Abstract
TWEAK [TNF (tumour necrosis factor)-like weak inducer of apoptosis] is a member of the TNF superfamily of cytokines. TWEAK binds with high affinity to a single TNF receptor super-family member, Fn14 (fibroblast growth factor-inducible 14). This interaction can stimulate a variety of biological responses, depending on the cell type analysed. The murine Fn14 extracellular region is only 53 amino acids in length and primarily consists of a CRD (cysteine-rich domain) containing three disulphide bonds. In the present study, we investigated whether TWEAK binding to this CRD was dependent on selected evolutionarily conserved amino acid residues by using a site-specific mutagenesis approach and several different ligand-binding assays. Our results indicate that three residues within the predicted Fn14 CRD A1 module (Asp45, Lys48 and Met50) and one residue within the predicted D2 module (Asp62) are each critical for high-affinity TWEAK binding. Mutation of the three charged polar residues Asp45, Lys48 and Asp62 had the greatest deleterious effect, suggesting that electrostatic interactions between TWEAK and Fn14 residues may be particularly important for complex formation or stability. To determine whether the four critical residues were likely to be located on the Fn14 CRD surface, we made an Fn14 homology model based on a previously derived X-ray structure for the B-cell maturation antigen receptor, which also contains only one CRD. This model revealed that each of these critical residues were in areas of the receptor that are potentially capable of interacting with TWEAK. These results indicate that the TWEAK–Fn14 interaction is highly dependent on multiple Fn14 residues located in both CRD modules.
Keywords: cysteine-rich domain, fibroblast growth factor-inducible 14 (Fn14), mutagenesis, tumour necrosis factor (TNF), tumour necrosis factor-like weak inducer of apoptosis (TWEAK)
Abbreviations: APRIL, a proliferation-inducing ligand; BAFF, B-cell activation factor; BCA, bicinchoninic acid; BCMA, B-cell maturation antigen; BR3, BLys/BAFF receptor 3; CHO, Chinese-hamster ovary; CRD, cysteine-rich domain; FBS, fetal bovine serum; Fn14, fibroblast growth factor-inducible 14; HEK-293 cells, human embryonic kidney 293 cells; HUVEC, human umbilical-vein endothelial cell; TACI, transmembrane activator and CAML (calcium modulator and cyclophilin ligand) interactor; TNF, tumour necrosis factor; TNFR, TNF receptor; TWEAK, TNF-like weak inducer of apoptosis
INTRODUCTION
TNF (tumour necrosis factor) superfamily members bind specific cell-surface receptors and thereby play important roles in many biological processes, including host defence, autoimmunity, organogenesis, inflammation and apoptosis [1,2]. Most of these superfamily members are initially synthesized as type II transmembrane proteins containing a relatively short intracellular N-terminal domain and an approx. 150-amino-acid extracellular C-terminal TNF homology domain. The TNF homology domains of these ligands have only approx. 25% overall sequence identity but they all fold into a β-sandwich jelly roll structure that interacts with adjacent monomers to form a bell-shaped homotrimer [3]. Many TNF superfamily members can be cleaved, either intracellularly or at the cell surface, to produce a soluble, trimeric, biologically active cytokine [3].
TWEAK (TNF-like weak inducer of apoptosis) was initially described as a member of the TNF superfamily with weak apoptotic activity [4]. It is now recognized that, although soluble TWEAK is indeed an apoptotic factor for certain tumour cell lines [5], it more frequently induces other biological effects when it is added to cells in culture (reviewed in [6,7]). For example, TWEAK has been reported to stimulate cell proliferation [8–12], survival [13], migration [9,10,14] and differentiation [15]. In addition, TWEAK treatment of several different cell types activates the NF-κB (nuclear factor κB) signalling pathway [10,16–22] and induces the expression of pro-inflammatory molecules [4,9,19,21,23,24]. It has also been reported that TWEAK can induce blood vessel formation (angiogenesis) in vivo [8,25].
TWEAK activity is mediated via binding to Fn14 (fibroblast growth factor-inducible 14), a plasma membrane-anchored protein initially discovered in a differential display cloning project [26] and then subsequently identified as a TWEAK-binding TNFR (TNF receptor) superfamily member by Wiley et al. [27]. Most of the TNFR superfamily members are type I transmembrane proteins and all of these proteins contain an extracellular domain that is structurally characterized by the presence of one to six CRDs (cysteine-rich domains) [1,3,28]. The canonical CRD is approx. 40 amino acids in length and contains six conserved cysteine residues that form three intrachain disulphide bridges [1,3]. The CRD itself is typically composed of two distinct structural modules [29]. Fn14 contains a 53-amino-acid extracellular domain with one canonical CRD. There have been no previous biochemical or structural studies describing amino acid residues within the Fn14 CRD that are critical for high-affinity TWEAK binding. Therefore, in the present study, we used a site-specific mutagenesis approach to determine whether several selected evolutionarily conserved residues within the Fn14 CRD sequence were required for TWEAK binding to this region. We found that three charged amino acid residues within this CRD were particularly critical for an effective TWEAK–Fn14 interaction.
MATERIALS AND METHODS
Cell culture
HUVECs (human umbilical-vein endothelial cells; Cambrex) and HEK-293 (human embryonic kidney 293) and HEK-293T cells (A.T.C.C., Manassas, VA, U.S.A.) were grown as described previously [10].
Xenopus Fn14 cDNA sequence analysis
A Xenopus laevis heart cDNA library EST (expressed sequence tag) sequence (GenBank® accession no. BG371238) with sequence similarity to mammalian Fn14 sequences was identified and the corresponding plasmid clone was obtained from the IMAGE consortium (Integrated Molecular Analysis of Genomes and their Expression consortium) (clone ID 4406996). The approx. 1.3 kb insert was sequenced in its entirety using a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems), appropriate oligonucleotide primers (IDT, Coralville, IA, U.S.A.) and an Applied Biosystems model 3100 Avant DNA sequencer. The Xenopus Fn14 nucleotide and deduced amino acid sequences have been deposited in the GenBank® Nucleotide Sequence Database (accession no. AY458020).
Construction of the Fn14–Fc and Fc expression plasmids and isolation of stably transfected HEK-293 cell lines
Stable HEK-293T cell lines transfected with the plasmid pSecTag2/Fn14-Fc-myc (encoding the wild-type murine Fn14 extracellular domain fused to murine Fc) or the plasmid pSecTag2/Fc-myc (encoding murine Fc) have been described previously [10,30]. The PCR overlap extension method was used to make mutations in the murine Fn14 CRD nucleotide sequence [31]. Briefly, PCR was performed in a GeneAmp PCR System 9700 thermocycler (Applied Biosystems) using the pSecTag2/Fn14-Fc-myc plasmid as the template, Vent polymerase (New England Biolabs) and mutagenic oligonucleotides. The PCR products were isolated and ligated into pCMV-Script (Invitrogen) according to the manufacturer's instructions. All constructs were verified by DNA sequence analysis. These constructs encoded ten Myc epitope-tagged Fn14 CRD mutants (denoted C36S, C52S, C55S, S38A, D45A, K48A, M50A, P56A, D62A and D62E). Subconfluent HEK-293 cells were transfected with each of these plasmids using Lipofectamine™ Plus (Invitrogen) according to the manufacturer's instructions. Cells were cultured in standard growth medium containing 600 μg/ml G418 (Mediatech) and stably transfected pooled cell lines were isolated.
Western blot and ligand blot analyses
Cell lines expressing the various Fn14–Fc proteins or the control Fc protein were cultured in a growth medium containing 10% (v/v) Ultra-low FBS (fetal bovine serum) (Invitrogen) until they reached a similar confluency. The conditioned media were collected and centrifuged at 2000 g for 5 min at room temperature to remove any contaminating cells. The cells were washed with PBS, harvested and lysed in HNTG buffer (50 mM Hepes, pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100 and 10% glycerol) [25]. Protein concentrations were determined using the BCA (bicinchoninic acid) protein assay (Pierce). For Western blot analysis, equivalent amounts of cell lysate protein and conditioned medium sample were subjected to SDS/PAGE under either reducing or non-reducing [no 2-mercaptoethanol in loading buffer (100 mM Tris/HCl, pH 6.8, 4% SDS, 20% glycerol and 0.2% Bromophenol Blue) and no heating at 95 °C prior to loading] conditions using 4–12% NuPAGE gradient gels (Invitrogen). Protein transfer to nitrocellulose membranes and protein detection using the anti-Myc antibody 9E10 were performed as described in [25]. For ligand blot analysis, equivalent amounts of cell lysate protein were subjected to SDS/PAGE under non-reducing conditions using 4–12% NuPAGE gradient gels and proteins were transferred to nitrocellulose membranes. Subsequent steps in the procedure were performed as described previously [16] except that, in some experiments, blots were incubated with CHO (Chinese-hamster ovary) cell-derived recombinant Fc–TWEAK fusion protein (a gift from Pascal Schneider, University of Lausanne, Epalinges, Switzerland) instead of insect-cell-derived recombinant TWEAK (a gift from Dr Tim Zheng, Biogen Idec, Cambridge, MA, U.S.A.).
Purification of the Fn14–Fc and Fc proteins and surface plasmon resonance assays
The Fn14–Fc and Fc proteins were first isolated from HEK-293 or HEK-293T conditioned media by affinity chromatography using Protein A–Sepharose (Amersham Biosciences) as described in [10]. This material was then further purified by chromatography on Ni2+-nitrilotriacetate–agarose (Qiagen) according to the manufacturer's instructions. The eluate was dialysed against PBS and protein concentrations were determined using the BCA protein assay (Pierce). Binding of the purified Fn14–Fc and Fc proteins to immobilized recombinant TWEAK was measured using a BIAcore 3000 instrument. For these studies, purified human TWEAK (PeproTech; 10 μg/ml in 10 mM sodium acetate, pH 4.0) was covalently coupled with the second flow path of a BIAcore CM5 sensor chip using the amine-coupling protocol supplied by the manufacturer (at a level of either 460 or 630 resonance units, depending on the experiment). The first flow path of the chip was activated and blocked with ethanolamine and used as a reference cell for each run. Various concentrations of the Fn14–Fc proteins or the Fc protein in 10 mM Hepes (pH 7.4), 150 mM NaCl, 3 mM EDTA and 0.005% surfactant P20 (HBS-EP) were injected for 3 min at a flow rate of 30 μl/min and the binding kinetics was recorded. HBS-EP was applied for 2.5 min to dissociate bound protein and the flow paths were regenerated with 10 mM glycine (pH 2.5). Using BIAcore software (BIAevaluation 3.0), the data were globally fitted to a 1:1 binding model to obtain values for the forward (ka) and reverse (kd) reactions. The equilibrium dissociation constants (Kd) were then calculated as Kd=kd/ka.
HUVEC proliferation assays
Cells were seeded in triplicate in 24-well plates at a density of 2000 cells/well in endothelial cell basal medium-2 supplemented with 5% FBS and 1× ascorbic acid (Cambrex). On days 1, 3 and 5, cells were either left untreated or treated with fresh medium containing TWEAK (100 ng/ml) alone or TWEAK pre-incubated with the various Fn14–Fc proteins or the Fc protein (1.0 μg/ml; all purified as described above). On day 6, cell numbers were determined using the CyQuant Cell Proliferation assay (Molecular Probes). Statistical analysis was performed using Student's t test, and cell number differences were considered to be statistically significant at P <0.05.
CRD alignment and Fn14 structure model
The human and mouse Fn14 CRD sequences were aligned to the human TACI [transmembrane activator and CAML (calcium modulator and cyclophilin ligand) interactor] membrane-proximal CRD (denoted TACI_d2) and the human BCMA (B-cell maturation antigen) CRD sequences using the sequence analysis program Xsae and manually optimized based on structure-assisted alignment of BCMA and TACI_d2 (Protein Data Bank codes 1XU2 and 1XU1 respectively). Based on this sequence alignment, a homology model of both human and murine Fn14 was constructed using the X-ray structure of APRIL (a proliferation-inducing ligand)–BCMA complexes [32] as a template with the program Xsae. The structure was minimized using the program MOLOC and Figure 6(B) was made with the program PyMOL (Delano Scientific). Cysteine connectivity and loop topology is expected to be accurate, but precise backbone and side chain conformations may differ from a structure determined using experimental data.
Figure 6. Sequence alignment of the TACI_d2, BCMA and Fn14 CRDs and predicted structure of the Fn14 CRD.
(A) Alignment of the indicated human (h) or mouse (m) CRD sequences. Cysteine residues are shown in boldface and the TACI_d2 and BCMA cysteine connectivity is shown above the alignment. Amino acid residues (non-cysteine residues) critical for APRIL binding to TACI_d2 [32] or BCMA [39] and for TWEAK binding to Fn14 (the present study) are underlined. Fn14 residues that are dispensable for TWEAK binding (the present study) are marked with a dot. (B) Cα trace (left) of a homology model of mFn14 CRD based on the X-ray crystal structure of the APRIL–BCMA complex [32]. The cysteine residues (yellow) and the non-cysteine residues shown to be important (red) or dispensable (green) for TWEAK binding to Fn14 are labelled and rendered as sticks. The approximate boundaries of the A1 and D2 modules are marked. On the right, the mFn14 homology model is shown as a molecular surface. The orientation and colour scheme are identical with those on the left.
RESULTS
XenopusFn14 cDNA sequence analysis
The predicted human [33], murine [26] and rat [34] Fn14 protein sequences, each 129 amino acids in length, have been reported previously. We obtained an X. laevis Fn14 cDNA and sequenced it in its entirety in order to compare the three different mammalian Fn14 sequences with that of a non-mammalian vertebrate. The predicted full-length XenopusFn14 protein is 120 amino acids in length with a molecular mass of 13295 Da and a pI of 6.80. This protein is predicted to contain a 22-amino-acid signal peptide sequence, a 50-amino-acid extracellular domain, a 21-amino-acid transmembrane domain and a 27-amino-acid cytoplasmic tail (Figure 1). The human and XenopusFn14 proteins have 41% overall amino acid sequence identity with highest sequence conservation in the extracellular domain and in the TNFR-associated factor binding region within the cytoplasmic tail [16,17,27]. The mammalian and Xenopus Fn14 extracellular domains each contain one canonical CRD with the consensus sequence CX12CX2CX2CX8CX2C, where C is cysteine and Xn is the number of intervening amino acids. The CRDs of the four receptors contained 15 evolutionarily conserved residues, including the six cysteine residues.
Figure 1. Comparison of the human, murine, rat and toad Fn14 deduced amino acid sequences.
The human (Homo sapiens; GenBank® accession no. NM_016639), murine (Mus musculus; NM_013749), rat (Rattus norvegicus; NM_181086) and toad (X. laevis; AY458020) Fn14 sequences are aligned. Conserved residues are boxed and the numbers to the right refer to the last amino acids in each row. The six cysteine residues within the extracellular domain are shown in boldface, the signal peptidase (SP) cleavage site is indicated, and the transmembrane (TM) domain is boxed with a dotted line. The nine murine Fn14 residues that were mutated in the present study are indicated with a black box above the alignment.
Mutagenesis of selected Fn14 CRD residues and Fn14–Fc protein characterization
We decided to identify TWEAK-binding residues in the Fn14 CRD using a site-specific mutagenesis strategy. To facilitate detection and purification of the various mutants, we expressed them as soluble extracellular domain fusion proteins linked at their C-terminus to the hinge and Fc portion of IgG1 heavy chain. The divalent Fn14–Fc proteins also contained a c-Myc epitope and a polyhistidine tag (Figure 2A). We made the amino acid substitutions in the murine Fn14 CRD sequence, not the human Fn14 CRD sequence, because we had already constructed an expression plasmid encoding the wild-type murine Fn14 extracellular domain fused to the Fc portion of IgG1 [10,30] and this construct could serve as our mutagenesis template.
Figure 2. Structural properties and characterization of the murine Fn14–Fc and Fc proteins.
(A) Schematic representation of the wild-type (WT) Fn14–Fc protein is shown. The Ig κ chain signal peptide (SP), the Fn14 extracellular domain (Fn14-EC), the IgG hinge and Fc domain, and the region containing the Myc epitope and polyhistidine tags are indicated. The CRDs are represented by loops and the bars represent cysteine residues. The Fc protein is similar, except that the Fn14-EC region is absent. (B, C) Stable cell lines expressing the indicated Fn14–Fc proteins or the Fc protein as well as untransfected parental cells (‘ND’, no DNA) were grown in culture. Conditioned medium samples were collected, cells were harvested and lysed, and equal amounts of cellular protein were subjected to SDS/PAGE under reducing conditions. Western blot analysis was then performed using an anti-Myc antibody. Upper panels: the cell lysate samples; lower panels: the conditioned medium samples. Molecular-mass sizes given in kDa.
We initially made seven mutant constructs to probe different portions of the CRD. The Fn14 CRD probably has an A1–D2 modular structure with the disulphide cross-links Cys36–Cys49, Cys52–Cys64 and Cys55–Cys67 ([35] and see below). Therefore three cysteine-to-serine mutations (denoted C36S, C52S and C55S) were generated in order to disrupt formation of each predicted disulphide bond. In the predicted A1 module, we mutated the conserved charged residues Asp45 and Lys48 and the conserved non-polar residue Met50 to alanine residues (denoted D45A, K48A and M50A). In the predicted D2 module, we mutated the conserved charged residue Asp62 to alanine (denoted D62A). Stable HEK-293 cell lines expressing these various Fn14–Fc proteins were established and we first investigated whether the cells expressed and secreted the various proteins by Western blot analysis of both cell lysates and conditioned media. Wild-type Fn14–Fc and the seven mutant Fn14–Fc proteins were expressed at comparable levels, but the Fn14–Fc C36S, C52S, C55S and D62A proteins were not secreted from the transfected cell lines, suggesting that these amino acid substitutions caused major perturbations in the Fn14 extracellular domain structure (Figure 2B, left- hand panels). When we mutated the Fn14 Asp62 residue to a glutamic residue, a more conservative substitution, the Fn14–Fc D62E protein was efficiently secreted (Figure 2B, right-hand panels).
We also decided to mutate the two non-conserved CRD residues Ser38 (in A1 module) and Pro56 (in D2 module) to alanine residues. We hypothesized that these amino acid replacements would serve as good ‘silent mutation’ controls to demonstrate that not all alanine substitutions within the CRD result in reduced TWEAK binding. These two Fn14–Fc proteins were efficiently expressed and secreted as predicted (Figure 2C). Finally, Western blot analysis was also performed using conditioned medium samples from the Fn14–Fc wild-type, S38A, D45A, K48A, M50A, P56A and D62E cell lines run under non-reducing conditions. This analysis confirmed that the Fn14–Fc molecules were secreted as disulphide-bonded dimers and no higher-molecular-mass species representing aggregated protein complexes were detected (results not shown).
Qualitative analysis of TWEAK binding to the mutant Fn14–Fc proteins by ligand blot analysis
As an initial binding assay, we determined whether soluble human TWEAK could bind to the various Fn14–Fc proteins when they were immobilized on a nitrocellulose membrane (i.e. ligand blot analysis). Cell lysates were prepared from the stable cell lines and protein was subjected to SDS/PAGE under non-reducing conditions. Duplicate blots were prepared and one was used for Western blot analysis. All 11 Fn14–Fc proteins migrated at the appropriate apparent molecular masses (Figure 3, lower panels). In the case of the Fn14–Fc C36S, C52S and C55S mutants, higher-molecular-mass species were evident, suggesting some aggregation. The second blot was incubated sequentially with recombinant TWEAK and then with anti-TWEAK antibodies. We demonstrated above that the Fn14–Fc C36S, C52S, C55S and D62A mutants were clearly misfolded and therefore, as expected, there was no TWEAK binding to these four proteins (Figure 3, upper left panel). These four proteins were not examined further. The Fn14–Fc D45A, K48A, M50A and D62E proteins, which do not exhibit obvious structural abnormalities, displayed either an intermediate level of TWEAK binding (M50A), a very low level of TWEAK binding (K48A) or no detectable TWEAK binding (D45A and D62E) relative to the Fn14–Fc wild-type protein (Figure 3, upper left panel). The Fn14–Fc S38A and P56A proteins, which were also efficiently secreted and thus do not exhibit obvious structural abnormalities, bound TWEAK as well as (S38A) or slightly better than (P56A) the Fn14–Fc wild-type protein in this assay (Figure 3, upper right panel). These two proteins were not examined further.
Figure 3. Ligand blot analysis of TWEAK binding to the Fn14–Fc and Fc proteins.
Cell lysates were prepared from cell lines expressing the indicated Fn14–Fc proteins or the Fc protein as well as from untransfected parental cells (‘ND’, no DNA) and equal amounts of cellular protein were analysed by SDS/PAGE under non-reducing conditions. Ligand blot analysis was performed using recombinant TWEAK and anti-TWEAK antibodies (upper panels) and Western blot analysis was performed using an anti-Myc antibody (lower panels). WT, wild-type. Molecular-mass sizes are given in kDa.
Quantitative analysis of TWEAK binding to the mutant Fn14–Fc proteins by BIAcore analysis
Surface plasmon resonance measurements on a BIAcore 3000 instrument were then performed to measure the binding constants of wild-type and mutant Fn14–Fc proteins for TWEAK. In these experiments, TWEAK was coupled with a sensor chip and the soluble Fn14–Fc wild-type, D45A, K48A, M50A and D62E proteins, as well as the control Fc protein, were injected in the mobile phase. When increasing concentrations of the Fn14–Fc wild-type protein were injected, dose-dependent binding to the immobilized TWEAK was observed and a Kd of 0.8 nM was calculated from the kinetic binding data (Figure 4A). Injection of the Fn14–Fc wild-type, D45A, K48A, M50A and D62E proteins (and the control Fc protein, all at 10 nM) revealed that the four mutants and the Fc protein had significantly reduced binding affinity for TWEAK (Figure 4B). Indeed, a Kd of 1.3 nM was calculated from the kinetic binding data for wild-type Fn14–Fc protein, but a Kd for the Fn14–Fc mutants and the Fc protein could not be determined because they displayed negligible binding. When the six proteins were injected at a 10-fold higher concentration, some binding to the Fn14–Fc M50A protein was detected (Figure 4C), consistent with the ligand blot results described above. A Kd for wild-type Fn14–Fc (4.7 nM) and for the Fn14–Fc M50A mutant (71 nM) could be calculated from a global fit of this binding data. Data for the other three mutants and the Fc protein were not fitted because they still displayed negligible binding.
Figure 4. Surface plasmon resonance analysis of Fn14–Fc and Fc binding to immobilized TWEAK.
(A) Sensorgrams showing real-time binding of soluble wild-type (WT) Fn14–Fc protein (0.2–5.0 nM) to TWEAK immobilized on a CM5 chip. Flow cells without any immobilized protein were used as the controls for non-specific binding and were subtracted from the presented data. (B) Sensorgrams showing real-time binding of soluble wild-type (WT) Fn14–Fc protein, four different Fn14–Fc mutant proteins and control Fc protein (all at 10 nM) to TWEAK immobilized on a CM5 chip. (C) Sensorgrams showing real-time binding of soluble wild-type (WT) Fn14–Fc protein, four different Fn14–Fc mutant proteins and control Fc protein (all at 100 nM) to TWEAK immobilized on a CM5 chip.
Analysis of TWEAK binding to the mutant Fn14–Fc proteins in solution
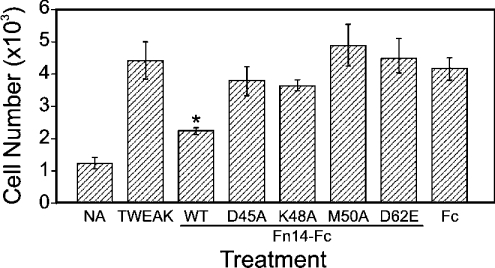
We have previously shown that the soluble Fn14–Fc wild-type protein can function as a decoy receptor and inhibit TWEAK-stimulated cellular responses in vitro [10,14,22]. For example, when TWEAK is pre-incubated with the Fn14–Fc wild-type protein prior to its addition to quiescent HUVECs in culture, TWEAK mitogenic activity on these cells is significantly reduced [10]. We used this same HUVEC proliferation assay to determine whether the Fn14–Fc D45A, K48A, M50A and D62E proteins, like the wild-type protein, can bind TWEAK in solution and act as decoy receptors. TWEAK was pre-incubated with a 10-fold molar excess of the Fn14–Fc wild-type, D45A, K48A, M50A and D62E proteins (and the control Fc protein) and was added to cells. After 6 days of incubation, the cells were collected and cell numbers were determined. TWEAK increased HUVEC growth 3.6-fold under these experimental conditions, and this effect was significantly inhibited only when the cytokine was pre-incubated with the wild-type Fn14–Fc fusion protein (Figure 5).
Figure 5. Effect of the Fn14–Fc and Fc proteins on TWEAK-stimulated HUVEC proliferation.
HUVECs were seeded at low density, placed into growth-factor-depleted medium for 1 day and then either left unstimulated (NA, no addition) or stimulated with TWEAK alone or TWEAK pre-incubated with the indicated Fn14–Fc or Fc protein. The assay was terminated on day 6 and cell numbers were calculated. Cell numbers are expressed as the means±S.D. for triplicate wells representative of two independent experiments. *P<0.05 compared with TWEAK treatment alone.
Predicted structure of the Fn14 CRD
High-resolution solution or crystal structures for the TWEAK receptor-binding domain, the Fn14 extracellular domain (or CRD) or TWEAK–Fn14 complexes are currently unavailable. However, both biochemical and structural studies have been performed to investigate the ligand-binding properties of the three other TNFR superfamily members that have only one or two CRDs [32,35–39]. These three receptors are (i) BR3 [BLys/BAFF (B-cell activation factor) receptor 3], which contains a single partial CRD (only four cysteine residues) and binds the ligand BAFF, (ii) BCMA, which contains a single canonical CRD and binds both BAFF and APRIL, and (iii) TACI, also a receptor for APRIL and BAFF, which contains two canonical CRDs. Interestingly, although TACI contains two CRDs, the membrane-proximal CRD (denoted TACI_d2) is the primary determinant for high-affinity ligand binding [32]. The disulphide-bonding patterns of the TACI_d2 and BCMA CRDs have been determined and each of these domains has an A1–D2 modular structure [32,35]. In addition, investigators have used combinatorial alanine-scanning mutagenesis to identify amino acid residues within these two CRDs that are critical for binding of the shared ligand APRIL [32,39]. Therefore, as an initial approach to determine why TWEAK had reduced binding affinity for the Fn14–Fc D45A, K48A, M50A and D62E mutant receptors, we first compared the human TACI_d2, BCMA and Fn14 CRD sequences and then made an Fn14 homology model. In the CRD sequence alignment, there are seven conserved residues: the six cysteine residues and a leucine residue (Fn14 position 46) (Figure 6A). However, both the human and murine Fn14 CRDs contain an aromatic tryptophan residue at position 42 that aligns with a TACI_d2 tyrosine residue and a BCMA phenylalanine residue. In Xenopus, this Fn14 residue is a tyrosine. It has been postulated that these particular TACI and BCMA aromatic side-chain residues stabilize the A1 module β-hairpin region [32], so the Fn14 tryptophan (or tyrosine) may have a similar function. This sequence comparison indicates that the overall structure of the Fn14 CRD may be similar to that of the BCMA and TACI_d2 CRDs (i.e. an A1–D2 modular structure).
We next made a homology model of the Fn14 CRD using this alignment and the X-ray crystal structure of APRIL–BCMA complexes as a template [32]. This model is compatible with Fn14 possessing the same cysteine connectivity pattern (Cys1–Cys2, Cys3–Cys5, Cys4–Cys6) and modular structure (A1–D2) as reported for the BCMA and TACI_d2 CRDs [32,35]. Also, the model indicates that Asp45, Lys48, Met50 and Asp62 are in areas of the receptor that are potentially capable of interacting with TWEAK (Figure 6B).
DISCUSSION
TWEAK, a member of the TNF superfamily, acts on responsive cells via binding to Fn14, the smallest TNFR superfamily member identified to date (reviewed in [6,7]). Indeed, the human Fn14 extracellular domain is only 53 amino acids in length and contains one canonical CRD. In the present study, we used a site-specific mutagenesis approach to investigate TWEAK binding to the Fn14 CRD. The various CRD mutants were expressed in stably transfected HEK-293 cells as secretable Fn14 extracellular domain–Fc fusion proteins with Myc and polyhistidine tags in order to rapidly identify those mutations that significantly altered CRD structure and to simplify mutant protein identification and purification.
We first compared three different mammalian (human, murine and rat) and one non-mammalian (X. laevis, an African clawed toad) Fn14 CRD sequences in order to identify evolutionarily conserved residues that might be critical sites for TWEAK binding. There were 15 conserved residues within the 32-amino-acid CRD, including the six cysteine residues. Seven of the conserved residues were selected for mutation. Specifically, we made three cysteine-to-serine mutations (C36S, C52S and C55S) in order to test whether each of the three predicted disulphide bonds contributed to the overall structural integrity of the CRD. We also replaced four conserved non-cysteine residues – the three charged amino acids (Asp45, Lys48 and Asp62) and the single sulphur-containing residue (Met50) – with alanine or, in the case of Asp62, with both alanine and glutamic residues. Two non-conserved, non-cysteine residues (Ser38 and Pro56) were also selected for alanine replacement. All of the mutations were made in the murine Fn14 CRD. Since the human and murine Fn14 CRDs have 94% sequence identity (two amino acid differences) and human TWEAK can bind with high affinity to murine Fn14 [16,40], it is likely that our findings studying the murine receptor are applicable to the human receptor as well.
The Fn14–Fc C36S, C52S, C55S and D62A mutant proteins were expressed in HEK-293 cells but were not secreted, presumably due to major structural perturbations. Indeed, we detected higher-molecular-mass species when the three cysteine mutants were examined under non-reducing conditions, suggesting that unpaired cysteine residues were promoting the formation of disulphide-linked aggregates. The D62A protein was also not secreted, therefore, in order to investigate the importance of this particular aspartic acid residue, we made the D62E mutant, which was efficiently secreted. This result demonstrates that correct Fn14 CRD folding is highly dependent on the presence of a charged residue at position 62. As expected, the Fn14–Fc C36S, C52S, C55S and D62A proteins did not bind TWEAK in the ligand blot assay, and, since they were probably misfolded, they were not examined further.
The Fn14–Fc S38A, D45A, K48A, M50A, P56A and D62E proteins were efficiently expressed and secreted from the stably transfected cell lines, suggesting that these mutations did not significantly impair CRD folding. TWEAK binding to these six mutant receptors was first tested using a ligand blot assay. We detected normal TWEAK binding to the S38A and P56A proteins. Some TWEAK binding was also detected to the M50A protein and, to a lesser extent, the K48A protein. No TWEAK binding to D45A or D62E was detected. TWEAK binding to the four binding-deficient receptors was then analysed using the BIAcore surface plasmon resonance assay. In these assays, TWEAK bound to the Fn14–Fc wild-type receptor with an affinity constant (Kd) of 0.8, 1.3 or 4.7 nM, depending on the experimental conditions. The first two values are probably the most accurate affinity estimates, since the last value was obtained using a very high Fn14–Fc concentration (100 nM). Nevertheless, this Kd range is consistent with previous studies measuring TWEAK–Fn14 binding using either an ELISA with immobilized Fn14–Fc protein (1.1 nM; [10]) or by adding 125I-Fn14–Fc to cells expressing membrane-bound TWEAK (2.3 nM; [27]). Some TWEAK binding to the M50A mutant was also detected using the BIAcore assay (Kd of 71 nM) but no binding was detected to the D45A, K48A or D62E mutant receptors. Finally, we confirmed that TWEAK binding to the Fn14–Fc D45A, K48A, M50A and D62E mutants was significantly impaired by showing that these fusion proteins could not act as decoy receptors and inhibit TWEAK proliferative activity on endothelial cells.
In addition to Fn14, three other TNFR superfamily members contain either one (BR3 and BCMA) or two (TACI) CRDs. Therefore we compared the Fn14 CRD sequence with the membrane-proximal TACI_d2 CRD sequence and the BCMA CRD sequence in order to gain insight into the Fn14 CRD folding pattern. The TACI_d1 CRD is not required for high-affinity ligand binding [32] and the BR3 CRD is atypical (it contains four instead of six cysteine residues), so these domains were not included in the analysis. The CRD alignment indicated that, although there is minimal sequence identity between these three CRDs, the overall structure and topology of the Fn14 CRD will probably resemble that of BCMA and TACI_d2 (i.e. an A1–D2 module pair). This conclusion is based on the conservation of the cysteine residues, the length of the two modules and the presence of an aromatic residue at Fn14 CRD position 42. However, it is clear that the Fn14 CRD loop and side-chain conformation will differ significantly from these two receptors. For example, the BCMA and TACI (and BR3) CRDs all share a common six-residue sequence element, the DXL motif, which adopts a type I β-turn structure that is essential for BAFF and APRIL binding [32,35–39]. This motif is not present in the Fn14 CRD and therefore it is not unexpected that BAFF and APRIL do not bind the Fn14 receptor.
We found that the three evolutionarily conserved charged CRD residues Asp45, Lys48 and Asp62 are particularly important for high-affinity TWEAK binding. Structures of the TWEAK receptor-binding domain, the Fn14 CRD or TWEAK–Fn14 complexes have not been reported, therefore we are unable to fully explain at this time why these charged residues strongly influence ligand binding. It is possible that substitution of these residues triggers structural changes that indirectly compromise TWEAK binding activity. However, since the Fn14 homology model indicates that the Asp45, Lys48 and Asp62 residues are likely to be located on the Fn14 CRD surface, the charged residues may directly participate in TWEAK binding; for example, they may form salt bridges with oppositely charged TWEAK residues. In the case of the Asp62 residue within the D2 module, even a conservative substitution mutation (D62E) prevented TWEAK binding, thus Asp62 must make some exquisite interaction that does not tolerate a methylene group at this position. In summary, our results indicate that the Fn14 CRD is likely to be a compact highly ordered structure and that TWEAK binding to this domain is dependent on charged residues located in each submodule. Additional mutagenesis and structural studies are required to further explore how individual Fn14 CRD amino acid residues influence ligand-binding affinity and specificity.
Acknowledgments
We thank Dr Sarah Hymowitz (Genentech, South San Francisco, CA, U.S.A.) for helpful comments and for the Fn14 structure model. We also thank Dr Tim Zheng for providing the insect cell-derived recombinant TWEAK and the anti-TWEAK antibody, and Dr Pascal Schneider for providing the CHO cell-derived recombinant Fc–TWEAK. This study was supported in part by NIH (National Institutes of Health, Bethesda, MD, U.S.A.) grant HL-39727 (J. A. W.).
References
- 1.Locksley R. M., Killeen N., Lenardo M. J. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 2.Hehlgans T., Pfeffer K. The intriguing biology of the tumour necrosis factor/tumour necrosis factor receptor superfamily: players, rules and the games. Immunology. 2005;115:1–20. doi: 10.1111/j.1365-2567.2005.02143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodmer J., Schneider P., Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem. Sci. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 4.Chicheportiche Y., Bourdon P. R., Xu H., Hsu Y., Scott H., Hession C., Garcia I., Browning J. L. TWEAK, a new secreted ligand in the tumor necrosis factor family that weakly induces apoptosis. J. Biol. Chem. 1997;272:32401–32410. doi: 10.1074/jbc.272.51.32401. [DOI] [PubMed] [Google Scholar]
- 5.Nakayama M., Ishidoh K., Kojima Y., Harada N., Kominami E., Okumura K., Yagita H. Fibroblast growth factor-inducible 14 mediates multiple pathways of TWEAK-induced cell death. J. Immunol. 2003;170:341–348. doi: 10.4049/jimmunol.170.1.341. [DOI] [PubMed] [Google Scholar]
- 6.Wiley S. R., Winkles J. A. TWEAK, a member of the TNF superfamily, is a multifunctional cytokine that binds the TweakR/Fn14 receptor. Cytokine Growth Factor Rev. 2003;14:241–249. doi: 10.1016/s1359-6101(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 7.Campbell S., Michaelson J., Burkly L., Putterman C. The role of TWEAK/Fn14 in the pathogenesis of inflammation and systemic autoimmunity. Front. Biosci. 2004;9:2273–2284. doi: 10.2741/1395. [DOI] [PubMed] [Google Scholar]
- 8.Lynch C. N., Wang Y. C., Lund J. K., Chen Y., Leal J. A., Wiley S. R. TWEAK induces angiogenesis and proliferation of endothelial cells. J. Biol. Chem. 1999;274:8455–8459. doi: 10.1074/jbc.274.13.8455. [DOI] [PubMed] [Google Scholar]
- 9.Harada N., Nakayama M., Nakano H., Fukuchi Y., Yagita H., Okumura K. Pro-inflammatory effect of TWEAK/Fn14 interaction on human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2002;299:488–493. doi: 10.1016/s0006-291x(02)02670-0. [DOI] [PubMed] [Google Scholar]
- 10.Donohue P. J., Richards C. M., Brown S. A., Hanscom H. N., Buschman J., Thangada S., Hla T., Williams M. S., Winkles J. A. TWEAK is an endothelial cell growth and chemotactic factor that also potentiates FGF-2 and VEGF-A mitogenic activity. Arterioscler. Thromb. Vasc. Biol. 2003;23:594–600. doi: 10.1161/01.ATV.0000062883.93715.37. [DOI] [PubMed] [Google Scholar]
- 11.Jakubowski A., Ambrose C., Parr M., Lincecum J. M., Wang M. Z., Zheng T. S., Browning B., Michaelson J. S., Baetscher M., Wang B., et al. TWEAK induces liver progenitor cell proliferation. J. Clin. Invest. 2005;115:2330–2340. doi: 10.1172/JCI23486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michaelson J. S., Cho S., Browning B., Zheng T. S., Lincecum J. M., Wang M. Z., Hsu Y. M., Burkly L. C. Tweak induces mammary epithelial branching morphogenesis. Oncogene. 2005;24:2613–2624. doi: 10.1038/sj.onc.1208208. [DOI] [PubMed] [Google Scholar]
- 13.Jakubowski A., Browning B., Lukashev M., Sizing I., Thompson J. S., Benjamin C. D., Hsu Y., Ambrose C., Zheng T. S., Burkly L. C. Dual role for TWEAK in angiogenic regulation. J. Cell Sci. 2002;115:267–274. doi: 10.1242/jcs.115.2.267. [DOI] [PubMed] [Google Scholar]
- 14.Tran N. L., McDonough W. S., Donohue P. J., Winkles J. A., Berens T. J., Ross K. R., Hoelzinger D. B., Beaudry C., Coons S. W., Berens M. E. The human Fn14 receptor gene is up-regulated in migrating glioma cells in vitro and overexpressed in advanced glial tumors. Am. J. Pathol. 2003;162:1313–1321. doi: 10.1016/S0002-9440(10)63927-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polek T. C., Talpaz M., Darnay B. G., Spivak-Kroizman T. TWEAK mediates signal transduction and differentiation of RAW264.7 cells in the absence of Fn14/TweakR: evidence for a second TWEAK receptor. J. Biol. Chem. 2003;278:32317–32323. doi: 10.1074/jbc.M302518200. [DOI] [PubMed] [Google Scholar]
- 16.Brown S. A. N., Richards C. M., Hanscom H. N., Feng S. L., Winkles J. A. The Fn14 cytoplasmic tail binds tumour-necrosis-factor-receptor-associated factors 1, 2, 3 and 5 and mediates nuclear factor-κB activation. Biochem. J. 2003;371:395–403. doi: 10.1042/BJ20021730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han S., Yoon K., Lee K., Kim K., Jang H., Lee N. K., Hwang K., Young L. S. TNF-related weak inducer of apoptosis receptor, a TNF receptor superfamily member, activates NF-κB through TNF receptor-associated factors. Biochem. Biophys. Res. Commun. 2003;305:789–796. doi: 10.1016/s0006-291x(03)00852-0. [DOI] [PubMed] [Google Scholar]
- 18.Saitoh T., Nakayama M., Nakano H., Yagita H., Yamamoto N., Yamaoka S. TWEAK induces NF-κB2 p100 processing and long lasting NF-κB activation. J. Biol. Chem. 2003;278:36005–36012. doi: 10.1074/jbc.M304266200. [DOI] [PubMed] [Google Scholar]
- 19.Xu H., Okamoto A., Ichikawa J., Ando T., Tasaka K., Masuyama K., Ogawa H., Yagita H., Okumura K., Nakao A. TWEAK/Fn14 interaction stimulates human bronchial epithelial cells to produce IL-8 and GM-CSF. Biochem. Biophys. Res. Commun. 2004;318:422–427. doi: 10.1016/j.bbrc.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 20.Jin L., Nakao A., Nakayama M., Yamaguchi N., Kojima Y., Nakano N., Tsuboi R., Okumura K., Yagita H., Ogawa H. Induction of RANTES by TWEAK/Fn14 interaction in human keratinocytes. J. Invest. Dermatol. 2004;122:1175–1179. doi: 10.1111/j.0022-202X.2004.22419.x. [DOI] [PubMed] [Google Scholar]
- 21.Campbell S., Burkly L. C., Gao H., Berman J. W., Su L., Browning B., Zheng T., Schiffer L., Michaelson J. S., Putterman C. Proinflammatory effects of Tweak/Fn14 interactions in glomerular mesangial cells. J. Immunol. 2006;176:1889–1898. doi: 10.4049/jimmunol.176.3.1889. [DOI] [PubMed] [Google Scholar]
- 22.Tran N. L., McDonough W. S., Savitch B. A., Sawyer T. F., Winkles J. A., Berens M. E. The tumor necrosis factor-like weak inducer of apoptosis (TWEAK)-fibroblast growth factor-inducible 14 (Fn14) signaling system regulates glioma cell survival via NF-κB pathway activation and BCL-XL/BCL-W expression. J. Biol. Chem. 2005;280:3483–3492. doi: 10.1074/jbc.M409906200. [DOI] [PubMed] [Google Scholar]
- 23.Saas P., Boucraut J., Walker P. R., Quiquerez A., Billot M., Desplat-Jego S., Chicheportiche Y., Dietrich P. TWEAK stimulation of astrocytes and the proinflammatory consequences. Glia. 2000;32:102–107. [PubMed] [Google Scholar]
- 24.Chicheportiche Y., Chicheportiche R., Sizing I., Thompson J., Benjamin C. B., Ambrose C., Dayer J. Proinflammatory activity of TWEAK on human dermal fibroblasts and synoviocytes: blocking and enhancing effects of anti-TWEAK monoclonal antibodies. Arthritis Res. 2002;4:126–133. doi: 10.1186/ar388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho D. H., Vu H., Brown S. A. N., Donohue P. J., Hanscom H. N., Winkles J. A. Soluble tumor necrosis factor-like weak inducer of apoptosis overexpression in HEK293 cells promotes tumor growth and angiogenesis in athymic nude mice. Cancer Res. 2004;64:8968–8972. doi: 10.1158/0008-5472.CAN-04-1879. [DOI] [PubMed] [Google Scholar]
- 26.Meighan-Mantha R. L., Hsu D. K. W., Guo Y., Brown S. A. N., Feng S. Y., Peifley K. A., Alberts G. F., Copeland N. G., Gilbert D. J., Jenkins N. A., et al. The mitogen-inducible Fn14 gene encodes a type I transmembrane protein that modulates fibroblast adhesion and migration. J. Biol. Chem. 1999;274:33166–33176. doi: 10.1074/jbc.274.46.33166. [DOI] [PubMed] [Google Scholar]
- 27.Wiley S. R., Cassiano L., Lofton T., Davis-Smith T., Winkles J. A., Lindner V., Liu H., Daniel T. O., Smith C. A., Fanslow W. C. A novel TNF receptor family member binds TWEAK and is implicated in angiogenesis. Immunity. 2001;15:837–846. doi: 10.1016/s1074-7613(01)00232-1. [DOI] [PubMed] [Google Scholar]
- 28.Ware C. F. The TNF superfamily. Cytokine Growth Factor Rev. 2003;14:181–184. doi: 10.1016/s1359-6101(03)00032-7. [DOI] [PubMed] [Google Scholar]
- 29.Naismith J. H., Sprang S. R. Modularity in the TNF-receptor family. Trends Biochem. Sci. 1998;23:74–79. doi: 10.1016/s0968-0004(97)01164-x. [DOI] [PubMed] [Google Scholar]
- 30.Yepes M., Brown S. A. N., Moore E. G., Smith E. P., Lawrence D. A., Winkles J. A. A soluble Fn14–Fc decoy receptor reduces infarct volume in a murine model of cerebral ischemia. Am. J. Pathol. 2005;166:511–520. doi: 10.1016/S0002-9440(10)62273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 32.Hymowitz S. G., Patel D. R., Wallweber H. J., Runyon S., Yan M., Yin J., Shriver S. K., Gordon N. C., Pan B., Skelton N. J., et al. Structures of APRIL–receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding. J. Biol. Chem. 2005;280:7218–7227. doi: 10.1074/jbc.M411714200. [DOI] [PubMed] [Google Scholar]
- 33.Feng S. Y., Guo Y., Factor V. M., Thorgeirsson S. S., Bell D. W., Testa J. R., Peifley K. A., Winkles J. A. The Fn14 immediate-early response gene is induced during liver regeneration and highly expressed in both human and murine hepatocellular carcinomas. Am. J. Pathol. 2000;156:1253–1261. doi: 10.1016/S0002-9440(10)64996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller A. M., Pedre X., Kleiter I., Hornberg M., Steinbrecher A., Giegerich G. Targeting fibroblast growth factor-inducible-14 signaling protects from chronic relapsing experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2005;159:55–65. doi: 10.1016/j.jneuroim.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., Hong X., Kappler J., Jiang L., Zhang R., Xu L., Pan C. H., Martin W. E., Murphy R. C., Shu H. B., et al. Ligand–receptor binding revealed by the TNF family member TALL-1. Nature (London) 2003;423:49–56. doi: 10.1038/nature01543. [DOI] [PubMed] [Google Scholar]
- 36.Kayagaki N., Yan M., Seshasayee D., Wang H., Lee W., French D. M., Grewal I. S., Cochran A. G., Gordon N. C., Yin J., et al. BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kappaB2. Immunity. 2002;17:515–524. doi: 10.1016/s1074-7613(02)00425-9. [DOI] [PubMed] [Google Scholar]
- 37.Kim H. M., Yu K. S., Lee M. E., Shin D. R., Kim Y. S., Paik S. G., Yoo O. J., Lee H., Lee J. O. Crystal structure of the BAFF–BAFF-R complex and its implications for receptor activation. Nat. Struct. Biol. 2003;10:342–348. doi: 10.1038/nsb925. [DOI] [PubMed] [Google Scholar]
- 38.Gordon N. C., Pan B., Hymowitz S. G., Yin J., Kelley R. F., Cochran A. G., Yan M., Dixit V. M., Fairbrother W. J., Starovasnik M. A. BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site. Biochemistry. 2003;42:5977–5983. doi: 10.1021/bi034017g. [DOI] [PubMed] [Google Scholar]
- 39.Patel D. R., Wallweber H. J., Yin J., Shriver S. K., Marsters S. A., Gordon N. C., Starovasnik M. A., Kelley R. F. Engineering an APRIL-specific B cell maturation antigen. J. Biol. Chem. 2004;279:16727–16735. doi: 10.1074/jbc.M312316200. [DOI] [PubMed] [Google Scholar]
- 40.Potrovita I., Zhang W., Burkly L., Hahm K., Lincecum J., Wang M. Z., Maurer M. H., Rossner M., Schneider A., Schwaninger M. Tumor necrosis factor-like weak inducer of apoptosis-induced neurodegeneration. J. Neurosci. 2004;24:8237–8244. doi: 10.1523/JNEUROSCI.1089-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]