Abstract
Although the cold-shock response has now been studied in a number of different organisms for several decades, it is only in the last few years that we have begun to understand the molecular mechanisms that govern adaptation to cold stress. Notably, all organisms from prokaryotes to plants and higher eukaryotes respond to cold shock in a comparatively similar manner. The general response of cells to cold stress is the elite and rapid overexpression of a small group of proteins, the so-called CSPs (cold-shock proteins). The most well characterized CSP is CspA, the major CSP expressed in Escherichia coli upon temperature downshift. More recently, a number of reports have shown that exposing yeast or mammalian cells to sub-physiological temperatures (<30 or <37 °C respectively) invokes a co-ordinated cellular response involving modulation of transcription, translation, metabolism, the cell cycle and the cell cytoskeleton. In the present review, we summarize the regulation and role of cold-shock genes and proteins in the adaptive response upon decreased temperature with particular reference to yeast and in vitro cultured mammalian cells. Finally, we present an integrated model for the co-ordinated responses required to maintain the viability and integrity of mammalian cells upon mild hypothermic cold shock.
Keywords: cellular response, cold-shock protein, cold-shock response, control of gene expression, sub-physiological temperature, yeast
Abbreviations: CCT, chaperonin containing the T-complex polypeptide-1; CHO, Chinese-hamster ovary; Cirp, cold-inducible RNA-binding protein; CSP, cold-shock protein; EF1α, elongation factor 1α; eIF2α, eukaryotic initiation factor 2α; F-actin, filamentous actin; GST, glutathione S-transferase; hnRNA, heteronuclear RNA; Hog, high-osmolarity glycerol; HSP, heat-shock protein; IF, initiation factor; IRES, internal ribosome entry segment; MAPK, mitogen-activated protein kinase; miRNA, microRNA; ORF, open reading frame; PKA, protein kinase A; Rbm3, RNA-binding motif protein 3; STRE, stress-response element; STOP, stable tubule-only polypeptide; unr, upstream of N-ras; UTR, untranslated region
INTRODUCTION
A change in environmental temperature is one of the most common stresses experienced by a wide range of organisms from bacteria to plants and animals. Furthermore, temperature changes are utilized in the biotechnology industry for the culturing and preservation of cells, and in the medical field for the preservation of tissue and the treatment of brain damage [1]. The response of prokaryotic and eukaryotic systems to heat-shock stress has been investigated widely in a large number of organisms and model cell systems. The response generally invokes the up-regulation or induction of families of proteins termed HSPs (heat-shock proteins), whose functions are primarily to help deal with, and alleviate, the cellular stress imposed by heat stress [2]. On the other hand, although investigators have now been studying the cold-shock response in a variety of organisms for the last 2 decades or more, comparatively little is known about the molecular mechanisms that govern adaptation to cold stress, particularly in mammalian cells. Notably, all organisms from prokaryotes to plants and higher eukaryotes respond to cold shock in a comparatively similar manner. Generally, cells respond to cold stress by elite and rapid overexpression of a small group of proteins, the so-termed CSPs (cold-shock proteins) [3]. However, unlike HSPs, CSPs do not appear to be as rigorously conserved between prokaryotic and eukaryotic systems.
A number of plant genes are induced by low-temperature stress, and, in prokaryotes, cold stress induces several well-characterized CSPs [2]. In contrast, there is sparse information on the molecular mechanisms of the cold-shock response in mammalian cells; however, our current understanding suggests that the response involves a co-ordinated series of events involving modulation of transcription, translation, the cell cytoskeleton, the cell cycle and metabolic processes. Investigations to date have determined that, in response to cold stress, prokaryotic and eukaryotic cells generally suppress transcription and subsequent translation, except for a select number of CSPs whose synthesis continues or is up-regulated during cold shock [3]. In Escherichia coli, for example, approx. 27 CSPs have been identified hitherto [4]. The majority of these CSPs are regulated sequentially in response to temperature downshift and have fundamental functions that determine cell fate such as DNA replication, transcription, translation, RNA stabilization and ribosome assembly [4].
Various reports have now shown that when in vitro cultivation temperature is lowered, the rigidity of the cell membrane is increased which results in compromised membrane-associated cellular functions. Furthermore, cold stress dramatically hinders membrane-bound enzymes, slows down diffusion rates and induces cluster formation of integral membranous proteins [5]. Micro-organisms counteract the propensity for membranes to rigidify at lower temperature by adapting to the conditions in order to maintain a more-or-less constant degree of membrane fluidity (homoeoviscous adaptation). In order to achieve this, cold stress initiates a co-ordinated response whereby fatty acid desaturase and dehydrases are induced which in turn increases the ratio of polyunsaturated to saturated fatty acids and/or (but less frequently) actually decreases the length of the fatty acid chains in the membrane [6–9]. However, not all investigators consider changes in fatty acid composition strictly as a cold-shock response, as a number of bacterial strains are able to sustain growth under such stress without any change in the fatty acid composition of the membrane [10].
It is well documented that temperature downshift generally suppresses protein synthesis in both eukaryotic and prokaryotic cells [2,4,10–17]. This suppression of protein synthesis is associated with growth arrest and the expression of CSPs required for hypothermal adaptation [18]. Jones et al. [19] reported that E. coli actively arrests growth when subjected to temperature stress at 10 °C for 1–4 h (acclimation period), followed by a resumption of exponential growth although at a lower rate compared with that observed at 37 °C. Interestingly, during the growth lag period protein synthesis was repressed except for the predominant synthesis of 13 CSPs [19].
The expression of specific CSPs upon temperature downshift is thought to ensure accurate and enhanced translation of specific mRNAs at low temperatures [20]. Although studies have shown that mRNA translation appears to be a key control point in the cold-shock response, the overall mechanism of cold-shock-mediated inhibition of translation is yet to be fully elucidated. However, one direct consequence of cold stress is the suppression of translation initiation by the normal cap-dependent mechanism. It has been reported that the 5′-UTR (untranslated region) of mRNAs tend to form stable secondary structures upon cold stress, thereby masking the Shine–Dalgarno sequence of mRNAs from the ribosome or interfering with translation elongation steps [3]. A further mechanism by which translation may be inhibited in bacteria upon cold-shock is through ribosome-associated proteins that interfere with translation elongation. RaiA (ribosomal-associated inhibitor A) is a recently characterized bacterial CSP that interferes with the elongation cycle of translation by binding and blocking the ribosomal A site [21,22]. Furthermore, the trans-lational machinery of E. coli undergoes selective modifications during cold shock to allow for selective translation of cold-shock mRNAs, as described in more detail below.
It should be noted that cold stress exposes cells to two major stresses: those relating to changes in temperature and those related to changes in dissolved oxygen concentration at decreased temperature [23], and it is therefore necessary to consider potential responses to each, either independently or as part of a co-ordinated response. Separating the relative effects of temperature and oxygen as a result of decreased temperature is difficult and has not been extensively addressed to date. What is documented is that both changes in dissolved oxygen and temperature reduction result in a change in the metabolic state of in vitro cultured mammalian cells [24,25]. It is also suggested that the cold-stress-transduction pathways share common components with hypoxic transduction pathways and not hyperoxic pathways [26]. This is substantiated further by a recent report that the mammalian cold-shock-inducible proteins Rbm3 (RNA-binding motif protein 3) and Cirp (cold-inducible RNA-binding protein) are also up-regulated in response to hypoxia in a Hif (hypoxia-inducible factor)-independent manner [27]. Interestingly, it has been demonstrated that both oxygen uptake and ATP consumption are significantly decreased in CHO (Chinese-hamster ovary) cells grown at 30 °C, presumably because of decreased metabolism and not because of a direct response to changes in dissolved oxygen at lower temperatures [28].
THE COLD-SHOCK RESPONSE IN E. COLI AS A MODEL PROKARYOTIC SYSTEM
The cold-shock response has now been investigated in a number of bacteria, although the responses of the mesophilic bacterium E. coli upon temperature downshift from 37 to 10 °C and those in the Gram-positive soil bacterium Bacillus subtilis are by far the most well characterized to date. Although this review focuses upon the cold-shock response in yeast and mammalian cells, for completeness, we present the key responses upon temperature downshift in E. coli. The cold-shock response(s) in E. coli upon temperature downshift has been reviewed in detail elsewhere (for example, see [4,15,16]).
Upon a shift in temperature from 37 to 10 °C, E. coli selectively expresses approx. 27 CSPs over a period of approx. 4 h, while at the same time down-regulating global transcription and translation [4]. A number of these CSPs are absolutely essential for survival, but none have an exclusive role in cold adaptation, suggesting a co-operative mechanism. Although a precise role or function for these cold-shock-induced proteins has yet to be elucidated, the bacterial CSPs are highly conserved, and all have been demonstrated to exhibit single-stranded-nucleic-acid-binding activity, as described in detail elsewhere [4,15,16]. After the adaptation period, global protein synthesis is restarted, while the selective biosynthesis of CSPs is down-regulated [4].
The major CSP expressed under these conditions is CspA, an RNA-binding protein that is only synthesized under cold-shock conditions [10,19]. CspA is similar to the cold-shock domain of the eukaryotic Y-box factors and to the major B. subtilis CSP, CspB, which is known to bind preferentially to single-stranded nucleic acids [29]. The synthesis of CspA is dramatically induced upon cold shock in E. coli and reaches ∼13% of total protein synthesis at 10 °C [30]. CspA binds specifically to single-stranded RNA at low temperatures and may enhance translation by preventing the formation of mRNA secondary structures [31].
The ‘natural’ promoter of CspA does not appear to be required for cold-shock induction of CspA, and the major control mechanisms of CspA expression appear to exist at the post-transcriptional level [4]. CspA mRNA half-life is extremely short at 37 °C, but is increased markedly upon cold shock [32]. The unusually long 5′-UTR of CspA plays an important role in stabilizing CspA mRNA and enhancing translation efficiency at low temperature [33]. The conclusion is that the 5′-UTR of CspA mRNA harbours essential sequences that are required for mRNA stability [4,11,34]. Furthermore, CspA mRNA stability is transient, thus, once E. coli cells are adapted to cold temperature, CspA mRNA becomes highly unstable [35].
Thus, upon cold shock, E. coli dramatically reduces global protein synthesis, but selectively up-regulates the synthesis of CSPs. The reason for this discrepancy is both the change in mRNA stability of specific transcripts and the preferential translation of cold-shock mRNAs upon temperature downshift. Furthermore, preferential translation is due to the co-ordinated effect of several mechanisms which together result in the increased synthesis of key CSPs. These co-ordinated mechanisms include: (i) modification of the actual translational machinery of E. coli upon cold shock to facilitate selective mRNA translation, (ii) the presence of cis-elements within cold-shock mRNAs that improve translation under cold-shock conditions, and (iii) trans-acting factors that target cold-shock mRNAs and are found in increased abundance as part of the translational machinery of cold-shocked cells. In addition, in E. coli, three initiation factors (IF1, IF2 and IF3) form part of the translation-initiation mechanism step of protein synthesis. Upon cold shock, IF3 is considered to be the most important in terms of the selective translation of cold-shock mRNAs, although IF1 and CspA are also thought to be involved in preferential translation upon cold-shock, but without exhibiting mRNA selectivity [11]. Recently, two promoter regions, P1 and P2, have been identified in the InfA gene that encodes IF1, and cold shock results in the activation of the P1 promoter alone, while the transcripts from both promoters are reportedly stabilized upon cold shock, and this results in an approx. 30-fold increase in InfA mRNA levels at 10 °C compared with those observed at 37 °C [36].
The E. coli stress protein, protein Y, has also been shown to inhibit translation initiation at sub-physiological temperatures (i.e. during cold shock), but not at normal growth temperatures [37]. Two putative cold-induced DEAD (Asp-Glu-Ala-Asp) box RNA helicases, CshA and CshB, are also thought to work in a co-ordinated fashion with CSPs to rescue misfolded mRNA and help ensure that correct translation initiation occurs at decreased temperatures in B. subtilis [38]. A further DEAD box RNA helicase described in E. coli, CsdA, appears to accumulate during cold adaptation, but thereafter be assembled into degradosomes with RNase E after adaptation, suggesting that these proteins are important during the acclimatization phase of the cold-shock response, but are not required thereafter [39].
Cold shock also directly results in the induction of trehalose synthesis in E. coli via the up-regulation of the OtsA and OtsB genes upon temperature downshift to 16 °C. The proteins that these genes encode are responsible for the biosynthesis of trehalose. The induction of these genes is dependent upon the varsigma factor RpoS, via a CspA-independent mechanism. Furthermore, the biosynthesis of trehalose upon temperature shift to 16 °C in E. coli was shown to be crucial in cell survival when the temperature was decreased further to 4 °C [40], and there are close parallels to trehalose induction in yeast systems as discussed in more detail below.
In addition to those genes and mechanisms discussed above, there are a number of other partially characterized genes/proteins and mechanisms in bacteria and other unicellular micro-organisms that have been investigated as part of the cold-shock response [14,15], although it is beyond the scope of this review to detail these here. Of particular note is the work on the cold-shock responses in B. subtilis which has been reviewed previously [41]. Weber et al. [42] have suggested a direct link between CSPs, transcription and translation in B. subtilis, with CSPs localizing to areas of newly synthesized mRNA in the cytosolic space surrounding nucleoids and coupling transcription to translation initiation. In addition to the up-regulation of the classic CSPs CspB, CspC and CspD, recent transcriptomic and proteomic investigations into the adaptation of B. subtilis to growth at 15 °C confirm there are two phases of adaptation [43]. The major responses include almost complete induction of the SigB controlled stress regulon and partial induction of the SigE, SigF and SigG regulons [43]. Finally, other investigations have reported the importance of the induction of the lipid-modifying desaturase Des is induced upon cold-shock, increasing membrane fluidity at lower temperature [44–47].
THE COLD-SHOCK RESPONSE IN EUKARYOTIC SYSTEMS
In direct contrast with the molecular responses characterized in prokaryotic cells, the response of eukaryotic cells and systems to cold shock, and the biological mechanisms that govern the cellular responses to sub-physiological temperatures, are not well understood [14]. For the last two decades, there has been a relatively large volume of literature published on the cold-shock response in plants; however, these responses will not be discussed here. By far, the majority of work on plants has concentrated on seed storage, decreased temperature acclimatization and freeze tolerance [14]. Perhaps one of the greatest constraints upon our understanding of the cold-shock response in eukaryotic systems is the lack of agreement on what constitutes ‘cold shock’ in the literature, and the differential regulation observed between so called mild hypothermia and more extreme hypothermic temperatures. For example, studies on the responses of in vitro cultured mammalian cells to temperature downshift have ranged from a shift of a few degrees (to 32–35 °C) to a shift to more severe conditions (between 4 and 10 °C). However, although our understanding of the cold-shock response in eukaryotes is limited, several studies have demonstrated that, as in prokaryotes, induced CSPs are key determinants in the adaptation to growth and survival at lower temperatures [14,48,49]. What is clear is that the cold-shock response in eukaryotic cells involves a co-ordinated series of responses involving modulation of the cell cycle, metabolism, transcription, translation and the cell cytoskeleton.
THE YEAST COLD-SHOCK RESPONSE
In recent years, the response of yeast cells to cold shock has received attention, with a number of studies investigating the responses in Saccharomyces cerevisiae through the use of DNA microarray technology. It should be noted that yeast cells are adapted to lower temperatures and generally exhibit a more moderate cold-shock response than that observed in bacterial systems upon a shift from 30 to 10 °C [50]. A number of cold-shock- or low-temperature-induced genes have now been identified in yeast, including Tip1 and Tir1 [12]. However, yeast cells appear to initiate quite different responses at temperature falls in the region of 10–18 °C (cold-shock response) compared with those observed at temperatures of 10 °C or less (near-freezing response), as highlighted in Table 1 [51]. This may reflect the fact that yeast can actively grow at 10–18 °C, whereas growth stops at lower temperatures. Further studies have investigated more extreme cold and the stress response of cryopreservation and the recovery thereof from such storage [52].
Table 1. Yeast cells initiate different responses at temperature decreases to 10–18 °C (cold-shock response) compared with those observed at temperatures of 10 °C or less (near-freezing response), as highlighted below.
| Response | |
|---|---|
| Cold-shock (10–18 °C) | Near-freezing (<10 °C) |
| Active growth | Growth arrest |
| Three phases of adaptation | Induction of Tps1 and Tps2 trehalose-synthesizing enzymes and dramatic increase in the levels of trehalose |
| Response consists of ‘functional waves’, or a co-operative manner, to the general reduction in transcription and translation efficiency | Transcriptional regulation of Tps1 and Tps2 genes dependent on Msn2 and Msn4 transcription factors which bind STREs and up-regulate transcription |
| Early/mid phase | |
| Induction of Tip1, Tir1, Tir2 and Nsr1 | Induction of NTH1 mRNA, but increase in protein levels only upon rewarming |
| Up-regulation of genes associated with transcription followed by up-regulation of translational machinery | Increase in CCT mRNA levels, but not protein levels until rewarming |
| De novo synthesis of ribosomes | Increase in Hsp12, Hsp42, Hsp104 and SSA4 mRNA levels |
| Late phase | |
| Induction of metabolic, protein folding and signal-transduction genes | |
| Hsp12 and Hsp26 up-regulated | |
| Late induction of trehalose-synthesizing enzymes |
Perhaps the most dramatic temperature downshift response in yeast is the induction of the trehalose-synthesizing enzymes Tps1 and Tps2 in the near-freezing response, and the resulting production of large amounts of the chemical chaperone trehalose [13,50,51]. As discussed above, the induction of trehalose synthesis upon temperature downshift has also been reported previously in E. coli [40]. This effect is only observed at temperatures of 10 °C and below, and hence has been termed the ‘near-freezing response’. In fact, the accumulation of trehalose upon this severe cold shock has now been shown to correspond directly to the cells’ resistance to freezing and is absolutely critical for the maintenance of cell viability at 0 °C; however, the exact mechanism by which trehalose protects the cell remains to be elucidated. Upon return to normal growth temperatures, trehalose levels are lowered rapidly, presumably as a result of degradation by the Nth1 enzyme whose gene expression is also marginally induced upon cold shock, but induced further upon a return to 30 °C [51].
The mechanism by which this increase in trehalose concentration is controlled in response to cold shock at temperatures of 10 °C or less has been elucidated recently [51]. In exponentially growing cells at 30 °C, the mRNA levels of Tps1 and Tps2 are barely detectable; however, upon temperature downshift to 10, 4 or 0 °C, the levels increase dramatically so that at 0 °C there is a 20-fold increase in the levels of Tps1 and Tps2 mRNA 15–20 h after temperature shift [51]. The reason for this dramatic increase appears to be both transcriptional regulation and increased mRNA stabilization as Tps1 and Tps2 mRNA is reported to be extremely stable at 0 °C, but extremely unstable at 30 °C, whereby the mRNA levels are returned to basal levels in less than 5 min [51]. The actual mechanism of Tps1 and Tps2 mRNA stabilization has not yet been elucidated. Transcriptional regulation and activation of the Tps1 and Tps2 genes at 0 °C is dependent on the Msn2,4 pathway, whereby Msn2 and Msn4 are transcription factors which bind STREs (stress-response elements) and up-regulate transcription [51]. STREs are reportedly present in the promoter regions of the Tps1 and Tps2 genes, further supporting this mechanism [51].
In addition to the trehalose response, the near-freezing response in yeast selectively induces a number of classic stress-response genes including Hsp12, Hsp42, Hsp104 and Ssa4, although many heat-shock genes were not induced [13,51]. These results contrast markedly with the cold-shock response in yeast and E. coli when the temperature is shifted to 10–20 °C, as most heat-shock genes are down-regulated under these conditions. Thus it appears that the co-ordinated induction of trehalose biosynthesis and specific stress proteins is required for the adaptation and survival of yeast cells at near-freezing temperatures. In addition, at 4 °C, genes that are involved in glycogen biosynthesis are reportedly induced along with increased levels of phospholipids, mannoproteins and CSPs, consistent with roles and changes to membrane maintenance and the permeability of the cell wall [13].
A number of approaches, including DNA microarray-based studies, have shown additional or different responses at temperatures above 10 °C compared with those initiated at near-freezing temperatures. The use of such approaches to investigate the response of yeast to decreased temperatures has shown that S. cerevisiae overexpresses a set of genes including Nsr1, Tip1, Tir1 and Tir2 under these higher-temperature cold-stress conditions [20]. The products of these genes are involved in pre-rRNA processing, ribosome biogenesis and maintenance of the cell wall respectively [53,54], suggesting that such functions and components are crucial in the cellular adaptation to, and recovery from, cold shock. Furthermore, several DNA microarray-based studies have now identified a number of additional genes whose expression is induced upon cold shock or temperature downshift [12,13,20,55,56].
Of particular interest is one such DNA microarray-based study that investigated the time-dependent response of S. cerevisiae upon cold shock at 10 °C. This investigation showed that, under these conditions, cold stress altered directly the transcription of approx. 25% of all S. cerevisiae genes that were investigated [20]. However, a large number of these changes can be attributed to the general reduction in both the transcription observed and the activity of fundamental metabolic pathways [55]. More interestingly, this study showed that S. cerevisiae responds to temperature stress at 10 °C in ‘sequential waves’, or a co-operative manner, to the general reduction in transcription and translation efficiency in contrast with that observed in the near-freezing response as outlined in Table 1. The responses can be delineated into three phases, whereby the early and mid phases are characterized by the initial up-regulation of a number of genes that are associated with the transcriptional machinery, which is then followed by an up-regulation of the translational machinery in the mid phase [20].
As described above, translational efficiency is compromised and decreased upon temperature downshift because of the formation of mRNA secondary structures and an increase in the proportion of inactivated ribosomes in yeast. At 10 °C, yeast cells would appear to, at least partially, compensate for this and allow translation to proceed at a rate sufficient to synthesize essential proteins by synthesizing ribosomes de novo. Concurrently, and in the third phase, genes that are involved in the cellular processes of metabolism, protein folding and signal transduction are also up-regulated [20]. This co-ordinated response involving the regulation of ‘sequential waves’ of genes encoding modules of cellular machinery that are involved in particular cellular processes has also been observed by others [12,55].
In contrast with the rapid increase in trehalose observed in the near-freezing response, trehalose biosynthesis appears to increase only in the late-phase response at 10 °C [20]. It is interesting to note that, in these studies, the majority of HSP genes were down-regulated and only two, Hsp12 and Hsp26 genes, were up-regulated in response to cold stress at 10 °C in direct contrast with the up-regulation of HSP genes observed in the near-freezing response [20]. Furthermore, the authors of this study suggest that the cAMP/PKA (protein kinase A) pathway plays a central role in the control of gene expression in yeast upon temperature down-shift to 10 °C [20]. It is suggested that, in the early to mid phase of the response, the cAMP/PKA pathway is involved in the upregulation of genes that encode translational machinery, while in the late-phase response, the pathway is involved in the down-regulation of general stress-response genes [20].
A more recent study has shown that, upon temperature down-shift to 12 or 4 °C, the Hog (high-osmolarity glycerol) pathway is activated in yeast via activation and phosphorylation of MAPK (mitogen-activated protein kinase) Hog1p. It is thought that Hog activation occurs due to changes in the fluidity of the membrane as a result of cold shock. In the absence of Hog1p, the induction of those genes required for the biosynthesis of trehalose and glycerol was not observed. Although both 12 and 4 °C resulted in the up-regulation of glycerol synthesis, deletion of the Hog1 gene had no effect on cells grown at 12 °C, but decreased tolerance to freezing [57]. Cold shock has also been shown to induce a protein-kinase-mediated response in the fission yeast Schizosaccharomyces pombe [58].
A number of additional genes have also been implicated in the cold-shock response in yeast, including Sec11, involved in protein transport [55], and Bfr2, involved in protein secretion [59]. In S. cerevisiae, the levels of mRNA of the essential gene Bfr2 have been shown to increase rapidly over 5-fold in response to cold shock [59]. The reason for, or mechanism of, this induction is not currently known, although the protein product of Bfr2, Bfr2p, involved in protein trafficking to the Golgi, is thought to be essential for mass growth and/or cell proliferation in S. cerevisiae [59]. Similarly, the mRNA levels of Cer1p, an Hsp70-related protein involved in the translocation of a subset of proteins into the endoplasmic reticulum in S. cerevisiae, increases upon cold shock, and it has been suggested that there is enhanced demand for the chaperone activity of Cer1p at lower temperatures [60].
Other key genes which are thought to be regulated in response to cold shock in yeast include Nsr1, involved in ribosome synthesis [53], and the genes encoding subunits of CCT (chaperonin containing the T-complex polypeptide-1) [12,61]. The role of CCT in cold shock is currently unknown; however, the client proteins of this chaperonin include tubulin and actin, suggesting a link between the cytoskeleton and the cold-shock response. Furthermore, CCT appears to be involved in the recovery from cold shock and the transition back to higher temperatures rather than in survival or adaptation to lower temperatures. Thus, upon temperature downshift to 4 °C, mRNA levels of CCTα have been shown to increase 3–4-fold, but an increase in the protein level was observed only upon a shift from 4 to 10 °C [61]. It therefore appears that transcriptional up-regulation at the near-freezing response ensures an increase in the levels of mRNA which are then presumably sequestered or stabilized until conditions are more favourable when this is converted into protein. CCT is also implicated in the progression of the cell cycle in addition to the folding of actin and tubulin [62–64], and it is tempting to speculate that the sequestered mRNA is preserved so that, upon an increase in temperature, CCT protein is immediately available to aid re-entry into the cell cycle.
Further studies have recently reported the up-regulation of a subunit of CCT in Delia antiqua (onion maggot) upon temperature reduction, the first such report outside of yeast [65]. This suggests that CCT may play a more conserved role in the cold-shock response across a range of organisms. However, as outlined below, the response of the CCT subunits to cold stress in mammalian cells remains to be elucidated. Finally, it should be noted that the majority of studies reported to date on the response of S. cerevisiae to cold shock have investigated changes at the mRNA level. Others have shown that there is often a poor correlation between mRNA and protein levels in yeast systems [66,67], and therefore the response at the protein level (the proteome) largely remains unknown.
THE COLD-SHOCK RESPONSE IN MAMMALIAN CELLS
As stated above, the response to heat shock has been investigated extensively in mammalian cells as well as prokaryotic organisms and plants, and involves a rapid induction of a family of highly conserved proteins known as HSPs [68]. The molecular mechanisms by which HSP induction is controlled, and the molecular and biochemical functions of HSPs are well illustrated in the literature [48]. On the other hand, little is known about the biological and molecular mechanisms that are involved in the control and induction of the cold-shock response in mammalian cells, despite its potential importance in organ and tissue storage, hiber-nation, and the fact that the testis and the skin are exposed to varying temperatures. Most of the cold-shock data published on mammalian systems to date has emerged from studies on adaptive thermogenesis, cold tolerance, the use of decreased temperature culture of mammalian cells for bioprocessing and the production of recombinant proteins, and from medical clinics investigating the effects of cooling on organ transplants. The use of cold-shock or decreased-temperature cultivation for the improvement of recombinant protein yields from in vitro cultured mammalian cells, and the mechanisms at work, have been reviewed recently [1].
Comparative studies of hibernating mammals and in vitro cultured cells have shown that animal cells generally respond to cold temperature by a profound reduction of metabolism, reduction in glucose and glutamine consumption, inhibition of ATP expenditure, a decrease in free radical oxygen species, inhibition of metabolic waste release, a reduction in protease activity, arrest of the cell cycle (mainly at G1 phase), increased viability, disassembly of the cell cytoskeleton, inhibition of translation (at the levels of both initiation and elongation), transcription attenuation, delayed apoptosis and increased resistance against shear stress [69]. Thus, as in prokaryotes, many of the observed responses upon cold shock are akin to those observed upon heat stress. At extreme low temperatures, mammalian cells die via necrosis owing to the formation of ice crystals and the resultant damage that this imparts upon membranes and organelles. Furthermore, as described for yeast systems above, cold shock results in changes to the lipid bilayer and composition of the membrane in mammalian cells [48]. Unsurprisingly, as in yeast, it appears that mammalian cells undergo different responses at moderate hypothermic temperatures (25–35 °C), whereby cells can proliferate and grow, as opposed to more severe temperature downshifts (0–10 °C), whereby growth is arrested [1]. However, it has been demonstrated that CHO cells may be ‘paused’ at temperatures between 6 and 24 °C for up to 3 weeks and still exhibit high viabilities and exponential growth upon return to more physiological temperatures [70].
Previous reports have suggested there are five mechanisms by which cold-shock-induced changes occur in gene expression in mammalian cells. The five proposed mechanisms are: (i) a general reduction in transcription and translation, (ii) inhibition of RNA degradation, (iii) increased transcription of specific target genes via elements in the promoter region of such genes, (iv) alternative pre-mRNA splicing, and (v) via the presence of cold-shock-specific IRESs (internal ribosome entry segments) in mRNAs that result in the preferential and enhanced translation of such mRNAs upon cold shock [48]. Furthermore, decreased temperature not only results in temperature stress, but also potentially in oxygen stress, which is also due to the fact that oxygen dissolves at higher concentrations as the temperature is decreased [23]. Thus one might expect to observe responses to changes in oxygen concentration or availability upon cold shock that have been characterized previously. Indeed, this is the case, and it is thought that the hypoxic signal transduction pathway and the cold-shock response share common components [26]. Further support for this lies in the fact that expression of the two major mammalian CSPs, Rbm3 and Cirp, which are discussed in detail below, is regulated in response to hypoxic conditions [27]. It should be noted that, upon temperature reduction, dissolved oxygen concentrations actually increase, suggesting that a hypoxic response may be invoked either upon rewarming, whereby dissolved oxygen levels would be expected to fall again, or as a result of intracellular changes in oxygen levels or reactive oxygen species.
As in prokaryotes, the cold-shock-mediated non-specific loss of protein-synthesis machinery has been reported in various mammalian cell lines [71]. However, it appears that the mechanism of protein synthesis inhibition upon cold shock is quite different in mammalian cells from those reported in prokaryotes. It has been suggested that cytoskeleton disassembly upon cold shock might account for the suppression of protein synthesis in mammalian cells [2]. Furthermore, much evidence now suggests that the translation apparatus in eukaryotic cells forms a permanently and spatially orchestrated structure [72]. Additionally, translation components such as polyribosomes, mRNAs and IFs co-localize with the cytoskeleton [73]. However, at mild-hypothermic temperatures, the cell cytoskeleton does not disassemble, and translation is most likely to be limited via the well-characterized de/phosphorylation of key initiation and elongation factors, a process which is known to limit global translation initiation under conditions of environmental stress [74,75]. Increased mRNA stability and the formation of stable secondary structures are also likely to contribute to compromised global translation efficiency at mild-hypothermic temperatures.
The findings discussed above suggest that the cytoskeleton may well function as an organizer of the protein-synthesis machinery [73]. Further support for this theory is presented in a study by Liu et al. [76] who demonstrated that EF1α (elongation factor 1α) acts as a linker between the mRNA translation machinery and the cytoskeleton. EF1α facilitates non-selective association of mRNA to the cytoskeleton by binding F-actin (filamentous actin) and mRNAs concurrently [76]. EF1α also binds and transports aminoacyl-tRNA to the A site of the ribosome in a GTP-dependent manner [77]. Moreover, several studies on intact and permeabilized CHO cell models have shown that cold shock can initiate microtubule disassembly. Microtubule disassembly is associated with alterations to the supramolecular organization of the translation machinery and reduction of F-actin owing to disruption of actin filaments [73,78]. Associated with this, cold shock results in changes to cellular membrane permeability, leading to a reduction in cellular pH. Consequently, cold shock can alter the interaction of EF1α with the actin cytoskeleton via pH control, and therefore affect translation directly [73].
Perturbations of the cytoskeleton due to cold shock may therefore cause disruption of the translational machinery and consequently inhibit protein biosynthesis at low temperatures. Such a link between the regulation of the cytoskeleton, actin assembly and translation efficiency upon stress conditions would not be unique to cold-shock conditions. Previous studies have shown that, under glucose-limiting conditions, yeast cells independently, yet simultaneously, regulate the organization of the actin cytoskeleton and translation initiation [79]. Furthermore, as discussed above, the role of CCT upon cold shock in mammalian cells has not yet been elucidated, and, in view of the possible role of this chaperonin (whose client proteins include actin and tubulin) in yeast cells upon temperature downshift, and the obvious importance of cell cytoskeleton integrity upon cold-shock, it would be surprising if CCT was not also involved in modulating this response. A recent paper has suggested that the eukaryotic chaperone machinery, which includes CCT, has two distinct functions in eukaryotic cells, including the presence of so-termed stress-repressed chaperones that are transcriptionally, functionally and physically linked to the translational apparatus [80]. The cytoskeleton may therefore provide a ‘work bench’ for a multitude of processes and control mechanisms, bringing the translational machinery and specific mRNAs together and allowing for the translation of specific mRNAs at local sites. Such functions may be of enhanced importance upon temperature reduction.
The induction of STOPs (stable-tubule-only polypeptides) that stabilize microtubules at decreased temperatures [81,82] also suggests that cell cytoskeleton components play an important role in the cold-shock response, or at least that the maintenance of the cytoskeleton is crucial. Although the disassembly of microtubules and actin filaments is well documented at sub-physiological temperatures [73], the microtubule network is stabilized upon cold-shock due to the association of STOPs with the network. The role of STOPs and their mode of action has been well reviewed elsewhere [82]. Others have reported that protein kinase CK2 also protects and stabilizes polymerized microtubules against cold-induced depolymerization [83]. In addition to the stabilization of microtubules, it has been suggested that, in the case of F-actin microfilaments, low temperature actually increases the formation of these microfilaments [84], and an intact F-actin system is absolutely required for efficient mammalian protein synthesis [73]. Regulation and maintenance of the cell cytoskeleton would therefore appear to be crucial in the cold-shock response in in vitro cultured mammalian cells at temperatures of moderate hypothermia, and may be closely linked to maintaining translational efficiency at such temperatures.
MAMMALIAN CSPs
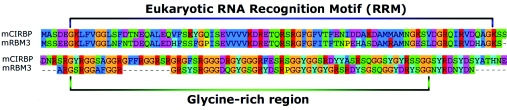
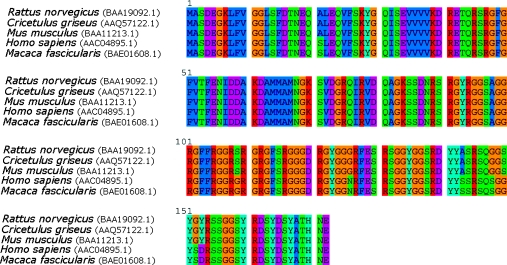
As in the case of prokaryotic and yeast systems, mammalian cells express several CSPs upon exposure to mild hypothermia. The only two well-characterized mammalian CSPs reported to date are Cirp and Rbm3. It is widely believed that the Cirp and Rbm3 proteins are involved in modulation of transcription and translation. Both proteins are highly similar and belong to the glycine-rich RNA-binding protein family (see Figure 1), although the exact function of these two proteins has yet to be elucidated [85]. Furthermore, the Cirp protein appears to be highly conserved across those organisms studied to date, as shown in Figure 2. Members of the glycine-rich RNA-binding protein family are characterized by a consensus sequence RNA-binding domain at the N-terminus and a glycine-rich domain at the C-terminus [86], as depicted in Figure 1. Moreover, both Cirp and Rbm3 are thought to be modulators of gene expression during mild hypothermic conditions by functioning as RNA chaperones that facilitate translation [2]. Thus, as observed in E. coli, cold shock appears to result in the selective stabilization of specific mRNAs that are required for survival and subsequently enhanced expression of CSPs. Furthermore, such proteins share the common activity of binding mRNAs, although the exact mechanism by which they do this, or the function of this activity, remains uncharacterized.
Figure 1. Predicted amino acid sequence alignment of the N- and C-terminal regions of the CSPs Cirp (CIRBP) and Rbm3 (RBM3) in mouse.
The two proteins are highly similar and belong to the glycine-rich RNA-binding protein family, which are characterized by a consensus sequence RNA-binding domain or eukaryotic RNA-recognition motif (RRM) at the N-terminus and a glycine-rich domain at the C-terminus.
Figure 2. Predicted amino acid sequence alignment of Cirp from various species.
The sequence data for each species were obtained from the NCBI (National Center for Biotechnology Information) protein database and the appropriate accession numbers are shown in parentheses. The predicted amino acid sequence across all species shown is virtually entirely conserved.
The Rbm3 gene resides on the Xp11.2 region of the human chromosome and was initially predicted to encode a 157-amino-acid protein (17 kDa) that closely resembles human RNA-binding proteins [87]. Mouse Rbm3 cDNA encodes an 18 kDa protein with 94% identity in amino acid sequence with that of human Rbm3. Danno et al. [88] have reported that the Rbm3 transcript is markedly increased in HepG2, NC65, HeLa, T24, K562 and TAMA26 cells when culture temperature is shifted from 37 to 32 °C [88]. Additionally, Northern blot analysis has shown that the tissue distribution of Rbm3 is limited to the pancreas, adrenal gland, placenta and testis, and is not expressed in the heart or thyroid [86,88]. The precise physiological and biochemical functions of Rbm3 are poorly understood; however, it has been reported that Rbm3 significantly suppressed polyglutamine-induced cell death in neuronal cells (SK-N-SH) by an unknown mechanism [89]. Furthermore, Rbm3 has been shown to be involved in cytokine-dependent proliferation. Interestingly, from a cold-shock perspective whereby oxygen levels may change, Rbm3 transcription is up-regulated in response to hypoxia. Cirp is also induced to a similar extent in response to hypoxia [27].
The regulatory elements within the UTRs of the Rbm3 transcript have been studied extensively. Chappell et al. [90] isolated a 1.4 kb Rbm3 cDNA clone from a mouse neonatal brain cDNA library. The 5′ leader sequence of this cDNA clone was long at 720 nt, and is significantly longer than the mouse and human sequences that had been reported previously [90]. Analysis of the Rbm3 5′ leader sequence revealed 12 upstream ORFs (open reading frames), none of which overlapped the Rbm3 main ORF. Furthermore, by cloning the 5′-UTR directly upstream of a reporter gene, it was shown that the Rbm3 5′-UTR mediates high levels of translation at 32 °C [90]. Several deletion and mutational analysis experiments on a putative IRES within the 720 bp 5′ leader sequence of the Rbm3 mRNA revealed at least nine discrete cis-acting sequences including a 22-nt IRES module, a 10-nt enhancer and two inhibitory sequences. Binding assays suggested that four cis-acting sequences probably bind specifically to different cytoplasmic proteins [91]. As a result of these findings, it is thought that the 5′-UTR of Rbm3 mRNA contains a number of specialized sequences that facilitate cap-independent translation to ensure mRNA translation upon cold shock, despite general (cap-dependent) mRNA translation being compromised [90,91].
As mammalian cells are able to grow and proliferate at mild-hypothermic temperatures, global translation must still be able to proceed at a rate sufficient to sustain growth and proliferation, albeit at a decreased rate. There is now evidence that the CSP Rbm3 itself is involved in the regulation of global protein synthesis by altering miRNA (microRNA) levels upon exposure of mammalian cells to conditions of mild-hypothermia [85]. miRNAs are short single-stranded RNA hairpins that interact with target mRNAs at specific sites and either induce cleavage of the target message or inhibit translation with the assistance of the multiprotein RISC (RNA-induced silencing complex) [92]. Dresios et al. [85] have suggested that Rbm3 may alter global translation upon cold shock by counteracting the effect of miRNAs under these conditions, thus removing an obstacle that might otherwise compromise mRNA translation further.
The CSP Cirp was the first identified CSP in mammalian cells. Far less is known about the molecular mechanisms of Cirp induction in comparison with Rbm3. What is evident is that the predicted amino acid sequence of Cirp is highly conserved across those organisms studied to date (Figure 2). It has been reported that Cirp is expressed ubiquitously in different tissues at 37 °C, and, unlike bacterial CspA, Cirp mRNA stability is not enhanced upon temperature reduction [93,94]. Moreover, Cirp transcription and translation undergoes marked induction in response to sub-physiological temperatures [2].
Mouse Cirp cDNA isolated from adult mouse testis encodes a 172-amino-acid 18 kDa protein [95]. Analysis of the predicted amino acid sequence of Cirp has determined that Cirp belongs to the glycine-rich RNA-binding protein family and is likely to be similar in structure to human Rbm3 (see Figure 1) and plant glycine-rich RNA-binding proteins such as Brassica napus BnGRP10 and Arabidopsis thaliana Ccr1 [96]. Subsequently, Cirp has been identified in rat, Mexican axolotl, Xenopus and bullfrogs [97–100]. The human Cirp gene is located at the chromosomal locus 19p13.3 and encodes an 18 kDa protein with 95.3% identity in an amino acid sequence with mouse Cirp [95]. Mouse Cirp is constitutively and weakly expressed in the testis, lung, heart and tests of the adult mouse at 37 °C [93,94]. Similarly, human Cirp mRNA is constitutively expressed in K562, HepG2, NC65, HeLa, T24 and NEC8 human cells [95]. However, Northern and Western blot analysis of lysates from mouse and human cells have shown that Cirp mRNA and protein levels are increased markedly in response to mild cold treatment (32–25 °C), but not to harsh cold treatment (15 °C and below). Cirp is also a nuclear protein, and is expressed ubiquitously in different tissues [97]. Nuclear localization was confirmed when GST (glutathione S-transferase)-tagged Cirp cDNA was transfected into COS-7 cells and a fluorescence signal was found only in the nucleoplasm [96].
Analysis of the binding activity of GST-tagged Cirp revealed that Cirp binds to all RNA homopolymers at low NaCl concentrations; however, at higher NaCl concentrations, Cirp shows higher binding affinity to polyadenylated RNAs only [96]. Consequently, Fujita [2] and others (reviewed in [4]) have proposed that Cirp may function as an RNA chaperone, preventing the formation of mRNA secondary structure at low temperatures. However, unlike the mRNA of the bacterial CSP CspA, Cirp mRNA stability is not affected by decreasing temperature. Cirp gene expression therefore appears to be controlled both transcriptionally by cold-responsive elements within its promoter [2] and possibly translationally by elements within the transcript. Additionally, it has been proposed that Cirp plays an essential role in hypothermia-induced suppression of cell proliferation. When the culture temperature of BALB/3T3 cells was shifted from 37 to 32 °C, growth was severally impaired. Concomitantly, growth inhibition at 32 °C was partially reversed when the induction of Cirp was inhibited by antisense oligonucleotides to Cirp mRNA. On the other hand, overexpression of Cirp in BALB/3T3 cells at 37 °C caused prolongation of doubling time and accumulation of cells at the G1 phase of the cell cycle [96]. Finally, beside the predicted role of Cirp as a molecular chaperone upon cold shock, emerging evidence strongly suggests that Cirp seems to play major roles in differentiation and during embryonic development [2].
The actual molecular mechanisms governing transcriptional regulation of mammalian cold-shock genes are currently unknown; however, many have speculated that cold-shock genes might be regulated at the transcription level by cis-regulatory elements within their promoters [48]. Previous reports have suggested that the transcription of the mammalian cold-shock-responsive genes may be regulated by specific upstream elements in response to a specific signal (i.e. mild hypothermia); however, if such elements do exist, they remain to be characterized [2]. A well-illustrated example of this kind of regulated transcription is the regulation of Hsp70. Deletion analysis of sequences upstream of the transcription start site of human Hsp70 have revealed a heat-shock element which is required to induce Hsp70 mRNA upon heat shock [101]. The induction of HSPs upon rewarming may actually play a crucial part in readjusting to warmer temperatures as transient cold shock appears to induce a heat-shock response once cells are returned to physiological temperatures [102].
ADDITIONAL RESPONSES TO COLD SHOCK IN MAMMALIAN CELLS
In view of the proposed role of Hog1p in yeast cells upon cold shock and recovery, it is interesting to note that its mammalian orthologue, p38, has also been implicated in the cold-shock response in mammalian cell systems. Phosphorylation of p38 MAPK upon temperature reduction did not actually appear to play a role in the cold-shock response itself, but rather seems to be involved in the response upon recovery from low temperatures, inducing the up-regulation of several key proteins [26]. Phosphorylation of JNK (c-Jun N-terminal kinase) is also reportedly increased upon cold shock and then increased further by rewarming [23].
A limited number of transcriptomic and proteomic investigations into mild hypothermia cold-shock responses in mammalian cells have been published recently. One of the few transcriptomic investigations to date into the effect of cold shock on in vitro cultured mammalian cells reported that, in 3T3 cells, more than 10% of those genes investigated (1176 cellular genes) showed changes in expression levels upon temperature shift from 37 to 32 °C. Of particular note, a number of genes from the cell cytoskeleton family were observed to change [103]. Several other transcriptome and proteome studies into the response of CHO cells to cold shock have also established that CHO cells respond actively to sub-physiological temperatures (33 and 30 °C) by overexpressing a defined set of CSPs [104,105]; however, once again, the significance of such findings remains to be elucidated. Both transcriptional and translational control have also been investigated upon mammalian hibernation, whereby body temperature falls to 0–5 °C. Such studies have once again confirmed a general reduction and suppression of global translation, with selective translation of specific mRNAs and the induction of hypoxia-related genes [106] in agreement with those cold-shock-induced changes observed in model in vitro cultured mammalian cell systems.
Numerous studies have now shown that heterologous protein expression from mammalian cells may be increased at sub-physiological temperatures [103–105,107–117]. However, this effect is variable and appears to be temperature-, recombinant-protein- and cell-line-dependent [1]. Despite this, a number of investigations have reported that increases in mRNA stability and concentration at lower temperature account for any observed increase in heterologous protein production [114]. Thus, despite the likely formation of potentially translational-limiting mRNA secondary structures at lower temperatures, increased stability of mRNAs combined with a reduction in global translation allows for the increased synthesis of those mRNAs present in relatively large concentrations.
Finally, apoptosis (programmed cell death) is a genetically regulated process, which is essential for appropriate development, differentiation, tissue homoeostasis and defence against pathogens [118]. Apoptosis is induced by an array of different signals, including radiation, growth factor withdrawal, and cytotoxic agents like chemotherapeutic drugs [119]. Previous studies have shown that when the cultivation temperature of CHO cells is shifted from 37 to 30 °C, up to 87% of cells arrest growth at G1 and the onset of apoptosis is delayed significantly compared with cells grown at 37 °C. One could therefore argue that the delay in apoptosis upon cold shock is to some extent due to the reduction of cellular metabolism, although other complicated defensive mechanisms cannot be ruled out [28]. Zhang et al. [120] have reported that the expression of the anti-apoptotic protein Bcl-2 is enhanced in response to mild hypothermia. It should be noted, however, that the hypothermia-associated anti-apoptotic response varies from one cell line to another. For instance, the rat histiocytic cell line, BC-8 (originated from AK-5 tumour after single cell cloning), triggered apoptosis when cultivated at 30 °C and overexpression of exogenous recombinant Bcl-2 rescued cell death [121].
A MODEL FOR THE COLD-SHOCK RESPONSE IN MAMMALIAN CELLS UPON EXPOSURE TO MILD HYPOTHERMIA
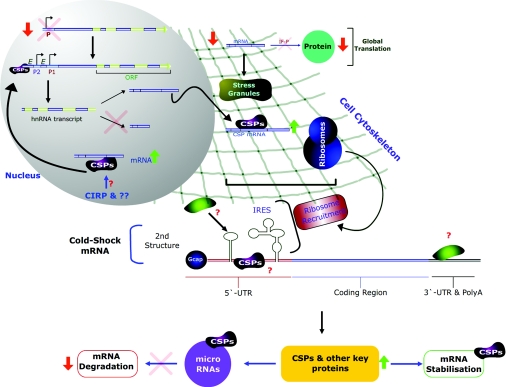
In considering all the available data, we propose a simple model for the major co-ordinated mechanisms and responses to mild hypothermic cold shock (25–35 °C) in mammalian cells. In this model, CSPs and the cell cytoskeleton play a central role, particularly at the translational level (Figure 3). Upon cold-shock perception, global transcription efficiency is decreased initially; however, both CSPs and other as-yet-unidentified proteins can bind to specific cis-elements in the promoter regions of target genes, up-regulating their transcription and leading to increased transcript levels. At the same time, alternative promoters may be activated/inactivated at decreased temperature, leading to alternative hnRNA (heteronuclear RNA) transcripts at lower temperatures. Either at the same time, or as a substitute for alternative promoters, we propose that alternative splicing of hnRNAs also results in the generation of cold-specific mRNAs which may have unique 5′- and/or 3′-UTRs and/or changes in the coding regions. Cold-specific mRNAs may then bind CSPs in the nucleus before transport to the cytoplasm, stabilizing the mRNA, or be transported directly to the cytoplasm without CSP chaperone interaction. Such alternative splicing has already been reported for heat-shock transcription factor 1 in Drosophila upon cold stress [122].
Figure 3. Proposed model for the co-ordinated cellular responses in mammalian cells upon exposure to mild hypothermia cold-shock (25–35 °C).
In the model, cold shock results in global transcription efficiency being decreased (designated by ↓ and X, top of nuclear compartment); however, both CSPs and other unidentified proteins bind to cis-elements (designated E) of target genes, up-regulating their transcription and resulting in increased hnRNA levels. Alternative promoters (designated P1 and P2) may also be activated in target genes at decreased temperature, leading to different hnRNA transcripts at lower temperatures. Alternative splicing (middle of nuclear compartment) also results in cold-specific mRNAs which may have unique 5′- and/or 3′-UTRs and/or changes in the coding regions. Cold-specific mRNAs may then bind CSPs in the nucleus before transport to the cytoplasm, stabilizing the mRNA, or be transported directly to the cytoplasm without CSP chaperones. In the cytoplasm, global mRNA translation is decreased (↓) via increased phosphorylation of IFs (IF-P) and the binding of specific, but currently unknown, proteins to ribosomes, preventing mRNA translation. Phosphorylation of eIF2α also results in the formation of mRNA stress granules, consisting of mRNAs that are required for recovery from cold shock upon rewarming. Central to translational control in this model is the interaction between CSPs, cold-specific mRNAs and the cell cytoskeleton. In the model, CSPs can ‘grab’ mRNAs as they emerge from the nucleus, or in the cytoplasm, and via interactions with the cell cytoskeleton (depicted as a ‘net’) form a ‘work bench’ for translation at local sites whereby ribosomes may be recruited. In this way, CSPs link transcription and translation via interactions with target mRNAs and the cytoskeleton. Furthermore, we predict that some cold-specific mRNAs, whose translation is required for cold-shock adaptation, contain IRES sequences, allowing the direct recruitment of ribosomes to the mRNA, circumventing the compromised cap-dependent initiation under cold-shock conditions. CSPs may also bind and stabilize their own or other mRNAs while undefined proteins (coloured green) also bind the 3′-UTR, all of which play a role in stabilizing cold-specific mRNAs at decreased temperatures. We also predict that trans-acting proteins bind secondary-structure elements in cold-shock mRNAs, stabilizing further such mRNAs and/or allowing more efficient scanning of the mRNA by ribosomes. All of these mRNA translation control mechanisms lead to an increase in the levels of CSPs and other key proteins required during cold-shock adaptation. The resulting newly synthesized CSPs (e.g. Cirp) can bind further mRNAs, stabilizing them, or be transported to the nucleus whereby they may act as transcription factors. Alternatively, CSPs may interact with specific miRNAs, preventing miRNAs from interacting with their target mRNAs and degrading them. The net effect is a decrease in mRNA degradation (↓) and thus an increase in mRNA levels. For further details see text.
In the cytoplasm, global mRNA translation is decreased via increased phosphorylation of IFs and the binding of specific, but currently unknown, proteins to mRNAs and ribosomes, preventing mRNA translation (Figure 3). In the model, the phosphorylation of eIF2α (eukaryotic initiation factor 2α) not only stimulates IRES-mediated translation as reported previously [123], but also triggers the formation of mRNA stress granules, sequestrating key mRNAs that are required for recovery from cold shock or upon adaptation to cold shock. Stress granules consist of accumulated untranslated mRNAs, they are found in the cytoplasm, and their formation is linked directly to eIF2α phosphorylation [124]. Furthermore, the assembly/disassembly of stress granules is thought to be involved in the control and regulation of the recovery of cells from environmental stresses [125], and we suggest that this is important upon recovery from mild-hypothermic temperatures.
Central to translational control under mild-hypothermia in this model is the interaction between CSPs, cold-specific mRNAs and the cell cytoskeleton (Figure 3). In the model, we propose that CSPs can ‘grab’ mRNAs as they emerge from the nucleus (or possibly in specific cases in the nucleus), and via interactions with the cell cytoskeleton form a ‘work bench’ for translation at local sites whereby ribosomes may be recruited and mRNA translation initiated. Furthermore, we predict that many cold-specific mRNAs, whose translation is required for cold-shock adaptation, contain IRES sequences allowing the direct recruitment of ribosomes to the mRNA, circumventing the compromised cap-dependent initiation under cold-shock conditions. CSPs may also bind and stabilize their own or other mRNAs, while as-yet-unidentified proteins may also bind the 5′/3′-UTR, all of which play a role in stabilizing cold-specific mRNAs at decreased temperatures. We also predict that trans-acting proteins bind secondary structure cis-elements in the 5′-UTR of cold-shock mRNAs. In the model, this stabilizes such mRNAs further and/or allows more efficient scanning of the mRNA by ribosomes, possibly by changing the structure of the mRNA or by ‘melting’ the RNA and allowing scanning to proceed. This is analogous to rhinovirus IRES-mediated RNA translation which has been shown to be dependent upon the presence of trans-acting factors upstream of N-ras (unr). Unr is predicted to act as an RNA chaperone, interacting with the RNA and allowing the IRES to obtain the correct conformation for ribosome binding [126]. All of these mRNA translation control mechanisms ultimately lead to an increase in the levels of CSPs and other key proteins that are required during cold-shock adaptation or more efficient mRNA translation at lower temperatures. The resulting CSPs may then bind further mRNAs, stabilizing them, or be transported to the nucleus whereby they may act as transcription factors. We also propose that particular CSPs interact with specific miRNA targets, preventing miRNAs from interacting with their target mRNAs and degrading them. The net effect is a decrease in mRNA degradation and thus an increase in mRNA levels.
Conclusions
It remains to be seen whether this rather simple model accurately reflects the major controls and the complex nature of the cold-shock response in mammalian cells. Regardless, advancements in our fundamental understanding of the molecular responses and mechanisms at play during cold-shock adaptation, survival and upon rewarming are likely to continue to be important in the areas of cold adaptation, tissue and organ storage, bioprocessing, therapeutic treatment of brain damage and our general understanding of stress responses. Interest in the cold-shock response is therefore really heating up.
Acknowledgments
We thank Dr Martin Carden, Department of Biosciences, University of Kent, for useful discussions during the preparation of this manuscript.
References
- 1.Al-Fageeh M. B., Marchant R. J., Carden M. J., Smales C. M. The cold-shock response in cultured mammalian cells: harnessing the response for the improvement of recombinant protein production. Biotechnol. Bioeng. 2006;93:829–835. doi: 10.1002/bit.20789. [DOI] [PubMed] [Google Scholar]
- 2.Fujita J. Cold shock response in mammalian cells. J. Mol. Microbiol. Biotechnol. 1999;1:243–255. [PubMed] [Google Scholar]
- 3.Ermolenko D. N., Makhatadze G. I. Bacterial cold-shock proteins. Cell. Mol. Life Sci. 2002;59:1902–1913. doi: 10.1007/PL00012513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gualerzi C. O., Giuliodori A. M., Pon C. L. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 2003;331:527–539. doi: 10.1016/s0022-2836(03)00732-0. [DOI] [PubMed] [Google Scholar]
- 5.Hazel J. R. Thermal adaptation in biological membranes: is homeoviscous adaptation the explanation? Annu. Rev. Physiol. 1995;57:19–42. doi: 10.1146/annurev.ph.57.030195.000315. [DOI] [PubMed] [Google Scholar]
- 6.Avery S. V., Lloyd D., Harwood J. L. Temperature-dependent changes in plasma-membrane lipid order and the phagocytotic activity of the amoeba Acanthamoeba castellanii are closely correlated. Biochem. J. 1995;312:811–816. doi: 10.1042/bj3120811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carty S. M., Sreekumar K. R., Raetz C. R. Effect of cold shock on lipid A biosynthesis in Escherichia coli: induction at 12 °C of an acyltransferase specific for palmitoleoyl-acyl carrier protein. J. Biol. Chem. 1999;274:9677–9685. doi: 10.1074/jbc.274.14.9677. [DOI] [PubMed] [Google Scholar]
- 8.Cossins A. R., Macdonald A. G. The adaptation of biological membranes to temperature and pressure: fish from the deep and cold. J. Bioenerg. Biomembr. 1989;21:115–135. doi: 10.1007/BF00762215. [DOI] [PubMed] [Google Scholar]
- 9.Dickens B. F., Thompson G. A., Jr Rapid membrane response during low-temperature acclimation: correlation of early changes in the physical properties and lipid composition of Tetrahymena microsomal membranes. Biochim. Biophys. Acta. 1981;644:211–218. doi: 10.1016/0005-2736(81)90377-1. [DOI] [PubMed] [Google Scholar]
- 10.Golovlev E. L. Bacterial cold shock response at the level of DNA transcription, translation and chromosome dynamics. Mikrobiologiia. 2003;72:5–13. [PubMed] [Google Scholar]
- 11.Giuliodori A. M., Brandi A., Gualerzi C. O., Pon C. L. Preferential translation of cold-shock mRNAs during cold adaptation. RNA. 2004;10:265–276. doi: 10.1261/rna.5164904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homma T., Iwahashi H., Komatsu Y. Yeast gene expression during growth at low temperature. Cryobiology. 2003;46:230–237. doi: 10.1016/s0011-2240(03)00028-2. [DOI] [PubMed] [Google Scholar]
- 13.Murata Y., Homma T., Kitagawa E., Momose Y., Sato M. S., Odani M., Shimizu H., Hasegawa-Mizusawa M., Matsumoto R., Mizukami S., et al. Genome-wide expression analysis of yeast response during exposure to 4 °C. Extremophiles. 2006;10:117–128. doi: 10.1007/s00792-005-0480-1. [DOI] [PubMed] [Google Scholar]
- 14.Phadtare S., Alsina J., Inouye M. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 1999;2:175–180. doi: 10.1016/S1369-5274(99)80031-9. [DOI] [PubMed] [Google Scholar]
- 15.Phadtare S. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 2004;6:125–136. [PubMed] [Google Scholar]
- 16.Weber M. H., Marahiel M. A. Bacterial cold shock responses. Sci. Prog. 2003;86:9–75. doi: 10.3184/003685003783238707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulusu N. N., Tezcan E. F. Cold shock proteins. Turk. J. Med. Sci. 2001;31:283–290. [Google Scholar]
- 18.Rieder C. L., Cole R. W. Cold-shock and the mammalian cell cycle. Cell Cycle. 2002;1:169–175. [PubMed] [Google Scholar]
- 19.Jones P. G., VanBogelen R. A., Neidhardt F. C. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 1987;169:2092–2095. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahara T., Goda T., Ohgiya S. Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. J. Biol. Chem. 2002;277:50015–50021. doi: 10.1074/jbc.M209258200. [DOI] [PubMed] [Google Scholar]
- 21.Agafonov D. E., Kolb V. A., Nazimov I. V., Spirin A. S. A protein residing at the subunit interface of the bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12345–12349. doi: 10.1073/pnas.96.22.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agafonov D. E., Kolb V. A., Spirin A. S. Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep. 2001;2:399–402. doi: 10.1093/embo-reports/kve091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohsaka Y., Ohgiya S., Hoshino T., Ishizaki K. Phosphorylation of c-Jun N-terminal kinase in human hepatoblastoma cells is transiently increased by cold exposure and further enhanced by subsequent warm incubation of the cells. Cell. Physiol. Biochem. 2002;12:111–118. doi: 10.1159/000063787. [DOI] [PubMed] [Google Scholar]
- 24.Chuppa S., Tsai Y. S., Yoon S., Shackleford S., Rozales C., Bhat R., Tsay G., Matanguihan C., Konstantinov K., Naveh D. Fermenter temperature as a pool for control of high-density perfusion cultures of mammalian cells. Biotechnol. Bioeng. 1997;55:328–338. doi: 10.1002/(SICI)1097-0290(19970720)55:2<328::AID-BIT10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 25.Jan D. C. H., Petch D. A., Huzel N., Butler M. The effect of dissolved oxygen on the metabolic profile of a murine hybridoma grown in serum-free medium in continuous culture. Biotechnol. Bioeng. 1997;54:153–164. doi: 10.1002/(SICI)1097-0290(19970420)54:2<153::AID-BIT7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 26.Gon Y., Hashimoto S., Matsumoto K., Nakayama T., Takeshita I., Horie T. Cooling and rewarming-induced IL-8 expression in human bronchial epithelial cells through p38 MAP kinase-dependent pathway. Biochem. Biophys. Res. Commun. 1998;249:156–160. doi: 10.1006/bbrc.1998.9115. [DOI] [PubMed] [Google Scholar]
- 27.Wellmann S., Buhrer C., Moderegger E., Zelmer A., Kirschner R., Koehne P., Fujita J., Seeger K. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J. Cell Sci. 2004;117:1785–1794. doi: 10.1242/jcs.01026. [DOI] [PubMed] [Google Scholar]
- 28.Moore A., Mercer J., Dutina G., Donahue C. J., Bauer K. D., Mather J. P., Etcheverry T., Ryll T. Effects of temperature shift on cell cycle, apoptosis and nucleotide pools in CHO cell batch cultures. Cytotechnology. 1997;23:47–54. doi: 10.1023/A:1007919921991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schindelin H., Jiang W., Inouye M., Heinemann U. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1994;91:5119–5123. doi: 10.1073/pnas.91.11.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein J., Pollitt N. S., Inouye M. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang W., Hou Y., Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 32.Brandi A., Pietroni P., Gualerzi C. O., Pon C. L. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol. Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]
- 33.Mitta M., Fang L., Inouye M. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol. Microbiol. 1997;26:321–335. doi: 10.1046/j.1365-2958.1997.5771943.x. [DOI] [PubMed] [Google Scholar]
- 34.Fang L., Jiang W., Bae W., Inouye M. Promoter-independent cold-shock induction of cspA and its derepression at 37 °C by mRNA stabilization. Mol. Microbiol. 1997;23:355–364. doi: 10.1046/j.1365-2958.1997.2351592.x. [DOI] [PubMed] [Google Scholar]
- 35.Goldenberg D., Azar I., Oppenheim A. B. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol. Microbiol. 1996;19:241–248. doi: 10.1046/j.1365-2958.1996.363898.x. [DOI] [PubMed] [Google Scholar]
- 36.Ko J. H., Lee S. J., Cho B., Lee Y. Differential promoter usage of infA in response to cold shock in Escherichia coli. FEBS Lett. 2005;580:539–544. doi: 10.1016/j.febslet.2005.12.066. [DOI] [PubMed] [Google Scholar]
- 37.Vila-Sanjurjo A., Schuwirth B. S., Hau C. W., Cate J. H. Structural basis for the control of translation initiation during stress. Nat. Struct. Mol. Biol. 2004;11:1054–1059. doi: 10.1038/nsmb850. [DOI] [PubMed] [Google Scholar]
- 38.Hunger K., Beckering C. L., Wiegeshoff F., Graumann P. L., Marahiel M. A. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J. Bacteriol. 2006;188:240–248. doi: 10.1128/JB.188.1.240-248.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prud'homme-Genereux A., Beran R. K., Iost I., Ramey C. S., Mackie G. A., Simons R. W. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome’. Mol. Microbiol. 2004;54:1409–1421. doi: 10.1111/j.1365-2958.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- 40.Kandror O., DeLeon A., Goldberg A. L. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9727–9732. doi: 10.1073/pnas.142314099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber M. H., Marahiel M. A. Coping with the cold: the cold shock response in the Gram-positive soil bacterium Bacillus subtilis. Philos. Trans. R. Soc. London Ser. B. 2002;357:895–907. doi: 10.1098/rstb.2002.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber M. H., Volkov A. V., Fricke I., Marahiel M. A., Graumann P. L. Localization of cold shock proteins to cytosolic spaces surrounding nucleoids in Bacillus subtilis depends on active transcription. J. Bacteriol. 2001;183:6435–6443. doi: 10.1128/JB.183.21.6435-6443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Budde I., Steil L., Scharf C., Volker U., Bremer E. Adaptation of Bacillus subtilis to growth at low temperature: a combined transcriptomic and proteomic appraisal. Microbiology. 2006;152:831–853. doi: 10.1099/mic.0.28530-0. [DOI] [PubMed] [Google Scholar]
- 44.Kaan T., Homuth G., Mader U., Bandow J., Schweder T. Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response. Microbiology. 2002;148:3441–3455. doi: 10.1099/00221287-148-11-3441. [DOI] [PubMed] [Google Scholar]
- 45.Mansilla M. C., de Mendoza D. The Bacillus subtilis desaturase: a model to understand phospholipid modification and temperature sensing. Arch. Microbiol. 2005;183:229–235. doi: 10.1007/s00203-005-0759-8. [DOI] [PubMed] [Google Scholar]
- 46.Cybulski L. E., del Solar G., Craig P. O., Espinosa M., de Mendoza D. Bacillus subtilis DesR functions as a phosphorylation-activated switch to control membrane lipid fluidity. J. Biol. Chem. 2004;279:39340–39347. doi: 10.1074/jbc.M405150200. [DOI] [PubMed] [Google Scholar]
- 47.Beckering C. L., Steil L., Weber M. H., Volker U., Marahiel M. A. Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis. J. Bacteriol. 2002;184:6395–6402. doi: 10.1128/JB.184.22.6395-6402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sonna L. A., Fujita J., Gaffin S. L., Lilly C. M. Effects of heat and cold stress on mammalian gene expression. J. Appl. Physiol. 2002;92:1725–1742. doi: 10.1152/japplphysiol.01143.2001. [DOI] [PubMed] [Google Scholar]
- 49.Fuller B. J. Gene expression in response to low temperatures in mammalian cells: a review of current ideas. CryoLetters. 2003;24:95–102. [PubMed] [Google Scholar]
- 50.Inouye M., Phadtare S. Cold shock response and adaptation at near-freezing temperature in microorganisms. Science STKE 2004. 2004:PE26. doi: 10.1126/stke.2372004pe26. [DOI] [PubMed] [Google Scholar]
- 51.Kandror O., Bretschneider N., Kreydin E., Cavalieri D., Goldberg A. L. Yeast adapt to near-freezing temperatures by STRE/Msn2,4-dependent induction of trehalose synthesis and certain molecular chaperones. Mol. Cell. 2004;13:771–781. doi: 10.1016/s1097-2765(04)00148-0. [DOI] [PubMed] [Google Scholar]
- 52.Odani M., Komatsu Y., Oka S., Iwahashi H. Screening of genes that respond to cryopreservation stress using yeast DNA microarray. Cryobiology. 2003;47:155–164. doi: 10.1016/j.cryobiol.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 53.Kondo K., Kowalski L. R., Inouye M. Cold shock induction of yeast NSR1 protein and its role in pre-rRNA processing. J. Biol. Chem. 1992;267:16259–16265. [PubMed] [Google Scholar]
- 54.Schade B., Jansen G., Whiteway M., Entian K. D., Thomas D. Y. Cold adaptation in budding yeast. Mol. Biol. Cell. 2004;15:5492–5502. doi: 10.1091/mbc.E04-03-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez-Vargas S., Estruch F., Randez-Gil F. Gene expression analysis of cold and freeze stress in Baker's yeast. Appl. Environ. Microbiol. 2002;68:3024–3030. doi: 10.1128/AEM.68.6.3024-3030.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L., Ohta A., Horiuchi H., Takagi M., Imai R. Multiple mechanisms regulate expression of low temperature responsive (LOT) genes in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2001;283:531–535. doi: 10.1006/bbrc.2001.4776. [DOI] [PubMed] [Google Scholar]
- 57.Panadero J., Pallotti C., Rodriguez-Vargas S., Randez-Gil F., Prieto J. A. A downshift in temperature activates the HOG pathway, which determines freeze tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:4638–4645. doi: 10.1074/jbc.M512736200. [DOI] [PubMed] [Google Scholar]
- 58.Soto T., Beltran F. F., Paredes V., Madrid M., Millar J. B., Vicente-Soler J., Cansado J., Gacto M. Cold induces stress-activated protein kinase-mediated response in the fission yeast Schizosaccharomyces pombe. Eur. J. Biochem. 2002;269:5056–5065. doi: 10.1046/j.1432-1033.2002.03214.x. [DOI] [PubMed] [Google Scholar]
- 59.Chabane S., Kepes F. Expression of the yeast BFR2 gene is regulated at the transcriptional level and through degradation of its product. Mol. Gen. Genet. 1998;258:215–221. doi: 10.1007/pl00008624. [DOI] [PubMed] [Google Scholar]
- 60.Hamilton T. G., Norris T. B., Tsuruda P. R., Flynn G. C. Cer1p functions as a molecular chaperone in the endoplasmic reticulum of Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:5298–5307. doi: 10.1128/mcb.19.8.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Somer L., Shmulman O., Dror T., Hashmueli S., Kashi Y. The eukaryote chaperonin CCT is a cold shock protein in Saccharomyces cerevisiae. Cell Stress Chaperones. 2002;7:47–54. doi: 10.1379/1466-1268(2002)007<0047:teccia>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu X., Lin C. Y., Lei M., Yan S., Zhou T., Erikson R. L. CCT chaperonin complex is required for the biogenesis of functional Plk1. Mol. Cell. Biol. 2005;25:4993–5010. doi: 10.1128/MCB.25.12.4993-5010.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camasses A., Bogdanova A., Shevchenko A., Zachariae W. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol. Cell. 2003;12:87–100. doi: 10.1016/s1097-2765(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 64.Yokota S. I., Yanagi H., Yura T., Kubota H. Upregulation of cytosolic chaperonin CCT subunits during recovery from chemical stress that causes accumulation of unfolded proteins. Eur. J. Biochem. 2000;267:1658–1664. doi: 10.1046/j.1432-1327.2000.01157.x. [DOI] [PubMed] [Google Scholar]
- 65.Kayukawa T., Chen B., Miyazaki S., Itoyama K., Shinoda T., Ishikawa Y. Expression of mRNA for the t-complex polypeptide-1, a subunit of chaperonin CCT, is upregulated in association with increased cold hardiness in Delia antiqua. Cell Stress Chaperones. 2005;10:204–210. doi: 10.1379/CSC-106R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Griffin T. J., Gygi S. P., Ideker T., Rist B., Eng J., Hood L., Aebersold R. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol. Cell. Proteomics. 2002;1:323–333. doi: 10.1074/mcp.m200001-mcp200. [DOI] [PubMed] [Google Scholar]
- 67.Gygi S. P., Rochon Y., Franza B. R., Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sieck G. C. Molecular biology of thermoregulation. J. Appl. Physiol. 2002;92:1365–1366. doi: 10.1152/japplphysiol.00003.2002. [DOI] [PubMed] [Google Scholar]
- 69.Van Breukelen F., Martin S. L. Molecular adaptations in mammalian hibernators: unique adaptations or generalized responses? J. Appl. Physiol. 2002;92:2640–2647. doi: 10.1152/japplphysiol.01007.2001. [DOI] [PubMed] [Google Scholar]
- 70.Hunt L., Hacker D. L., Grosjean F., De Jesus M., Uebersax L., Jordan M., Wurm F. M. Low-temperature pausing of cultivated mammalian cells. Biotechnol. Bioeng. 2005;89:157–163. doi: 10.1002/bit.20320. [DOI] [PubMed] [Google Scholar]
- 71.Burdon R. H. Temperature and animal cell protein synthesis. Symp. Soc. Exp. Biol. 1987;41:113–133. [PubMed] [Google Scholar]
- 72.Barbarese E., Koppel D. E., Deutscher M. P., Smith C. L., Ainger K., Morgan F., Carson J. H. Protein translation components are colocalized in granules in oligodendrocytes. J. Cell Sci. 1995;108:2781–2790. doi: 10.1242/jcs.108.8.2781. [DOI] [PubMed] [Google Scholar]
- 73.Stapulionis R., Kolli S., Deutscher M. P. Efficient mammalian protein synthesis requires an intact F-actin system. J. Biol. Chem. 1997;272:24980–24986. doi: 10.1074/jbc.272.40.24980. [DOI] [PubMed] [Google Scholar]
- 74.Wek R. C., Jiang H. Y., Anthony T. G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 75.Underhill M. F., Birch J. R., Smales C. M., Naylor L. H. eIF2α phosphorylation, stress perception, and the shutdown of global protein synthesis in cultured CHO cells. Biotechnol. Bioeng. 2005;89:805–814. doi: 10.1002/bit.20403. [DOI] [PubMed] [Google Scholar]
- 76.Liu G., Grant W. M., Persky D., Latham V. M., Jr, Singer R. H., Condeelis J. Interactions of elongation factor 1α with F-actin and beta-actin mRNA: implications for anchoring mRNA in cell protrusions. Mol. Biol. Cell. 2002;13:579–592. doi: 10.1091/mbc.01-03-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Day D. A., Tuite M. F. Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. J. Endocrinol. 1998;157:361–371. doi: 10.1677/joe.0.1570361. [DOI] [PubMed] [Google Scholar]
- 78.Weisenberg R. C. Microtubule formation in vitro in solutions containing low calcium concentrations. Science. 1972;177:1104–1105. doi: 10.1126/science.177.4054.1104. [DOI] [PubMed] [Google Scholar]
- 79.Uesono Y., Ashe M. P., Toh E. A. Simultaneous yet independent regulation of actin cytoskeletal organization and translation initiation by glucose in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:1544–1556. doi: 10.1091/mbc.E03-12-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Albanese V., Yam A. Y., Baughman J., Parnot C., Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 81.Slaughter T., Black M. M. STOP (stable-tubule-only-polypeptide) is preferentially associated with the stable domain of axonal microtubules. J. Neurocytol. 2003;32:399–413. doi: 10.1023/B:NEUR.0000011334.70648.87. [DOI] [PubMed] [Google Scholar]
- 82.Bosc C., Andrieux A., Job D. STOP proteins. Biochemistry. 2003;42:12125–12132. doi: 10.1021/bi0352163. [DOI] [PubMed] [Google Scholar]
- 83.Lim A. C., Tiu S. Y., Li Q., Qi R. Z. Direct regulation of microtubule dynamics by protein kinase CK2. J. Biol. Chem. 2004;279:4433–4439. doi: 10.1074/jbc.M310563200. [DOI] [PubMed] [Google Scholar]
- 84.Hoffmeister K. M., Falet H., Toker A., Barkalow K. L., Stossel T. P., Hartwig J. H. Mechanisms of cold-induced platelet actin assembly. J. Biol. Chem. 2001;276:24751–24759. doi: 10.1074/jbc.M011642200. [DOI] [PubMed] [Google Scholar]
- 85.Dresios J., Aschrafi A., Owens G. C., Vanderklish P. W., Edelman G. M., Mauro V. P. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1865–1870. doi: 10.1073/pnas.0409764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Danno S., Nishiyama H., Higashitsuji H., Yokoi H., Xue J. H., Itoh K., Matsuda T., Fujita J. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem. Biophys. Res. Commun. 1997;236:804–807. doi: 10.1006/bbrc.1997.7059. [DOI] [PubMed] [Google Scholar]
- 87.Derry J. M., Kerns J. A., Francke U. RBM3, a novel human gene in Xp11.23 with a putative RNA-binding domain. Hum. Mol. Genet. 1995;4:2307–2311. doi: 10.1093/hmg/4.12.2307. [DOI] [PubMed] [Google Scholar]
- 88.Danno S., Itoh K., Matsuda T., Fujita J. Decreased expression of mouse Rbm3, a cold-shock protein, in Sertoli cells of cryptorchid testis. Am. J. Pathol. 2000;156:1685–1692. doi: 10.1016/S0002-9440(10)65039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kita H., Carmichael J., Swartz J., Muro S., Wyttenbach A., Matsubara K., Rubinsztein D. C., Kato K. Modulation of polyglutamine-induced cell death by genes identified by expression profiling. Hum. Mol. Genet. 2002;11:2279–2287. doi: 10.1093/hmg/11.19.2279. [DOI] [PubMed] [Google Scholar]
- 90.Chappell S. A., Owens G. C., Mauro V. P. A 5′ leader of Rbm3, a cold stress-induced mRNA, mediates internal initiation of translation with increased efficiency under conditions of mild hypothermia. J. Biol. Chem. 2001;276:36917–36922. doi: 10.1074/jbc.M106008200. [DOI] [PubMed] [Google Scholar]
- 91.Chappell S. A., Mauro V. P. The internal ribosome entry site (IRES) contained within the RNA-binding motif protein 3 (Rbm3) mRNA is composed of functionally distinct elements. J. Biol. Chem. 2003;278:33793–33800. doi: 10.1074/jbc.M303495200. [DOI] [PubMed] [Google Scholar]
- 92.White-Grindley E., Si K. RISC-y memories. Cell. 2006;124:23–26. doi: 10.1016/j.cell.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 93.Nishiyama H., Danno S., Kaneko Y., Itoh K., Yokoi H., Fukumoto M., Okuno H., Millan J. L., Matsuda T., Yoshida O., Fujita J. Decreased expression of cold-inducible RNA-binding protein (CIRP) in male germ cells at elevated temperature. Am. J. Pathol. 1998;152:289–296. [PMC free article] [PubMed] [Google Scholar]
- 94.Nishiyama H., Xue J. H., Sato T., Fukuyama H., Mizuno N., Houtani T., Sugimoto T., Fujita J. Diurnal change of the cold-inducible RNA-binding protein (Cirp) expression in mouse brain. Biochem. Biophys. Res. Commun. 1998;245:534–538. doi: 10.1006/bbrc.1998.8482. [DOI] [PubMed] [Google Scholar]
- 95.Nishiyama H., Higashitsuji H., Yokoi H., Itoh K., Danno S., Matsuda T., Fujita J. Cloning and characterization of human CIRP (cold-inducible RNA-binding protein) cDNA and chromosomal assignment of the gene. Gene. 1997;204:115–120. doi: 10.1016/s0378-1119(97)00530-1. [DOI] [PubMed] [Google Scholar]
- 96.Nishiyama H., Itoh K., Kaneko Y., Kishishita M., Yoshida O., Fujita J. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J. Cell Biol. 1997;137:899–908. doi: 10.1083/jcb.137.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xue J. H., Nonoguchi K., Fukumoto M., Sato T., Nishiyama H., Higashitsuji H., Itoh K., Fujita J. Effects of ischemia and H2O2 on the cold stress protein CIRP expression in rat neuronal cells. Free Radical Biol. Med. 1999;27:1238–1244. doi: 10.1016/s0891-5849(99)00158-6. [DOI] [PubMed] [Google Scholar]
- 98.Bhatia R., Dube D. K., Gaur A., Robertson D. R., Lemanski S. L., McLean M. D., Lemanski L. F. Expression of axolotl RNA-binding protein during development of the Mexican axolotl. Cell Tissue Res. 1999;297:283–290. doi: 10.1007/s004410051356. [DOI] [PubMed] [Google Scholar]
- 99.Uochi T., Asashima M. XCIRP (Xenopus homolog of cold-inducible RNA-binding protein) is expressed transiently in developing pronephros and neural tissue. Gene. 1998;211:245–250. doi: 10.1016/s0378-1119(98)00102-4. [DOI] [PubMed] [Google Scholar]
- 100.Saito T., Sugimoto K., Adachi Y., Wu Q., Mori K. J. Cloning and characterization of amphibian cold inducible RNA-binding protein. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2000;125:237–245. doi: 10.1016/s0305-0491(99)00174-1. [DOI] [PubMed] [Google Scholar]
- 101.Wu B. J., Kingston R. E., Morimoto R. I. Human HSP70 promoter contains at least two distinct regulatory domains. Proc. Natl. Acad. Sci. U.S.A. 1986;83:629–633. doi: 10.1073/pnas.83.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu A. Y., Bian H., Huang L. E., Lee Y. K. Transient cold shock induces the heat shock response upon recovery at 37 °C in human cells. J. Biol. Chem. 1994;269:14768–14775. [PubMed] [Google Scholar]
- 103.Beer C., Buhr P., Hahn H., Laubner D., Wirth M. Gene expression analysis of murine cells producing amphotropic mouse leukaemia virus at a cultivation temperature of 32 and 37 °C. J. Gen. Virol. 2003;84:1677–1686. doi: 10.1099/vir.0.18871-0. [DOI] [PubMed] [Google Scholar]
- 104.Baik J. Y., Lee M. S., An S. R., Yoon S. K., Joo E. J., Kim Y. H., Park H. W., Lee G. M. Initial transcriptome and proteome analyses of low culture temperature-induced expression in CHO cells producing erythropoietin. Biotechnol. Bioeng. 2006;93:361–371. doi: 10.1002/bit.20717. [DOI] [PubMed] [Google Scholar]
- 105.Kaufmann H., Mazur X., Fussenegger M., Bailey J. E. Influence of low temperature on productivity, proteome and protein phosphorylation of CHO cells. Biotechnol. Bioeng. 1999;63:573–582. doi: 10.1002/(sici)1097-0290(19990605)63:5<573::aid-bit7>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 106.Storey K. B. Mammalian hibernation: transcriptional and translational controls. Adv. Exp. Med. Biol. 2003;543:21–38. [PubMed] [Google Scholar]
- 107.Ducommun P., Ruffieux P. A., Kadouri A., von Stockar U., Marison I. W. Monitoring of temperature effects on animal cell metabolism in a packed bed process. Biotechnol. Bioeng. 2002;77:838–842. doi: 10.1002/bit.10185. [DOI] [PubMed] [Google Scholar]
- 108.Fogolin M. B., Wagner R., Etcheverrigaray M., Kratje R. Impact of temperature reduction and expression of yeast pyruvate carboxylase on hGM-CSF-producing CHO cells. J. Biotechnol. 2004;109:179–191. doi: 10.1016/j.jbiotec.2003.10.035. [DOI] [PubMed] [Google Scholar]
- 109.Fox S. R., Yap M. X., Yap M. G., Wang D. I. Active hypothermic growth: a novel means for increasing total interferon-γ production by Chinese-hamster ovary cells. Biotechnol. Appl. Biochem. 2005;41:265–272. doi: 10.1042/BA20040067. [DOI] [PubMed] [Google Scholar]
- 110.Fox S. R., Patel U. A., Yap M. G., Wang D. I. Maximizing interferon-γ production by Chinese hamster ovary cells through temperature shift optimization: experimental and modeling. Biotechnol. Bioeng. 2004;85:177–184. doi: 10.1002/bit.10861. [DOI] [PubMed] [Google Scholar]
- 111.Fox S. R., Tan H. K., Tan M. C., Wong S., Yap M. G. S., Wang D. I. C. A detailed understanding of the enhanced hypothermic productivity of interferon-γ by Chinese-hamster ovary cells. Biotechnol. Appl. Biochem. 2005;41:255–264. doi: 10.1042/BA20040066. [DOI] [PubMed] [Google Scholar]
- 112.Furukawa K., Ohsuye K. Effect of culture temperature on a recombinant CHO cell line producing a C-terminal α-amidating enzyme. Cytotechnology. 1998;26:153–164. doi: 10.1023/A:1007934216507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schatz S. M., Kerschbaumer R. J., Gerstenbauer G., Kral M., Dorner F., Scheiflinger F. Higher expression of Fab antibody fragments in a CHO cell line at reduced temperature. Biotechnol. Bioeng. 2003;84:433–438. doi: 10.1002/bit.10793. [DOI] [PubMed] [Google Scholar]
- 114.Yoon S. K., Song J. Y., Lee G. M. Effect of low culture temperature on specific productivity, transcription level, and heterogeneity of erythropoietin in Chinese hamster ovary cells. Biotechnol. Bioeng. 2003;82:289–298. doi: 10.1002/bit.10566. [DOI] [PubMed] [Google Scholar]
- 115.Yoon S. K., Kim S. H., Lee G. M. Effect of low culture temperature on specific productivity and transcription level of anti-4-1BB antibody in recombinant Chinese hamster ovary cells. Biotechnol. Prog. 2003;19:1383–1386. doi: 10.1021/bp034051m. [DOI] [PubMed] [Google Scholar]
- 116.Yoon S. K., Choi S. L., Song J. Y., Lee G. M. Effect of culture pH on erythropoietin production by Chinese hamster ovary cells grown in suspension at 32.5 and 37.0 °C. Biotechnol. Bioeng. 2004;89:345–356. doi: 10.1002/bit.20353. [DOI] [PubMed] [Google Scholar]
- 117.Yoon S. K., Hwang S. O., Lee G. M. Enhancing effect of low culture temperature on specific antibody productivity of recombinant Chinese hamster ovary cells: clonal variation. Biotechnol. Progr. 2004;20:1683–1688. doi: 10.1021/bp049847f. [DOI] [PubMed] [Google Scholar]
- 118.Lindholm D., Arumae U. Cell differentiation: reciprocal regulation of Apaf-1 and the inhibitor of apoptosis proteins. J. Cell Biol. 2004;167:193–195. doi: 10.1083/jcb.200409171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Martin D. S., Stolfi R. L., Colofiore J. R. The chemotherapeutic relevance of apoptosis and a proposed biochemical cascade for chemotherapeutically induced apoptosis. Cancer Invest. 1997;15:372–381. doi: 10.3109/07357909709039742. [DOI] [PubMed] [Google Scholar]
- 120.Zhang Z., Sobel R. A., Cheng D., Steinberg G. K., Yenari M. A. Mild hypothermia increases Bcl-2 protein expression following global cerebral ischemia. Mol. Brain Res. 2001;95:75–85. doi: 10.1016/s0169-328x(01)00247-9. [DOI] [PubMed] [Google Scholar]
- 121.Khar A., Pardhasaradhi B. V., Ali A. M., Kumari A. L. Protection conferred by Bcl-2 expression involves reduced oxidative stress and increased glutathione production during hypothermia-induced apoptosis in AK-5 tumor cells. Free Radical Biol. Med. 2003;35:949–957. doi: 10.1016/s0891-5849(03)00469-6. [DOI] [PubMed] [Google Scholar]
- 122.Fujikake N., Nagai Y., Popiel H. A., Kano H., Yamaguchi M., Toda T. Alternative splicing regulates the transcriptional activity of Drosophila heat shock transcription factor in response to heat/cold stress. FEBS Lett. 2005;579:3842–3848. doi: 10.1016/j.febslet.2005.05.074. [DOI] [PubMed] [Google Scholar]
- 123.Fernandez J., Yaman I., Sarnow P., Snider M. D., Hatzoglou M. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2α. J. Biol. Chem. 2002;277:19198–19205. doi: 10.1074/jbc.M201052200. [DOI] [PubMed] [Google Scholar]
- 124.Kedersha N., Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 125.Kedersha N. L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell. Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brown E. C., Jackson R. J. All five cold-shock domains of unr (upstream of N-ras) are required for stimulation of human rhinovirus RNA translation. J. Gen. Virol. 2004;85:2279–2287. doi: 10.1099/vir.0.80045-0. [DOI] [PubMed] [Google Scholar]