Suk WA, Olden K, Yang RS. Chemical mixtures research: significance and future perspectives. Environ Health Perspect. 2002;110:891–892. doi: 10.1289/ehp.110-1241268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant RW. The National Center for Toxicogenomics: using new technologies to inform mechanistic toxicology. Environ Health Perspect. 2002;110:A8–10. doi: 10.1289/ehp.110-a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Blomme EA, Waring JF. Toxicogenomics in drug discovery: from preclinical studies to clinical trials. Chem Biol Interact. 2004;150:71–85. doi: 10.1016/j.cbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lord PG. Progress in applying genomics in drug development. Toxicol Lett. 2004;149:371–375. doi: 10.1016/j.toxlet.2003.12.045. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, Zacharewski TR. Toxicogenomics in risk assessment: applications and needs. Toxicol Sci. 2006;89:352–360. doi: 10.1093/toxsci/kfj018. [DOI] [PubMed] [Google Scholar]
- Abbott A. Animal testing: more than a cosmetic change. Nature. 2005;438:144–146. doi: 10.1038/438144a. [DOI] [PubMed] [Google Scholar]
- Boess F, Kamber M, Romer S, Gasser R, Muller D, Albertini S, Suter L. Gene expression in two hepatic cell lines, cultured primary hepatocytes, and liver slices compared to the in vivo liver gene expression in rats: possible implications for toxicogenomics use of in vitro systems. Toxicol Sci. 2003;73:386–402. doi: 10.1093/toxsci/kfg064. [DOI] [PubMed] [Google Scholar]
- Harris AJ, Dial SL, Casciano DA. Comparison of basal gene expression profiles and effects of hepatocarcinogens on gene expression in cultured primary human hepatocytes and HepG2 cells. Mutat Res. 2004;549:79–99. doi: 10.1016/j.mrfmmm.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Hartung T, Gribaldo L. New hepatocytes for toxicology? Trends Biotechnol. 2004;22:613–615. doi: 10.1016/j.tibtech.2004.10.010. discussion 615–616. [DOI] [PubMed] [Google Scholar]
- Burczynski ME, McMillian M, Ciervo J, Li L, Parker JB, Dunn RT, 2nd, Hicken S, Farr S, Johnson MD. Toxicogenomics-based discrimination of toxic mechanism in HepG2 human hepatoma cells. Toxicol Sci. 2000;58:399–415. doi: 10.1093/toxsci/58.2.399. [DOI] [PubMed] [Google Scholar]
- Luhe A, Hildebrand H, Bach U, Dingermann T, Ahr HJ. A new approach to studying ochratoxin A (OTA)-induced nephrotoxicity: expression profiling in vivo and in vitro employing cDNA microarrays. Toxicol Sci. 2003;73:315–328. doi: 10.1093/toxsci/kfg073. [DOI] [PubMed] [Google Scholar]
- Poland A, Knutson JC. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu Rev Pharmacol Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Denison MS, Heath-Pagliuso S. The Ah receptor: a regulator of the biochemical and toxicological actions of structurally diverse chemicals. Bull Environ Contam Toxicol. 1998;61:557–568. doi: 10.1007/PL00002973. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Seidel SD, Denison MS. Differential gene expression in wild-type and arnt- defective mouse hepatoma (Hepa1c1c7) cells. Toxicol Sci. 1999;52:217–225. doi: 10.1093/toxsci/52.2.217. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Chittim B, Harkema JR, Jump DB, Zacharewski TR. Temporal and Dose-Dependent Hepatic Gene Expression Patterns in Mice Provide New Insights into TCDD-Mediated Hepatotoxicity. Toxicol Sci. 2005;85:1048–1063. doi: 10.1093/toxsci/kfi162. [DOI] [PubMed] [Google Scholar]
- Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. The aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin- independent gene batteries. Mol Pharmacol. 2005 doi: 10.1124/mol.105.018705. [DOI] [PubMed] [Google Scholar]
- Felsher DW, Zetterberg A, Zhu J, Tlsty T, Bishop JM. Over expression of MYC causes p53-dependent G2 arrest of normal fibroblasts. Proc Natl Acad Sci U S A. 2000;97:10544–10548. doi: 10.1073/pnas.190327097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, Nilsson LM, Neale G, Kramer DL, Porter CW, Cleveland JL. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell. 2005;7:433–444. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Wolfle D, Marotzki S, Dartsch D, Schafer W, Marquardt H. Induction of cyclooxygenase expression and enhancement of malignant cell transformation by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Carcinogenesis. 2000;21:15–21. doi: 10.1093/carcin/21.1.15. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Petersen DD, Fornace AJ., Jr Cellular responses to oxidative stress: the [Ah] gene battery as a paradigm. Environ Health Perspect. 1990;88:13–25. doi: 10.1289/ehp.908813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- Lee TC, Li L, Philipson L, Ziff EB. Myc represses transcription of the growth arrest gene gas1. Proc Natl Acad Sci USA. 1997;94:12886–12891. doi: 10.1073/pnas.94.24.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel AL, Shchors K. Mechanisms of c-myc-mediated transcriptional repression of growth arrest genes. Exp Cell Res. 2003;283:17–21. doi: 10.1016/S0014-4827(02)00020-4. [DOI] [PubMed] [Google Scholar]
- Packham G, Cleveland JL. Induction of ornithine decarboxylase by IL-3 is mediated by sequential c-Myc-independent and c-Myc-dependent pathways. Oncogene. 1997;15:1219–1232. doi: 10.1038/sj.onc.1201273. [DOI] [PubMed] [Google Scholar]
- Ryu MS, Lee MS, Hong JW, Hahn TR, Moon E, Lim IK. TIS21/BTG2/PC3 is expressed through PKC-delta pathway and inhibits binding of cyclin B1-Cdc2 and its activity, independent of p53 expression. Exp Cell Res. 2004;299:159–170. doi: 10.1016/j.yexcr.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Barnes-Ellerbe S, Knudsen KE, Puga A. 2,3,7,8-Tetrachlorodibenzo-p-dioxin blocks androgen-dependent cell proliferation of LNCaP cells through modulation of pRB phosphorylation. Mol Pharmacol. 2004;66:502–511. doi: 10.1124/mol.104.000356. [DOI] [PubMed] [Google Scholar]
- Wang W, Smith R, 3rd, Safe S. Aryl hydrocarbon receptor-mediated antiestrogenicity in MCF-7 cells: modulation of hormone-induced cell cycle enzymes. Arch Biochem Biophys. 1998;356:239–248. doi: 10.1006/abbi.1998.0782. [DOI] [PubMed] [Google Scholar]
- Dodd LE, Korn EL, McShane LM, Chandramouli GV, Chuang EY. Correcting log ratios for signal saturation in cDNA microarrays. Bioinformatics. 2004;20:2685–2693. doi: 10.1093/bioinformatics/bth309. [DOI] [PubMed] [Google Scholar]
- Hsiao LL, Jensen RV, Yoshida T, Clark KE, Blumenstock JE, Gullans SR. Correcting for signal saturation errors in the analysis of microarray data. Biotechniques. 2002;32:330–332, 334, 336. doi: 10.2144/02322st06. [DOI] [PubMed] [Google Scholar]
- Yuen T, Wurmbach E, Pfeffer RL, Ebersole BJ, Sealfon SC. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 2002;30:e48. doi: 10.1093/nar/30.10.e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhe A, Suter L, Ruepp S, Singer T, Weiser T, Albertini S. Toxicogenomics in the pharmaceutical industry: hollow promises or real benefit? Mutat Res. 2005;575:102–115. doi: 10.1016/j.mrfmmm.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Pennie WD, Tugwood JD, Oliver GJ, Kimber I. The principles and practice of toxigenomics: applications and opportunities. Toxicol Sci. 2000;54:277–283. doi: 10.1093/toxsci/54.2.277. [DOI] [PubMed] [Google Scholar]
- Darlington GJ, Bernhard HP, Miller RA, Ruddle FH. Expression of liver phenotypes in cultured mouse hepatoma cells. J Natl Cancer Inst. 1980;64:809–819. [PubMed] [Google Scholar]
- Bernhard HP, Darlington GJ, Ruddle FH. Expression of liver phenotypes in cultured mouse hepatoma cells: synthesis and secretion of serum albumin. Dev Biol. 1973;35:83–96. doi: 10.1016/0012-1606(73)90008-0. [DOI] [PubMed] [Google Scholar]
- Ng HH, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/S0959-437X(99)80024-0. [DOI] [PubMed] [Google Scholar]
- Clark SJ, Melki J. DNA methylation and gene silencing in cancer: which is the guilty party? Oncogene. 2002;21:5380–5387. doi: 10.1038/sj.onc.1205598. [DOI] [PubMed] [Google Scholar]
- Jin B, Kim G, Park DW, Ryu DY. Microarray analysis of gene regulation in the Hepa1c1c7 cell line following exposure to the DNA methylation inhibitor 5-aza-2'-deoxycytidine and 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol In Vitro. 2004;18:659–664. doi: 10.1016/j.tiv.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Huang G, Elferink CJ. Multiple mechanisms are involved in Ah receptor-mediated cell cycle arrest. Mol Pharmacol. 2005;67:88–96. doi: 10.1124/mol.104.002410. [DOI] [PubMed] [Google Scholar]
- Ge NL, Elferink CJ. A direct interaction between the aryl hydrocarbon receptor and retinoblastoma protein. Linking dioxin signaling to the cell cycle. J Biol Chem. 1998;273:22708–22713. doi: 10.1074/jbc.273.35.22708. [DOI] [PubMed] [Google Scholar]
- Levine-Fridman A, Chen L, Elferink CJ. Cytochrome P4501A1 promotes G 1 phase cell cycle progression by controlling aryl hydrocarbon receptor activity. Mol Pharmacol. 2004;65:461–469. doi: 10.1124/mol.65.2.461. [DOI] [PubMed] [Google Scholar]
- Marlowe JL, Knudsen ES, Schwemberger S, Puga A. The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S phase-specific gene expression. J Biol Chem. 2004;279:29013–29022. doi: 10.1074/jbc.M404315200. [DOI] [PubMed] [Google Scholar]
- Yang YH, Speed T. Design issues for cDNA microarray experiments. Nat Rev Genet. 2002;3:579–588. doi: 10.1038/nrg863. [DOI] [PubMed] [Google Scholar]
- dbZach: Toxicogenomic Database http://dbzach.fst.msu.edu
- Michigan State University's Research Technology Support Facility http://www.genomics.msu.edu
- Burgoon LD, Eckel-Passow JE, Gennings C, Boverhof DR, Burt JW, Fong CJ, Zacharewski TR. Protocols for the assurance of microarray data quality and process control. Nucleic Acids Res. 2005;33:e172. doi: 10.1093/nar/gni167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel JE, Gennings C, Therneau TM, Burgoon LD, Boverhof DR, Zacharewski TR. Normalization of two-channel microarray experiments: a semiparametric approach. Bioinformatics. 2005;21:1078–1083. doi: 10.1093/bioinformatics/bti105. [DOI] [PubMed] [Google Scholar]
- Eckel JE, Gennings C, Chinchilli VM, Burgoon LD, Zacharewski TR. Empirical bayes gene screening tool for time-course or dose-response microarray data. J Biopharm Stat. 2004;14:647–670. doi: 10.1081/BIP-200025656. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, Bussey KJ, Riss J, Barrett JC, Weinstein JN. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer 3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hepalclc7 TCDD time course microarray data. Ratios represent expression relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene and time-point basis using the model-based t-value.
Hepalclc7 TCDD dose-response microarray data. Ratios represent expression relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene and dose basis using the model-based t-value.
C57BL/6 mice hepatic tissue TCDD time course microarray data. Ratios represent expression relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene and time-point basis using the model-based t-value.
C57BL/6 mice hepatic tissue TCDD dose-response microarray data. Ratios represent expression relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene and dose basis using the model-based t-value.
Untreated Hepalclc7 and C57BL/6 sample microarray data. Ratios represent basal expression of Hepalclc7 cells relative to hepatic tissue from C5VBL/6 mice. P1(t)-values represent posterior probabilities of activity on a per gene and dose basis using the model-based t-value.
Gene names and primer sequences (5'-3') for transcripts verified by QRTPCR. Primer pair sequences used to verify in vitro and in vivo microarray results using QRTPCR.
Hepa1c1c7 TCDD treatment and harvesting regimen. For the time course study, wild-type and ARNT-deficient c4 mutant cells were treated with 10 nM TCDD or 0.1% DMSO vehicle and harvested at 1, 2, 4, 8, 12, 24, or 48 hrs post-treatment. Untreated controls were harvested at 0 hrs (as indicated by *). The dose-response study was done performed with Hepa1c1c7 wild-type cells and treated with 0.001, 0.01, 0.1, 1.0, 10, 100 nM TCDD or 0.1% DMSO vehicle and harvested 12 hrs post-treatment (as indicated by ‡).
Microarray experimental designs for A) temporal, B) dose-response and C) basal expression studies. A) Temporal gene expression patterns were analyzed by an independent reference design in which cells treated with TCDD (T) were co-hybridized to time-matched vehicle controls (V). This design involves two independent labelings per sample for a total of 14 arrays per replicate. Four biological replicates were conducted for a total of 56 microarrays. Numbers indicate time points for comparison. B) Dose-dependent changes in gene expression were analyzed 12 hrs after treatment using a common reference design in which cells treated with TCDD were co-hybridized with a common vehicle control. This design involves two independent labelings per sample for a total of 12 arrays per replicate. Four biological replicates were conducted for a total of 48 microarrays. Numbers indicate TCDD concentration in nM units. C) Comparative basal gene expression levels between untreated in vitro and in vivo samples were analyzed by an independent reference design. Four biological replicates of untreated Hepa1c1c7 cells and hepatic tissue from C57BL/6 mice harvested at 0 hrs were co-hybridized and two independent labelings were performed per sample for a total of 8 arrays. Double-headed arrows indicate dye swaps (each sample labeled with Cy3 and Cy5 on different microarrays).