Abstract
Recently we discovered a previously uncharacterized gene with the characteristics of a membrane progestin receptor (mPR) in a fish model, spotted seatrout. Here, we report the identification, cloning, and characteristics of other members of this hitherto unknown family of putative mPRs from several vertebrate species, including human, mouse, pig, Xenopus, zebrafish, and Fugu, with highly conserved nucleotide and deduced amino acid sequences and similar structures to the spotted seatrout mPR. The 13 vertebrate genes identified seem to belong to an unknown gene family. Phylogenetic analysis indicates these cDNAs comprise three distinct groups (named α, β, and γ) within this gene family. Structural analyses of the translated cDNAs suggest they encode membrane proteins with seven transmembrane domains. The transcript sizes of the human α, β, and γ putative mPR mRNAs varied from 2.8 to 5.8 kb and showed distinct distributions in reproductive, neural, kidney and intestinal tissues, respectively. Recombinant human α, γ, and mouse β proteins produced in an Escherichia coli expression system demonstrated high affinity (Kd = 20–30 nM) saturable binding for progesterone. Further analysis of binding to the γ-subtype revealed binding was specific for progestins and was displaceable, with rapid rates of association and dissociation (t1/2 = 2–8 min). These results suggest this is a new family of steroid receptors unrelated to nuclear steroid receptors, but instead having characteristics of G protein-coupled receptors.
Although the existence of specific receptors on the surface of target cells mediating rapid nongenomic actions of steroids was recognized 20 years ago (1, 2), efforts to determine the structures of steroid membrane receptors have been unsuccessful until now (3–5). In the accompanying paper in this issue of PNAS (6), we described a gene discovered in a teleost species, spotted seatrout, whose protein fulfils the criteria for its designation as a steroid membrane receptor, including structural plausibility, specific tissue and plasma membrane localization, steroid binding characteristic of steroid and progestin receptors, coupling to second messenger pathways, regulation by steroid hormones, and biological relevance. Evidence was obtained that this progestin membrane receptor (mPR) is the intermediary in progestin induction of oocyte meiotic maturation in teleost fishes and activates an inhibitory G protein (Gi/o), which suggests it may be a G protein-coupled receptor (GPCR).
The aims of this study were to search for related cDNA fragments of other vertebrates in the genomic databases, clone and sequence their full-length cDNAs, and partially characterize these genes and their recombinant proteins. This paper reports the identification of 13 additional vertebrate genes closely related to the spotted seatrout mPR. Structural and phylogenetic evidence is presented that these vertebrate genes encode for membrane proteins with seven or occasionally six transmembrane domains that can be classified into three subtypes. Hybridization results indicate that the cDNAs of the three subtypes have different distributions in human tissues. In addition, initial information on the steroid binding characteristics of three recombinant proteins representing each of the mammalian subtypes suggests that they are also mPRs.
Materials and Methods
Identification and Sequence Analysis of Putative mPRs.
A BLAST search of the human genome database was conducted by using the seatrout mPR coding sequence to identify homologous human cDNAs. Two human sequence fragments with 54% and 50% amino acid sequence identity to the seatrout mPR were discovered in the database. These two human genomic sequence fragments were subsequently used for BLAST searches of EST databases. Six cDNA clones were identified, two each from human, mouse, and pig. The identified clones were isolated, fully sequenced in both directions, and the sequences were deposited into GenBank (accession nos. AF313615–AF313620). Intriguingly, two additional cDNA sequences with 29% amino acid identity to that of seatrout mPR were also identified in human and mouse databases. Thereafter, we searched for cognate cDNAs in other vertebrates by using cloning techniques and database mining. The cDNA sequences of mammals and seatrout were used to design degenerate primers to obtain cognate cDNA sequences from zebrafish and Xenopus (see Supporting Text, Fig. 6, and Table 2, which are published as supporting information on the PNAS web site, www.pnas.org). Total RNA was isolated from ovarian and brain tissues of Xenopus and zebrafish by using TRIzol extraction reagent (Invitrogen). The mRNA was isolated from the total RNA with magnetic-oligo (dT) particles by using a Straight A mRNA Isolation System kit (Novagen). First-strand synthesis of cDNAs from the polyadenylated mRNA was performed by using a GeneRacer kit (Invitrogen). Cognate cDNA fragments were amplified from the synthesized cDNAs by using nested RT-PCR with two sets of primers. The amplified PCR products were cloned into a TA cloning vector (Invitrogen) and sequenced in both directions by using an automated DNA sequencer (Model 310, Perkin–Elmer). The sequences of the 5′ and 3′ ends of the cDNAs were obtained by using 5′- and 3′-RACE. Finally, the sequences of the full-length cDNAs were confirmed by using gene-specific primers based on the sequences of the 5′ and 3′ ends. Interestingly, multiple cognate sequence fragments were also identified from the Fugu (Japanese pufferfish) genome database.
Structural and Phylogenetic Analysis.
Computer analyses [DAS, HMMTOP, PREDICT PROTEIN, SOSUI (7), TMHMM, TMPRED, TOPPRED2] were used to predict the likely structure of the protein and its localization within the cell based on the deduced amino acid sequence. Multiple sequence alignments for phylogenetic analyses were constructed by using the PILEUP program of the Wisconsin Package (Genetics Computer Group, Madison, WI) based on a progressive alignment method (8). Sequence alignments were adjusted manually to minimize gaps with the aid of MACCLADE V.4.0 (9) to minimize gaps and to avoid introducing a gap in the middle of a codon. A hypervariable, and thus unalignable, 11-aa residue region (positions 237–247) was excluded from the alignments to perform optimal analyses. Phylogenetic analyses of the amino acid sequences using parsimony were conducted by using the computer program PAUP* V.4.0b10 (10). For the aligned 354 amino acid residues, 302 positions are variable, of which 283 are parsimony-informative. All characters were treated as unordered and given equal weights. Gaps were treated as missing characters. Relative branch support was evaluated by using nonparametric bootstrap (11) and decay index (12) analyses. Bootstrap values were based on 1,000 pseudoreplicates by using the heuristic search algorithm with the random stepwise addition of taxa (100 replicates), followed by tree bisection and reconnection (TBR) branch swapping. Decay values were computed by using a PAUP* decay index command file generated by MACCLADE and employing the default heuristic search strategy with TBR. Amino acid substitutions considered to be clade-diagnostic were determined by using ACCTRAN character optimization (13) in PAUP*. Character states optimized as synapomorphies for clades of interest were then checked against the data matrix to ensure that they were both invariant within the clade and absent in all other terminal units.
Northern Blot Analysis.
Multiple human tissue expression arrays and multiple tissue Northern blots were obtained from a commercial vendor (CLONTECH). Northern hybridization analysis was performed by using a Northern Max Kit (Ambion, Austin, TX). Receptor probes were prepared by digesting purified plasmids containing cDNA inserts with appropriate enzymes, and the desired fragments were purified by electrophoresis using low-melting agarose and a QIAquick Gel Extraction Kit (Qiagen, Chatsworth, CA). Thereafter, 20–25 ng of DNA were randomly labeled by using a Pharmacia Rediprime Kit and Amersham Pharmacia Redivue [α-32P]dCTP, and the labeled probe was purified by using a Pharmacia ProbeQuant G-50 microspin column. The incorporation rate of [α-32P]dCTP ranged from 50% to 80%, and the specific activity of the probes ranged from 1.2–1.6 × 109 cpm/μg. The membrane was hybridized overnight at 42°C with a labeled probe at a final concentration of ≈5 × 106 cpm/ml. Nonspecific binding was stripped off the membrane by washing twice with 20 ml of low-stringency buffer for 5 min at 42°C and with 20 ml of high-stringency buffer twice for 15 min at 42°C. The hybridized membrane was exposed to a Kodak Biomax MS film for 3 h to several days prior to film development.
Expression of Recombinant Protein in BL21(DE3) Escherichia coli cells.
The coding regions of the putative steroid membrane receptors were amplified by PCR from full-length cDNA plasmid clones. PCR was carried out in 100-μl aliquots containing 50 mM KCl, 10 mM Tris (pH 8.3), 1.5 mM MgCl2, 0.2 mM dNTP, a set of 5′ and 3′ primers (1 μM each), and 2.5 units of TaqDNA polymerase mix. After an initial 2-min denaturation at 94°C, the PCR cycle was repeated five times with denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and polymerization at 72°C for 2 min. Subsequently, 25 cycles of PCR were carried out under the same conditions, except annealing, which was conducted at 55°C. The PCR products were purified by electrophoresis using a low-melting agarose and a QIAquick Gel Extraction Kit. The purified PCR products were ligated into a pET-27b expression vector (Novagen). The pET vector was transformed into BL21(DE3) E. coli cells. The positive clones were purified and sequenced. Plasmids without an insert and with a reversed inserted mPR sequence were used as controls. Expression of recombinant proteins was induced with isopropyl β-d-thiogalactoside at 25°C. Soluble recombinant proteins were extracted from whole cells or whole-cell lysates and analyzed for progestin membrane receptor binding, as described (6).
Results
Phylogenetic and Structural Analyses of Cognate mPRs in Vertebrates.
Thirteen cDNAs similar to the seatrout mPR were identified in other vertebrate species. Computer structural analyses consistently predicted that all of these cDNAs encoded for plasma membrane proteins with seven, or occasionally six, transmembrane domains. The results of the phylogenetic analysis indicated that three major clades of mPRs, α, β, and γ, exist in two distantly related vertebrate classes, fishes and mammals (Fig. 1). The high bootstrap value of 100 strongly supports the divergence of mPRγ from the other two groups, α and β. The α and β nodes also have moderate to high bootstrap values of 67 and 83, respectively. The majority of the nodes in this analysis also have moderate to high decay support (Fig. 1). Maximum likelihood analyses (14) of aligned nucleotide sequences are congruent with the tree reported here (results not shown). There are 8, 16, and 38 amino acid substitutions that characterize and distinguish the mPRα, mPRβ, and mPRγ clades, respectively. The majority of the amino acid residues unique to mPRα are in the extracellular loops (63%, Fig. 2); those diagnostic of mPRβ are divided among the N-terminal (20%), transmembrane (40%), external loops (27%), and internal loops (13%; see Fig. 2 legend), whereas most of those unique to mPRγ are in the transmembrane domain (82%, data not shown). The human, pig, and mouse mPRαs have ORFs of 1038–1053 nt, encoding peptides with 346–350 amino acid residues, similar to the sizes of the mPRαs of spotted seatrout (352 aa) and zebrafish (354 aa). The ORFs of the mammalian, zebrafish, and Xenopus mPRβs (nucleotides 1064–1065; amino acids 352–354) are similar to those of mPRα, whereas the mPRγs are slightly shorter (330 aa).
Figure 1.
Unrooted phylogenetic tree (single most parsimonious tree, 932 steps; consistency index = 0.87; retention index = 0.86) showing the three clades of the putative mPRs. Branch lengths are proportional to the number of amino acid substitutions and bootstrap values (percentages); decay indexes are indicated on each branch.
Figure 2.
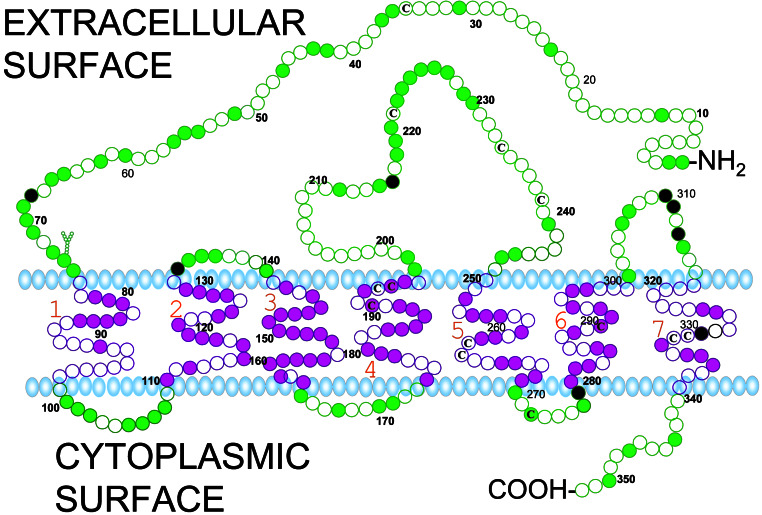
Proposed model for insertion of the seatrout putative mPR in the plasma membrane based on hydrophilicity and charges of the amino acid residues analyzed by sosui (8). Each circle represents one amino acid residue. Residues identical in seatrout mPRα and the five other vertebrate mPRαs are indicated by the filled colored circles. Amino acid residues diagnostic of this clade are at positions 68 (T), 133 (A), 216 (S), 279 (Q), 310 (Y), 311 (E), 313 (L), and 329 (T) (filled black circles). Y shows a potential site of N-linked glycosylation, a cysteine residue (C). The proposed models for the mPRβs and mPRγs are very similar, with large N-terminal and second extracellular loop domains, and a short C-terminal domain (not shown). Amino acid residues diagnostic of mPRβ are at positions 6 (L), 11 (T), 22 (L), 89 (V), 102 (L), 114 (L), 138 (S), 167 (S), 203 (R), 204 (P), 205 (Y), 206 (P), 213 (R), 266 (V), 326 (S), 328 (F) from the N-terminal end of human mPRβ.
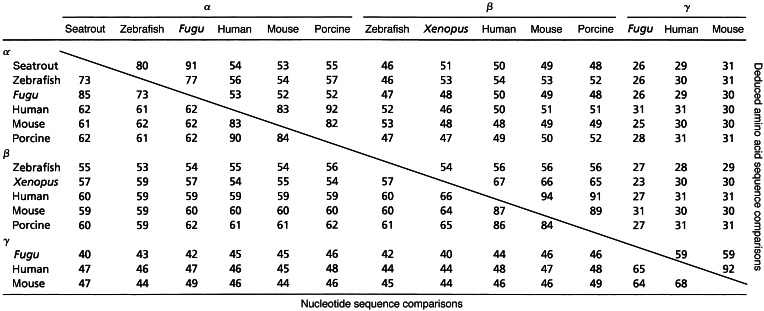
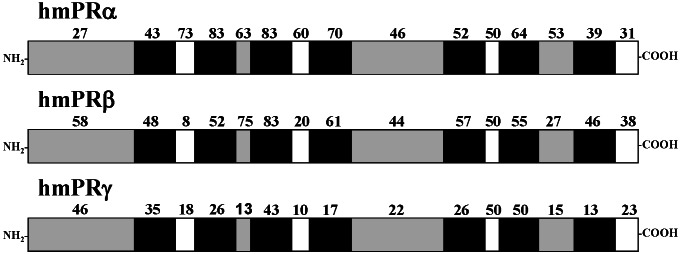
Phylogenetic analyses clearly classify the seatrout mPR into the α subgroup. Sequence identity comparisons also showed that seatrout mPR had a higher homology with the α group (Table 1; 52–91% for amino acids, 61–90% for nucleotides) compared with the β group (Table 1; 46–54% for amino acids, 53–62% for nucleotides), or γ group (25–31% for amino acids, 40–47% for nucleotides). The transmembrane domains of human mPRα and mPRβ show high amino acid sequence identities (typically 55–83%) with those of the seatrout mPRα (Fig. 3). Several of the intracellular domains also display high sequence identities with seatrout mPRα, especially those of the α-subtype. In contrast, the homology among extracellular domains is generally lower, with the exception of the second extracellular loop. Overall, seatrout mPRα shows lower homology to the various domains of human mPRγ (Fig. 3). A proposed model for the insertion of the seatrout membrane progesterone receptor in the plasma membrane based on hydrophobicity and charges of the amino acids analyzed by SOSUI (7) is shown in Fig. 2. A potential site of N-linked glycosylation is shown. Most of the transmembrane domains and portions of the N-terminal end and the large, second extracellular loops are highly conserved among all of the putative vertebrate mPRαs (shown as solid circles in Fig. 2). The positions of the eight residues unique to this clade are also shown (black solid circles).
Table 1.
Percent sequence identity between putative membrane progestin receptors
Figure 3.
Amino acid sequence comparison between seatrout mPRα and the three putative human mPRs. Numbers above the box indicate percent sequence identity of amino acids in each domain between the fish mPR and human putative mPRs. Extracellular, solid gray; transmembrane, solid black; cytoplasmic domains, white.
Tissue Distribution.
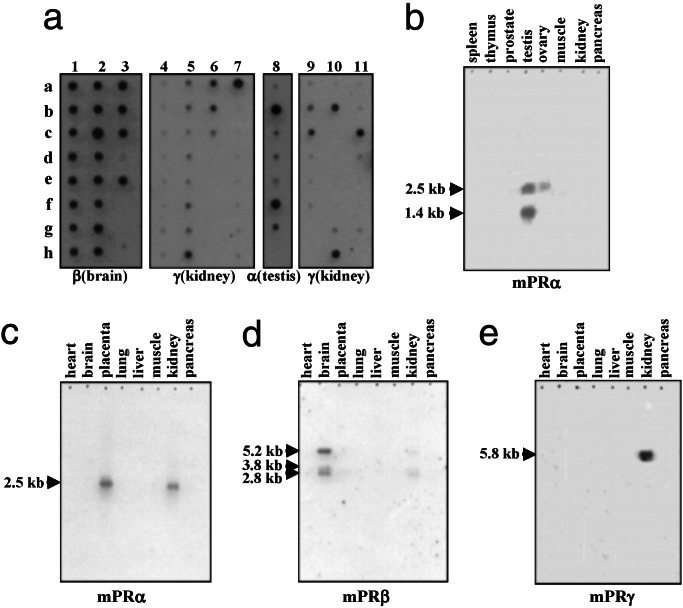
Dot blot analyses of human multiple tissue arrays with specific probes for human α, β, and γ showed distinct tissue localization of the three forms. The α form (from a testicular library) was localized in reproductive tissues, particularly in the placenta, testis, ovary, possibly in the uterus and bladder (Fig. 4a, lane 8), also in the kidney, and possibly in the adrenal (data not shown). The β form (from a brain library) is exclusively localized in neural tissues and was detected throughout the brain and in the spinal cord but not in the pituitary (Fig. 4a, lanes 1–3). The γ-subtype (from a colon library) is present in the kidney, fetal kidney, colon, a lung carcinoma, HeLa 53 cells, and possibly in the adrenal and lung (Fig. 4a, lanes 5–7, 10, 11). Multiple transcripts of each receptor subtype were detected in Northern blot analyses. Dot blot analysis does not reveal which transcript is recognized by the mPR probe. Two α transcripts in the testis, with a major band at 1.4 kb and a minor one at 2.5 kb, were detected by Northern blot analysis of multiple tissues (Fig. 4b). The larger (2.5-kb) band was also detected in the ovary, placenta, and kidney (Fig. 4 b and c). Three β transcripts were obtained in brain, with a major signal at 5.2 kb and two minor signals at 3.2 and 2.8 kb (Fig. 4d). A positive signal for γ was detected in the kidney at 5.8 kb (Fig. 4e). It is noteworthy that tissue distributions of the three human mPR subtypes revealed by Northern blot analysis are distinct, with significant coexpression in only a few tissues such as the kidney.
Figure 4.
(a) Dot blot hybridization of human mPR α (testicular), β (brain), and γ (kidney) mRNA probes with human multiple tissue arrays (CLONTECH). Only lanes displaying strong hybridizations with each of the probes are shown. Lanes 1–3 (various regions of CNS): b1 and g1, cerebral cortex; a2 and b2, cerebellum; e2, caudate nucleus; c3, thalamus; d3, pituitary gland; e3, spinal cord. Lane 4: regions of heart. Lanes 5 and 6 (gastrointestinal tract): h5, a6, and b6, colon. Lane 7 (kidney, hemopoietic, and lymphatic tissues): a7, kidney. Lane 8 (reproductive tissues): b8, placenta; d8, uterus; f8, testis; g8, ovary. Lane 9: liver and endocrine glands. Lane 10: lymphomas and carcinomas. Lane 11 (fetal tissues): c11, fetal kidney. (b–e) Northern hybridization of human α, β, and γ mRNA probes with human multiple tissue Northern blots (CLONTECH).
Steroid Binding Characteristics.
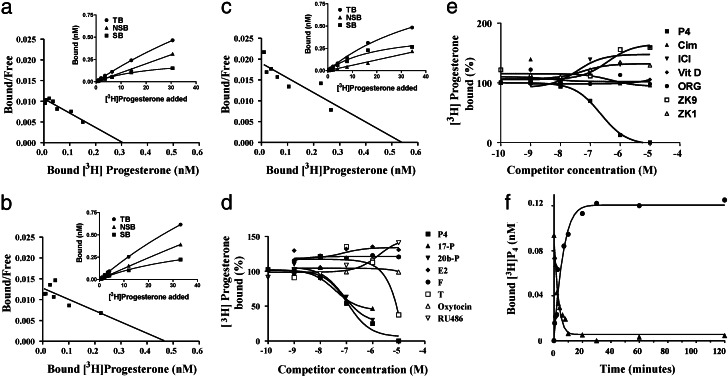
Recombinant proteins were produced in the E. coli expression system for each of the three subtypes. Saturation analyses and Scatchard plots of [3H]progesterone binding to the recombinant proteins showed the presence of a single class of high affinity (Kd = 28–39 nM), saturable (Bmax = 0.3–0.5 nM) progesterone binding sites for mouse β mPR (Fig. 5a), human α mPR (Fig. 5b), and human γ mPR (Fig. 5c). The control vector proteins without cDNA inserts demonstrated no specific binding to [3H]progesterone. Steroid competition studies with the human γ recombinant protein showed that binding is specific for progesterone and several of its hydroxylated derivatives, whereas an estrogen, androgen, and corticosteroid had low or no affinity for the receptor (Fig. 5d). Several synthetic progestins and antiprogestins also showed no affinity for the human γ protein (Fig. 5e). Association and dissociation kinetics of [3H]progesterone binding to the human γ protein were rapid, with t1/2 s = 2–4 min (Fig. 5f). These results demonstrate that the recombinant proteins of mammalian representatives of each of the three mPR subtypes have several binding characteristics typical of steroid receptors.
Figure 5.
Steroid binding characteristics of recombinant mammalian proteins produced in E. coli. Saturation (a–c Insets) and Scatchard analyses of [3H]progesterone binding to recombinant mouse β (a), human α (b), and human γ (c) proteins. (d) Competition curves of steroid binding to the recombinant human mPRγ protein. Binding is expressed as a percentage of maximum specific binding. P4, progesterone; 17-P, 17α-hydroxyprogesterone; 20b-P, 20β-hydroxyprogesterone; E2, estradiol-17β; T, testosterone; F, cortisol; RU486, synthetic antiprogestin. (e) Competition curves of binding of synthetic antihormones to the recombinant human mPRγ protein. Vit D: 1α, 25(OH)-vitamin D. Antiprogestins: ZK9, ZK98299; ZK1, ZK112983; ORG, ORG31710. Antiandrogen: Cim, cimetidine. Antiestrogen: ICI, ICI182,780. (f) Time course of association and dissociation of specific progesterone binding to human mPRγ.
Discussion
Structural and phylogenetic analyses show the 13 additional genes identified in other vertebrate species are closely related to the seatrout mPR described (6) and can be classified into three subtypes. Computer analyses using a variety of programs that predict protein structure (DAS, HMMTOP, PREDICT PROTEIN, SOSUI, TMHMM, TMPRED, TOPPRED2) indicate that all 14 genes encode proteins that are located in the plasma membrane and typically have seven transmembrane domains as well as both extracellular N-terminal and intracellular C-terminal domains. These protein structures are typical of GPCRs. Although pharmacological and biological evidence indicates that seatrout mPRα activates an inhibitory G protein and, therefore, may be a GPCR (6), equivalent information is currently lacking for the other members of this protein family. However, the results of the present study clearly show that three mammalian genes, representing each of the three subtypes, encode proteins with binding characteristics typical of progestin membrane receptors. High affinity, saturable, single binding sites for progesterone can be demonstrated with the human α, mouse β, and human γ recombinant proteins produced in an E. coli expression system. Additional studies with the human γ protein show that steroid binding is specific for progestins and is readily displaceable. Moreover, the kinetics of association and dissociation are rapid, occurring within a few minutes, which is typical of steroid membrane receptors. Interestingly, several synthetic progestins and antiprogestins, which have relatively high binding affinities for nuclear progesterone receptors, display no binding affinity for the recombinant human γ protein. Lack of binding of synthetic progestins and antiprogestins to progestin membrane receptors has also been observed in biochemical binding studies (15). Finally, the human α, β, and γ transcripts are localized to specific tissues that are known targets of nongenomic progesterone actions in mammals (4, 16–18). Taken together, these results suggest that this is a hitherto unknown family of progestin membrane receptors that are unrelated to nuclear progesterone receptors.
The presence of the α-, β- and γ-subtypes both in a fish species, Fugu, and in several mammalian species, suggests they arose early in vertebrate evolution, before the divergence of the ray-finned (Actinopterygian) and lobe-finned (Sarcopterygian) fishes in the lower Devonian period, 400 million years ago (19). Information for hagfishes and lampreys may indicate whether the three subtypes could have arisen from a common ancestral gene by duplication during an earlier period of vertebrate evolution. Several such gene duplications have been proposed for nuclear steroid receptors based on the results of previous genetic analyses in these basal vertebrates (20, 21). One such duplication event in the Actinopterygian lineage, ≈200 million years ago (22), probably accounts for the presence of two paralogs of the ancestral nuclear estrogen receptor β gene in extant euteleosts (23). However, the similar branch lengths separating the three subtypes does not suggest further duplication and preservation of any of the mPR genes. The overall percent nucleotide and amino acid sequence identities among the vertebrate species for each of the putative mPR subtypes reflect their phylogenetic relationships, the phylogram showing the expected separate branches for the teleost and mammalian representatives of each subtype and the euteleost αs further branching into the Acanthopterygii (Fugu and spotted seatrout) and Ostariophysi (zebrafish). Interestingly, there is a marked divergence of the Fugu and mammalian mPRγs that may indicate modified functions within this subtype. More detailed comparisons of the sequence identities of various regions of the putative mPRs may be mented when information on their functional domains becomes available.
An interesting finding was that the Northern and dot blot hybridization analyses showed different tissue distributions for the mRNAs of the three human mPR subtypes, with only the kidney expressing more than one subtype in significant amounts. In contrast, multiple subtypes of nuclear steroid receptors are present in the brain and gonads and have overlapping distributions in certain regions (23, 24) and, therefore, have the potential to form heterodimers. GPCRs also form dimers that may be important for receptor activation (25, 26). However, the formation of heterodimers among the mPR subtypes is unlikely in most human tissues based on the distributions of their mRNAs. The significance of the distinct distributions of the three mPR subtypes in humans is unclear. One possible explanation for the apparent exclusive localization of the human mPRβ mRNA in neural tissues is that this subtype is characteristic of tissues of ectodermal origin. However, the mouse mPRβ was cloned from a testicular cDNA library, which suggests this subtype also occurs in tissues of mesodermal origin. Conversely, mouse mPRα was cloned from a brain cDNA library, which suggests that this subtype is also present in the brains of certain mammals. The low transcript levels of the α-subtype detected in the brains of spotted seatrout (6) are a further indication that mPRα may be involved in some cell surface–initiated progesterone actions in the vertebrate brain.
The distinct, nonoverlapping tissue distributions of the mRNAs of the human mPRα and -β subtypes indicate they regulate different physiological functions of progesterone. The presence of significant transcript levels of mPRα in the human testis, ovary, and placenta suggests it is the major subtype mediating the rapid, nongenomic actions of progesterone in reproductive tissues. There is considerable evidence that progesterone has rapid actions on human sperm to induce hyperactivity and the acrosome reaction by binding to an mPR (27, 28). Similarly, an mPR previously characterized biochemically on sperm in spotted seatrout is likely the intermediary in the rapid actions of progestins to increase sperm motility and fertilization success in this species (29). The finding that the mPRα protein is localized on seatrout sperm membranes suggests that it may be the mediator of this effect (6). Therefore, the mPRα is also a candidate, among others (30, 31), for the protein receptor involved in the rapid progesterone actions on mammalian sperm. Interestingly, subfertility and oligospermia in men have been associated with reduced responses to progesterone and decreased concentrations of the mPR (32–34). Thus, the human mPRα has potential medical significance in the diagnosis of male subfertility and as a target for a new generation of fertility drugs and contraceptives. Currently, evidence that mPRs have equivalent functions in female gametes in mammals is lacking. Although important roles for progestins and mPRs in the induction of meiotic maturation of oocytes have been clearly demonstrated in teleosts (35), there is no clear evidence progesterone may have similar functions in mammals. However, nongenomic progesterone actions and the presence of specific binding sites have been reported in the corpus luteum of mammals, which suggests mPRα may be involved in other ovarian functions controlled by somatic cells (17). Similarly, progesterone has been shown to bind to a specific mPR on Leydig cells in the testis to stimulate sodium influx and steroidogenesis (16). Nongenomic actions of progesterone have also been reported in several regions of the brain, including the hypothalamus (regulating LHRH secretion) and the ventral tegmental area (influencing sexual receptivity in rodents) (36, 37). The fact that one of the subtypes is present in the brain provides the first indication that progesterone and its metabolites may also exert nongenomic actions via specific mPRβ receptors, in addition to binding to modulatory sites on neurotransmitter receptors, such as the GABAa/benzodiazepine receptor Cl− channel complex (38, 39). There is currently less evidence for nongenomic actions of progesterone in tissues with significant transcript levels of mPRγ. However, progesterone has been shown to exert rapid, nongenomic actions in intestinal tissues, resulting in a reduction in smooth muscle calcium currents and, in renal cells, in an alteration of potassium conductance (18, 40). A variety of environmental toxicants could also potentially interfere with the nongenomic actions of progesterone in these target tissues in mammals by binding to their mPRs. This mechanism of endocrine disruption has previously been demonstrated for mPRα on the oocytes and sperm of spotted seatrout and a closely related species, Atlantic croaker, after exposure to organochlorines such as DDT that have estrogenic activities (41, 42).
The discovery of the molecular structure and likely orientation in the plasma membrane of a new class of steroid receptors, unrelated to nuclear receptors but, instead, characteristic of GPCRs, provides a plausible mechanistic explanation of how steroids acting at the cell surface can cause rapid intracellular responses. An understanding of the mechanism of nonclassical steroid action mediated by these receptors will provide a sound molecular basis for future studies in this emerging field of endocrinology. It will also enable investigations of the integration of rapid, nongenomic, and slower genomic responses to steroid hormones in target cells.
Supplementary Material
Acknowledgments
We thank John Stiller for his helpful discussions and skillful assistance regarding phylogenetic analyses. The human mPRγ clone was obtained from Dr. Sumio Sugano. The pig mPRβ EST clone was obtained from Dr. Merete Fredholm. This work was supported by National Institutes of Health Grant ESO4214 and Environmental Protection Agency STAR Grant R-82902401 (to P.T.), National Science Foundation Grant IBN-9980353 (to P.T. and Y.Z.), and by funds from the University of Texas at Austin and private donors.
Abbreviations
- mPR
membrane progestin receptor
- GPCR
G protein-coupled receptor
Footnotes
References
- 1.Pietras R, Szego C. Nature. 1975;253:357–359. doi: 10.1038/253357a0. [DOI] [PubMed] [Google Scholar]
- 2.Sadler S E, Maller J L. J Biol Chem. 1982;257:355–361. [PubMed] [Google Scholar]
- 3.Maller J L. Proc Natl Acad Sci USA. 2001;98:8–10. doi: 10.1073/pnas.98.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Revelli A, Massobrio M, Tesarik J. Endocr Rev. 1998;19:3–17. doi: 10.1210/edrv.19.1.0322. [DOI] [PubMed] [Google Scholar]
- 5.Watson C S, Gametchu B. Proc Soc Exp Biol Med. 1999;220:9–19. doi: 10.1046/j.1525-1373.1999.d01-2.x. [DOI] [PubMed] [Google Scholar]
- 6.Zhu Y, Rice C D, Pang Y, Pace M, Thomas P. Proc Natl Acad Sci USA. 2003;100:2231–2236. doi: 10.1073/pnas.0336132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirokawa A, Boon-Chieng S, Mitaku S. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 8.Feng D F, Doolittle R F. J Mol Evol. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 9.Maddison D R, Maddison W P. MacClade 4: Analysis of Phylogeny and Character Evolution. Sunderland, MA: Sinauer; 2000. , Version 4.0. [DOI] [PubMed] [Google Scholar]
- 10.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony. Sunderland, MA: Sinauer; 2000. , Version 4.0b10. [Google Scholar]
- 11.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 12.Bremer K. Evolution (Lawrence, Kans) 1988;42:795–803. doi: 10.1111/j.1558-5646.1988.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 13.Swofford D L, Maddison W P. Math Biosci. 1987;87:199–229. [Google Scholar]
- 14.Posada D, Crandall K A. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 15.Haukkamaa M. In: Steroid Hormone Receptors: Their Intracellular Location. Clark C R, editor. London: Harwood; 1987. pp. 155–169. [Google Scholar]
- 16.Rossato M, Nogara A, Merico M, Ferlin A, Foresta C. Steroids. 1999;64:168–175. doi: 10.1016/s0039-128x(98)00104-4. [DOI] [PubMed] [Google Scholar]
- 17.Rae M T, Menzies G S, McNeilly A S, Woad K, Webb R, Bramley T A. Biol Reprod. 1998;58:1394–1406. doi: 10.1095/biolreprod58.6.1394. [DOI] [PubMed] [Google Scholar]
- 18.Bielefeldt K, Waite L, Abboud F M, Conklin J L. Am J Physiol. 1996;271:G370–G376. doi: 10.1152/ajpgi.1996.271.2.G370. [DOI] [PubMed] [Google Scholar]
- 19.Colbert E H, Morales M. Evolution of the Vertebrates: A History of the Backboned Animals Through Time. 4th Ed. New York: Wiley; 1991. [Google Scholar]
- 20.Escriva H, Safi R, Hanni C, Langlois M C, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R, Laudet V. Proc Natl Acad Sci USA. 1997;94:6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thornton J W. Proc Natl Acad Sci USA. 2001;98:5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amores A, Force A, Yan Y-L, Joly L, Amemiya C, Fritz A, Ho R K, Langeland J, Prince V, Wang Y L, et al. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 23.Hawkins M B, Thornton J W, Crews D, Skipper J K, Dotte A, Thomas P. Proc Natl Acad Sci USA. 2000;97:10751–10756. doi: 10.1073/pnas.97.20.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shughrue P J, Komm B, Merchenthaler I. Steroids. 1996;61:678–681. doi: 10.1016/s0039-128x(96)00222-x. [DOI] [PubMed] [Google Scholar]
- 25.Hebert T E, Bouvier M. Biochem Cell Biol. 1998;76:1–11. doi: 10.1139/bcb-76-1-1. [DOI] [PubMed] [Google Scholar]
- 26.Rios C D, Jordan B A, Gomes I, Devi L A. Pharmacol Ther. 2001;92:71–87. doi: 10.1016/s0163-7258(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 27.Blackmore P F, Neulen J, Lattanzio F, Beebe S J. J Biol Chem. 1991;266:18655–18659. [PubMed] [Google Scholar]
- 28.Revelli A, Modotti M, Piffaretti-Yanez A, Masobrio M, Balerna M. Hum Reprod. 1994;9:760–766. doi: 10.1093/oxfordjournals.humrep.a138592. [DOI] [PubMed] [Google Scholar]
- 29.Thomas P, Breckenridge-Miller D, Detweiler C. Fish Physiol Biochem. 1997;17:109–116. [Google Scholar]
- 30.Luconi M, Bonaccorsi L, Bini L, Liberatori S, Pallini V, Forti G, Baldi E. Steroids. 2002;67:505–509. doi: 10.1016/s0039-128x(01)00173-8. [DOI] [PubMed] [Google Scholar]
- 31.Falkenstein E, Heck M, Gerdes D, Grube D, Christ M, Weigel M, Buddhikot M, Meizel S, Wehling M. Endocrinology. 1999;140:5999–6002. doi: 10.1210/endo.140.12.7304. [DOI] [PubMed] [Google Scholar]
- 32.Falsetti C, Baldi E, Krauz C, Casano R, Failli P, Forti G. J Androl. 1993;14:17–22. [PubMed] [Google Scholar]
- 33.Tesarik J, Mendoza C. Fertil Steril. 1992;58:793–797. doi: 10.1016/s0015-0282(16)55329-1. [DOI] [PubMed] [Google Scholar]
- 34.Kotwicka M, Warchol J B. Folia Histochem Cytobiol. 2001;39:139–140. [PubMed] [Google Scholar]
- 35.Thomas P, Zhu Y, Pace M. Steroids. 2002;67:511–517. doi: 10.1016/s0039-128x(01)00180-5. [DOI] [PubMed] [Google Scholar]
- 36.Tischkau S A, Ramirez V D. Proc Natl Acad Sci USA. 1993;90:1285–1289. doi: 10.1073/pnas.90.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frye C A. Horm Behav. 2001;40:226–233. doi: 10.1006/hbeh.2001.1674. [DOI] [PubMed] [Google Scholar]
- 38.Lan N C, Bolger M B, Gee K. Neurochem Res. 1991;16:347–356. doi: 10.1007/BF00966098. [DOI] [PubMed] [Google Scholar]
- 39.McEwen B S. Trends Pharmacol Sci. 1991;12:141–147. doi: 10.1016/0165-6147(91)90531-v. [DOI] [PubMed] [Google Scholar]
- 40.Steidl M, Pinggera G, Ritter M, Lang F. Am J Physiol. 1991;260:E743–E750. doi: 10.1152/ajpendo.1991.260.5.E743. [DOI] [PubMed] [Google Scholar]
- 41.Das S, Thomas P. Endocrinology. 1999;140:1953–1956. doi: 10.1210/endo.140.4.6781. [DOI] [PubMed] [Google Scholar]
- 42.Thomas P. In: Endocrine Disruptors: Effects on Male and Female Reproductive Systems. Naz R K, editor. Boca Raton, FL: CRC; 1999. pp. 3–38. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.