Abstract
Siglecs (sialic acid binding Ig-like lectins) are transmembrane receptors for sialylated glycoconjugates that modulate cellular interactions and signalling events in the haematopoietic, immune and nervous systems. Siglec-7 is a structural prototype for the recently described family of immune inhibitory CD33-related siglecs and is predominantly expressed on natural killer cells and monocytes, as well as subsets of CD8 T-cells. Siglec-specific inhibitors are desired for the detection of masked and unmasked forms of siglecs, to aid in dissection of signalling pathways and as tools to investigate siglecs as potential therapeutic targets. As a first step towards this end, we present the crystal structure of siglec-7 in complex with a sialylated ligand, the ganglioside analogue DSLc4 [α(2,3)/α(2,6) disialyl lactotetraosyl 2-(trimethylsilyl)ethyl], which allows for a detailed description of the binding site, required for structure-guided inhibitor design. Mutagenesis and binding assays were used to demonstrate a key structural role for Lys131, a residue that changes conformation upon sialic acid binding. Differences between the binding sites of siglec family members were then exploited using α-methyl Neu5Ac (N-acetylneuraminic acid) as a basic scaffold. A co-crystal of siglec-7 in complex with the sialoside inhibitor, oxamido-Neu5Ac [methyl α-9-(amino-oxalyl-amino)-9-deoxy-Neu5Ac] and inhibition data for the sialosides gives clear leads for future inhibitor design.
Keywords: ganglioside, immune tyrosine-based inhibitory motif (ITIM), leucocyte, sialic acid, sialic acid binding Ig-like lectin (siglec), x-ray crystallography
Abbreviations: CHO, Chinese hamster ovary; DSLc4, α(2,3)/α(2,6) disialyl lactotetraosyl 2-(trimethylsilyl)ether; ESRF, European Synchrotron Radiation Facility; Gal, galactose; GalNAc, N-acetyl galactosamine; Glc, glucose; ITIM, immune tyrosine-based inhibitory motif; MAG, myelin-associated glycoprotein; Neu5Ac, N-acetylneuraminic acid; NK, natural killer cell; OPD, O-phenylenediamine; oxamido-Neu5Ac, methyl α-9-(amino-oxalyl-amino)-9-deoxy-Neu5Ac; PEG, poly(ethylene glycol); RBC, red blood cell; rmsd, root mean square deviation; siglec, sialic acid binding Ig-like lectin
INTRODUCTION
The siglec (sialic acid binding Ig-like lectin) family is a group of transmembrane receptors with the ability to recognize sialic acids, a family of 9-carbon sugars which often cap the non-reducing ends of glycoconjugates in higher animals. Eleven siglecs have been described in humans, predominantly expressed on cells of the immune and haematopoietic systems [1,2]. Each siglec has a characteristic cell-type-dependent expression pattern and a distinct binding specificity for sialylated glycoconjugates [3]. Siglecs are often expressed in an overlapping manner on cells of the immune system and this may be important in allowing a given cell type the ability to respond to a spectrum of sialylated ligands.
With the exception of MAG [(myelin-associated glycoprotein)/siglec-4] and sialoadhesin (siglec-1), siglecs are thought to play a role in the suppression of immune cell activation [4], as they possess consensus ITIMs (immunoreceptor tyrosine-based inhibitory motifs) and ITIM-like motifs in their cytoplasmic tails. ITIMs have been described in a growing number of inhibitory receptors of the immune system [4a]. Their phosphorylation creates a high-affinity binding site for SH2 (Src-homology 2)-domain-containing phosphatases (SHP-1, SHP-2 and SHIP-1), and the subsequent antagonism of activating signals [5,6].
The human CD33-related siglecs (CD33 and siglec-5–siglec-11) and CD22 can interact with sialylated ligands presented on the same cell (cis interaction) [7], a phenomenon referred to as ‘masking’, as this prevents the binding of exogenously added ligands. For CD22 and some CD33-related siglecs expressed on blood leucocytes, B-cell activation results in partial unmasking [7,7a]. Cryptic sialic acid binding lectins on human blood leucocytes can be unmasked by sialidase treatment or cellular activation. However, for other siglecs such as siglec-7, which is expressed on NK (natural killer) cells, a variety of activation signals tested did not lead to unmasking [8]. The development of siglec-specific inhibitors based on a sialic acid template would provide useful tools for studying these receptors in their natural context. This has been achieved recently for CD22, by the modification of Neu5Ac (N-acetylneuraminic acid) at the C9 position with a biphenyl moiety, leading to a >200-fold improvement in relative inhibitory potency compared with the unmodified sugar [9]. The utility of this inhibitor was clearly demonstrated when it was used to reveal that cis interactions are important in the inhibitory function of CD22 during B-cell activation [10].
Siglec-7 represents a good candidate for structure-based inhibitor design. It is the only CD33-related siglec for which the structure has been determined so far and therefore provides a template on which other members of this family may be modelled [11]. It displays a unique ligand binding preference, binding internally branched α(2,6)-linked sialic acid and α(2,8)-linked disialic acids [12]. Siglec-7 is expressed predominantly on NK cells and it has been shown to inhibit NK cell cytotoxicity towards target cells over-expressing the α(2,8)-disialic acid-bearing ganglioside, GD3 [8]. The elevation of GD3 levels described for certain tumours (such as malignant melanoma and neuroblastoma) may, therefore, serve as an evasion strategy from NK killing [13]. A siglec-7-blocking compound could, therefore, have potential therapeutic value.
In the present study, we describe the first structural analysis of siglec-7 in complex with sialylated ligands: a ganglioside analogue, DSLc4 [α(2,3)/α(2,6) disialyl lactotetraosyl 2-(trimethylsilyl)ethyl] [14], and a derivative of sialic acid, oxamido-Neu5Ac [methyl α-9-(amino-oxalyl-amino)-9-deoxyNeu5Ac]. In combination with binding assays, these structures give us an insight into what governs ligand binding and possible routes for rational inhibitor design.
MATERIALS AND METHODS
Expression and purification of siglec-7
The siglec-7 V-set domain was expressed and purified as described by Alphey et al. [11]. Briefly, the region encoding the siglec-7 V-set domain was cloned into the pDEF expression vector and transfected into CHO (Chinese hamster ovary) Lec1 cells [20]. A stable cell line was established using selection with hygromycin B. Cells were cultured in α-MEM (α-modified Eagle's medium) containing 5% foetal calf serum and 1% penicillin/streptomycin mix (Life Technologies). Siglec-7 was purified from the medium using an anti-siglec-7 polyclonal antibody affinity column followed by size-exclusion chromatography. The protein was concentrated to 6 mg/ml in 25 mM Tris (pH 8.0) and 75 mM NaCl.
Synthesis of oxamido-Neu5Ac
Oxamido-Neu5Ac was prepared from the methyl α-glycoside of 9-amino-9-deoxy-Neu5Ac by reaction with activated oxamic acid (R. Brossmer, unpublished work). The analogue was characterized by high-resolution nuclear magnetic resonance spectroscopy and fast atom bombardment MS.
Crystallization and data collection
For co-crystallization experiments, siglec-7 was incubated on ice for 10 min with a 20-fold molar excess of ligand (either DSLc4 or oxamido-Neu5Ac). Crystallization was conducted by using sitting-drop vapour diffusion, using 0.5 μl of reservoir solution added to a 0.5 μl drop of protein–ligand complex, and equilibrated with 100 μl of reservoir solution at 20 °C. The DSLc4–siglec-7 co-crystals used for data collection were grown in crystal screen CS CRYO condition forty-one {8.5% iso-propanol, 0.095 M Hepes (pH 7.5), 17% PEG [poly(ethylene glycol)]4000, 15% glycerol}. A data set was collected to 1.90 Å (1 Å=0.1 nm) at the ESRF (European Synchrotron Radiation Facility; Grenoble, France), beamline ID14 EH4 at 100 K using an ADSC Q4 CCD detector.
Co-crystals for siglec-7 and oxamido-Neu5Ac grew in a screen containing 30% PEG 4000, 0.1 M sodium acetate (pH 4.6) and 0.2 M ammonium acetate. The crystal was cryogenically protected by soaking in the mother-liquor containing 10% glycerol before freezing in a stream of nitrogen gas. A dataset was collected on ESRF beamline ID14 EH4 using an ADSC Q4 CCD detector at 100 K. Data were processed and scaled using the HKL suite of programs [21].
Structure determination and refinement
The structures for siglec-7–DSLc4 and siglec-7–oxamido-Neu5Ac were solved by molecular replacement using the program AMoRe [22] with the siglec-7 V-set domain structure (PDB code 1o7S [11]) as a search model. For both, a single solution was found with R-factors of 0.33 and 0.32 and correlation coefficients of 0.70 and 0.72 respectively. The model phases were used in WarpNTrace, which constructed 102 and 95 out of 127 residues respectively [23]. The models were refined using CNS (crystallography and NMR system) [24] software interspersed with manual model building in the program O [25]. Final refinement was performed using the program Refmac 5 [26].
After model building and the inclusion of water molecules in the refinement, the terminal sialic acid residue was placed into the unbiased |Fo|−|Fc|,Φcalc electron density and further refinement was carried out. Strong connective |Fo|-|Fc|,Φcalc electron density was observed across the 2-fold symmetry axis between the sialic acids of adjacent asymmetric units (Figure 1). The mode of binding which could best account for this density is that the ligand bridges two siglec molecules. Although the DSLc4 ligand is not symmetrical, it does possess a degree of pseudosymmetry in the tetrasaccharide unit NeuAcα(2,3)Galβ(1,3)GlcNAcβ-[NeuAcα(2,6)], the plane of pseudosymmetry being about the Galβ(1,3)GlcNAcβ bond. The electron density was interpreted to consist of the DSLc4 tetrasaccharide, as described above, spanning two siglec-7 monomers without preferential orientation. The density between the sialic acid residues is, therefore, made up of both Gal and GlcNAc, which occupy the same space at 50% occupancy each. The tetrasaccharide was accommodated in the refinement by modelling it across the asymmetric unit at 50% occupancy, which allows the symmetry operation to generate the ligand in the opposite orientation. Thus there is a 2-fold disorder combined with a 2-fold crystallographic axis. As these two orientations are mutually exclusive, the van der Waals interactions were turned off between these molecules. This satisfied the electron density between the binding sites. Electron densities for the other sugar residues were not observed. For the oxamido-Neu5Ac–siglec-7 complex, after the inclusion of water molecules and model building, oxamido-Neu5Ac was modelled into the unbiased |Fo|−|Fc|,Φcalc density and was further refined.
Figure 1. The siglec-7–DSLc4 interaction.
(A) A schematic representation of the DSLc4 analogue. (B) The structure of the N-terminal, ligand-binding domain of siglec-7. β-strands are shown as blue arrows. The intrasheet disulphide bond is shown in green and the primary sialic acid binding Arg124 residue with carbons is in mauve. (C and D) show the DSLc4 ligand bridging two siglec-7 molecules related by a 2-fold crystallographic symmetry axis. Both orientations of the DSLc4 tetrasaccharide (green sticks) are shown modelled into an electron density that spans symmetry-related molecules. (C) The ligand is in the orientation: Neu5Acα(2,6)GlcNAcβ(3,1) Gal[Neu5Acα(2,3)], whereas in (D) the ligand is in the Neu5Acα(2,3)Galβ(1,3)GlcNAc[α(2,6)Neu5Ac] orientation. Unbiased (i.e. before inclusion of ligand) |Fo|-|Fc|,Φcalc electron density contoured at 2.25 σ is shown as a yellow meshwork. The side-chains of residues that interact with the ligand are labelled, and water molecules bridging the protein and the ligand are shown as magenta spheres. The potential hydrogen-bonding network is shown as black dotted lines.
Site-directed mutagenesis and expression of chimaeras
Mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) on a siglec-7–Fc chimaeric construct in a modified pEE14 plasmid. Successful mutagenesis was confirmed by sequencing. The plasmids were transiently transfected into COS-1 cells using FuGene 6 transfection reagent (Roche). The cells were cultured for a further 5 days in X-VIVO 10 serum-free medium (BioWhitaker) and the medium was harvested. The siglec-7–Fc concentration was assayed by ELISA and correct folding was confirmed by ELISA using anti-siglec-7 monoclonal antibodes (7.7a and 7.5a [27]).
RBC (red blood cell) binding assay
Siglec-7–Fc chimaeras were immobilized on microtitre plates to create a surface for human RBC capture [28]. For this procedure, a goat anti-(human Fc) antibody (Sigma) was coated on to immulon 4 HBX microtitre plates. The plates were washed with PBA (PBS, 0.1% BSA and 10 mM sodium azide) and blocked with 5% dried-milk powder. Serial dilutions of the siglec-7–Fc chimaera, 50 μl per well, were allowed to bind for 2 h at room temperature, followed by three washes with PBA. A 100 μl aliquot of an RBC suspension was added to each well and allowed to bind for 30 min at room temperature. Unbound cells were removed by gentle washing and the cell layer was allowed to air-dry, followed by methanol fixation and permeabilization. The extent of RBC binding was assayed using OPD (O-phenylenediamine peroxidase) as the substrate [28] and by measuring A450 with a Cytofluor (PerSeptive Biosysytems) plate reader.
Hapten inhibition assays with sialic acid derivatives
For hapten inhibition assays, Fc chimaeras containing the three N-terminal domains of siglec-7 were purified in Protein A–agarose from tissue culture supernatants of stably transfected CHO cells, as described by Yamaji et al. [12]. Microtitre plates with covalently linked Neu5Ac (GlycoWells from Lundonia, Lund, Sweden) were used as a binding target. Prior to the binding assay, the Fc-chimaeras (final concentration 200 ng/ml) were complexed with an anti-Fc antibody coupled to alkaline phosphatase (1:1000 dilution, supplied by Dianova, Hamburg, Germany) in HBS-T [10 mM Hepes, 150 mM NaCl (pH 7.4) and 0.05% Tween 20] for 15–30 min at room temperature. A 30 μl aliquot of this pre-complex was added per well and incubated for 4 h at 4 °C in the presence or absence of sialic acid derivatives. After five washes with HBS-T, the amount of bound alkaline phosphatase was quantified by determination of the initial reaction velocity using fluorescein disphosphate (15 μM) as substrate. To determine the concentration required for 50% inhibition (IC50), the inhibitor was used at seven different concentrations spanning two orders of magnitude. Each concentration experiment was carried out in triplicate and assays were repeated at least three times.
RESULTS AND DISCUSSION
The DSLc4 complex
Sparse matrix-sampling screening for siglec-7 co-crystallization with the DSLc4 ganglioside analogue (Figure 1A) yielded an unusually high number of hits: over 20 out of 144 conditions within a period of two days. All crystals were of a bi-pyramidal morphology, with a very similar appearance to that of an uncomplexed crystal form of siglec-7. Synchrotron diffraction data were collected to a resolution of 1.90 Å (Table 1). The unit cell dimensions were also similar to apo-siglec-7 and indexed in the same space group, P41212. Interpretation of the ligand density revealed that an ordered tetrasaccharide unit [Neu5-Acα(2,3)Galβ(1,3)GlcNAc[α(2,6)Neu5Ac] bridged the binding sites of siglec-7 in adjacent asymmetric units across a crystallographic 2-fold axis of rotation, with 2-fold disorder (Figures 1C and 1D). The net result is the stabilization of the same crystal packing structure, as observed for native siglec-7, and perhaps explains the unusual promotion of crystal formation. Comparison of the native and complexed form [rmsd (root mean square deviation)=0.73 Å on all Cα atoms] revealed that there is little deformation in crystal packing with only a 0.9 Å decrease in the c-axis of the unit cell.
Table 1. Data collection and refinement statistics.
The co-ordinates and structure factors have been deposited with the PDB (entries 2DF3 and 2G5R). Statistical analysis results for the highest resolution shell are shown in parentheses.
| Data Collection | Siglec-7–DSLc4 | Siglec-7–oxamido-Neu5Ac |
|---|---|---|
| Wavelength (Å) | 0.933 | 0.933 |
| Resolution (Å) | 25–1.90 (1.97–1.90) | 25–1.60 (1.66–1.60) |
| Space group | P41212 | P41212 |
| Unit cell (Å) | a=b=53.07, c=92.88 | a=b=52.93, c=93.24 |
| Reflections: | ||
| Observed | 46855 | 74536 |
| Unique | 11014 | 17748 |
| Redundancy | 4.3 (3.8) | 4.2 (2.2) |
| Rmerge | 0.078 (0.445) | 0.043 (0.338) |
| I/σI | 15.5 (3.1) | 21.5 (2.0) |
| Completeness (%) | 99.3 (94.0) | 97.5 (78.9) |
| Rfree reflections | 523 | 893 |
| Rcryst (%) | 19.7 | 21.4 |
| Rfree (%) | 23.7 | 22.8 |
| Total number of atoms | 1119 | 1053 |
| Protein | 944 | 904 |
| Water | 95 | 102 |
| Glycan | 14 | 14 |
| Ligand | 66 | 27 |
| Protein (Å2) | 20 | 18 |
| Water (Å2) | 27 | 27 |
| Glycan (Å2) | 33 | 27 |
| Ligand (Å2) | 24 | 44 |
| rmsd from ideal geometry | ||
| Bond lengths (Å) | 0.014 | 0.015 |
| Bond angles (°) | 1.65 | 1.35 |
| Main chain B (Å2) | 1.55 | 1.77 |
Within the ligand, intramolecular hydrogen-bonding between the Gal C4 hydroxy group and the Neu5Ac [α(2,3) linkage] C7 hydroxy group stabilizes the tetrasaccharide (Table 2), tethering it between the two symmetry-related siglec-7 molecules (Figures 1C and 1D). Although no direct contacts are made with the protein molecules beyond the terminal sugars, water-mediated hydrogen bonds are made between Asn133 and Lys135, and the C3 nitrogen of the GlcNAc and the oxygen of the glycosidic bond between Gal and GlcNAc [in the α(2,3) linkage orientation] (Figure 1D and Table 2).
Table 2. Hydrogen-bond network.
Potential hydrogen-bonds and the distances between atoms are indicated.
| Ligand–protein hydrogen-bonds | ||
|---|---|---|
| DSLc4 atom | Protein atom | Distance (Å) |
| Neu5Ac (α2,6) | ||
| O1A | Arg124 NH2 | 3.0 |
| O1B | Arg124 NH1 | 2.8 |
| N5 | Lys131 CO | 2.7 |
| O5 | Tyr26 OH | 3.4 |
| O8 | Asn133 N | 2.9 |
| O9 | Lys135 NZ | 2.8 |
| O9 | Asn133 CO | 2.8 |
| Neu5Ac (α2,3) | ||
| O1A | Arg124 NH2 | 2.7 |
| O1B | Arg124 NH1 | 2.9 |
| N5 | Lys131 CO | 3.3 |
| O5 | Tyr26 OH | 4.3 |
| O8 | Asn133 N | 3.0 |
| O9 | Lys135 NZ | 3.1 |
| O9 | Asn133 CO | 3.1 |
| DSLc4–protein hydrogen-bonds mediated by water | ||
| Atom | Water molecule | Distance (Å) |
| Gal (α2,3) 03 | 105 | 3.4 |
| GlcNAc (α2,3) N2 | 105 | 3.4 |
| Asn133 ND2 | 105 | 2.6 |
| Lys135 NZ | 105 | 2.7 |
| O4 Neu5Ac (α2,6) | 110 | 3.1 |
| O4 Neu5Ac (α 2,3) | 110 | 3.8 |
| Asn129 CO | 110 | 2.6 |
| Intramolecular hydrogen-bonds | ||
| DSLc4 atom | DSLc4 atom | Distance (Å) |
| Neu5Ac (α2,3) 07 | Gal 04 | 2.7 |
| Neu5Ac 08 (α2,6) | Neu5Ac 09 | 2.8 |
| Neu5Ac 08 (α2,3) | Neu5Ac 09 | 3.4 |
It is not clear whether the contacts observed outside the primary sialic acid binding site in this crystal structure represent ‘natural’ or artificial contacts which arise from crystal packing. Binding studies have shown that both the sialic acid linkage and the adjacent sub-terminal sugar are key contributors to siglec ligand specificity. Siglec-7 displays only a weak interaction with ligands bearing α(2,3)-linked sialic acids [12]. Comparison of the binding of siglec-7 with branched α(2,3)/α(2,6) disialyl ligands, such as disialylgalactosyl-globoside and disialyl Lewisa motifs and their monosialylated, α(2,3) sialic-acid-bearing equivalents, shows that it is the α(2,6) branch that is required for higher affinity binding [15,16]. Therefore we would expect to observe selective binding of the α(2,6) branch, rather than equal binding, as in this crystal structure. Furthermore, the C-C′ loop, which has been implicated in directing ligand binding specificity, is disordered in the siglec-7–DSLc4 complex. There are two possible explanations for the absence of direct contacts between side-chain and sub-terminal sugars in this crystal structure. First, regions of siglec-7, such as the C-C′ loop, may adopt different conformations in order to interact with the different linkages, and 2-fold disorder in the ligand may make such flexible protein interactions difficult to see in the electron density maps. Second, the interaction of the capping sialic acids drives cross-linking and stabilization of crystal packing, and the ligand is held in an artificial manner. Nevertheless, the water-mediated interaction between the ligand and residues Asn133 and Lys135, suggests that they may play a genuine role in ligand binding.
The ligand bridges two binding sites that are relatively close together, which possibly results in conformational strain in the ligand. This can be examined by comparing the dihedral bond angles of the DSLc4 ligand with those of the same linkages in other previously determined glycan structures. The ψ and Φ angles of Neu5Acα(2,3)Gal and Neu5Acα(2,6)GlcNAc linkages lie within regions common to other glycans deposited in the PDB (Table 3). The distortion lies in the Galβ(1,3)GlcNAc linkage. In structures deposited in the PDB, all ψ angles fall within the synclinal conformation, +28 to +68. In DSLc4 this lies in the opposite orientation at −41°. The Φ angle of this bond is again different, −59° (synclinal), compared with a synperiplanar conformation (−27 to +25°) adopted by other structures. The net result is that the ring-faces of Gal and GlcNAc in the DSLc4 glycan bound to siglec-7 lie at a 90° angle relative to each other, whereas in the other structures the rings are co-planar. The straining of the ligand is also reflected in the contacts made by each Neu5Ac group and, overall, the α(2,6)-linked Neu5Ac is more intimately associated with the binding site (Table 2).
Table 3. Glycosidic bond angles.
Dihedral angles for the glycosidic bonds of DSLc4 and equivalent linkages in structures deposited in the PDB were analysed using the Carbohydrate Ramachandran Plot (CARP) tool of the Carbohydrate Structure Suite (CSS) [29] (http://www.dkfz.de/spec/css/index.php). The dihedral angles for the Galβ(1,3)GlcNAc-linkage are defined as: Φ=O5-C1-O3′-C3′ and ψ=C1-O3′-C3′-C4′. For the Neu5Acα(2,3)Gal linkage: Φ=C1-C2-O3′-C3′ and ψ=C2-O2-C3′-C4′. For the Neu5Acα(2,6)GlcNAc linkage: Φ=C1-C2-O6′-C6′, ψ=C2-O2-C6′-C5′. The stereochemical conformation is given after the torsion angle, and is defined as: 0 to 30 synperiplanar (sp); 30° to 90° and -30° to -90° synclinal (sc); 90° to 150° and -90° to -150° anticlinal (ac); and±150° to 180° antiperiplanar (ap). No., number of ligand examples in the PDB.
| DSLc4 | Survey of PDB | ||||
|---|---|---|---|---|---|
| Linkage | Φ | Ψ | Φ | ψ | No. |
| Neu5Acα(2,6)GlcNAc | −84 (ac) | −169 (ap) | −85 to −82 (sc), +22 to +27 (sp) | +190 to +144 (ap) | 5 |
| Galβ(1,3)GlcNAc | −41 (sc) | −59 (sc) | +28 to +63 (sc) | −27 to +25 (sp) | 12 |
| Neu5Acα(2,3)Gal | −133 (ac) | −42 (sc) | −35 to −75 (sc), +175 to +190 (ap) | +26 to −44 (sp, sc) | 62 |
Sialic acid binding by siglec-7
Siglec-7 makes several contacts with the DSLc4 terminal sialic acid that are similar to those observed in the crystal structure of sialoadhesin in complex with 3′ sialyllactose (Figure 2). Arg124 forms a salt-bridge with the sialic acid carboxylate, an interaction that is essential for sialic acid binding in all siglecs [17,18] (Figure 2A). Hydrogen-bonds between the glycerol hydroxy groups at the C8 and C9 positions, and the nitrogen and protein backbone of the C5 N-acetyl group are conserved [19]. In both structures a tryptophan residue makes hydrophobic contacts with the glycerol moiety of the sialic acid. Sialoadhesin makes additional contacts with the primary sialic acid via a hydrogen-bond between the C4 hydroxy group and the carbonyl backbone of Ser103, and hydrophobic interactions between the N-acetyl methyl and Trp2 (Figure 2B). In the siglec-7–DSLc4 structure these contacts differ. An amino acid insertion in the F–G loop of sialoadhesin projects it towards the binding site, bringing the carbonyl oxygen of Ser103 to within hydrogen-bonding distance of the C4 hydroxy group. In siglec-7, the equivalent residue (Asn129), is 4.7 Å distant from the C4 hydroxy group. Instead, this interaction is replaced by a water-mediated hydrogen-bond. The tryptophan residue, which interacts with the N-acetyl methyl in sialoadhesin, is replaced with a tyrosine (Tyr26) residue in siglec-7. In the siglec-7–DSLc4 complex the orientation of the acetyl group is rotated 180°, which allows the oxygen to form a weak hydrogen-bond with the tyrosine hydroxy group.
Figure 2. Comparison of sialic-acid-binding by siglec-7 (A) with sialoadhesin (B).
An intra-sheet disulphide bond (green sticks) results in the widening of the β sandwich, exposing hydrophobic residues that contribute to the primary sialic acid binding site. In both structures the sialic acid carboxylate is anchored by an interaction with a conserved arginine residue. In siglec-7 the replacement of tryptophan (Trp2) with tyrosine (Tyr26), results in rotation of the C5 N-acetyl group to allow hydrogen-bonding with the side-chain hydroxy group.
Flexibility within the binding site
Comparison of the apo structures (PDB accession numbers, 1o7S and 1NKO; 1o7V is excluded from this analysis as a loop from a symmetry related molecule occupies the binding site) of siglec-7 with that of the complexed form revealed that there is little overall positional change in the residues that form the binding site (rmsd=1.2 Å on all atoms, Figure 3A). One notable difference is that in the apo structures the side-chain of Lys131 obscures access to the guanidinium group of Arg124. In the complexed form this residue moves away to reveal the primary arginine residue, which allows it to interact with the carboxy group of sialic acid (Figure 3A). A similar phenomenon is observed when comparing the apo (PDB accession number 1QFP) and bound (PDB accession number 1QFO) forms of sialoadhesin, in which the ‘essential’ arginine residue, Arg97, is masked by the side-chain of Arg105 (Figure 3B). Upon ligand-binding, Arg105 moves away (by 3.8 Å), and Arg97, no longer blocked by Arg105, moves 1.8 Å into a position where it can engage the carboxylate of sialic acid. With the exception of siglec-4/MAG, in all the human siglecs the residue equivalent to Lys131 in siglec-7 is either an arginine or a lysine residue (in siglec-4/MAG it is glutamine). When Lys131 was replaced with alanine by site-directed mutagenesis, this, unexpectedly, abolished binding of siglec-7–Fc to RBCs (Figure 3C), an effect comparable with that seen when the essential Arg124 was replaced with alanine (Figure 3C). Both mutant proteins appeared to be correctly folded as judged by their normal reactivity with anti-siglec-7 monoclonal antibodies (results not shown). These results suggest that rather than masking Arg124 from ligands, Lys131 is essential for ligand engagement. What biological significance this has is unclear; for example this residue may protect the primary arginine residue from non-productive or low-affinity interactions with organic or non-organic anions, which could disrupt binding-site integrity.
Figure 3. The role of Lys131.
The binding of sialic acid is accompanied by small, conserved conformational changes. Superposition of the apo and ligand-bound crystal structures of siglec-7 (A) and sialoadhesin (B), show that there is a conserved movement upon ligand binding. Side-chains which remain static are shown in cream (apo) and grey (bound ligand), whereas those that move are shown with orange (apo) and pink (ligand-bound) carbons. Arrows indicate the conformational change that accompanies ligand binding. A water molecule (green sphere), which hydrogen-bonds with the backbone nitrogen of Asn133 of siglec-7 is displaced by the sialic acid C8 hydroxy group. (C) An RBC-binding assay was used as a measure of sialic acid-dependent binding. Serial dilutions of siglec-7–Fc chimaeras, wild-type (■), and mutants R124A (◇) and K131A (●), were immobilized on microtitre plates and the extent of RBC binding was measured by an OPD assay.
Sialic-acid-based inhibitor design
Siglec-7 interacts only weakly with a Neu5Ac moiety. This has also been observed in hapten inhibition assays (Table 4) in which 3 mM methyl-α-Neu5Ac leads to only 35% inhibition. However, hydroxylation of the N-acetyl group to N-glycolyl enhances binding, since the inhibitory acitivity of methyl-α-Neu5Gc (methyl-α-5-glycolylneuraminic acid) is increased at least 2-fold. By contrast, this sialic acid is not bound by sialoadhesin or siglec-4/MAG (Kelm et al. [28]). Molecular modelling (results not shown) suggests that this additional hydroxy group could lie within hydrogen-bonding distance (between 2.7–2.6 Å) of the side-chain of Tyr26. For sialoadhesin or siglec-4, in which this position is occupied by a tryptophan residue, such a modification may be sterically unfavourable.
Table 4. Hapten inhibition assay data.
| Sialic acid derivative | IC50 (μM) |
|---|---|
| Me-α-Neu5Ac | >3000 (35%) |
| Me-α-oxamido-Neu5Ac | 1600 |
| Me-α-5-N-glycolyl-Neu | 2000 |
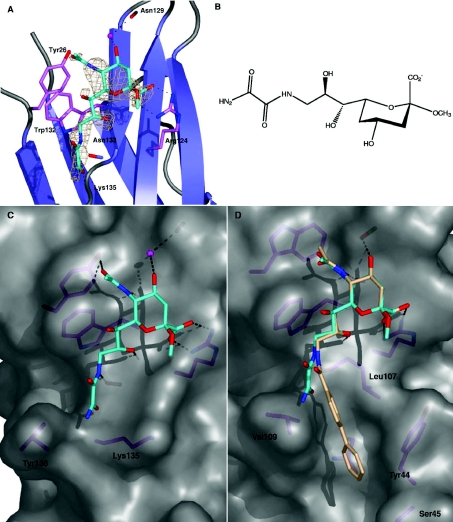
In hapten inhibition assays, Neu5Ac derivatized with an oxamido group at C9 of the glycerol side-chain (Figure 4B), was found to have at least 3-fold greater inhibitory activity than Neu5Ac (Table 4). By contrast, this sialic acid derivative inhibited sialoadhesin approx. 5-fold less compared with Neu5Ac (results not shown). To examine this differential binding, siglec-7 was co-crystallized with a 20-fold molar excess of oxamido-Neu5Ac, synchrotron diffraction data were collected to 1.6 Å, the structure was solved by molecular replacement and refined (rmsd=0.83 Å on all Cα atoms compared with the apo structure, Table 4). The structure of the siglec-7–oxamido-Neu5Ac complex revealed that many of the important contacts are maintained: salt-bridge formation between the carboxylate and Arg124, hydrogen-bonding between the Asn133 carbonyl and the C5 N-acetyl group, hydrophobic interactions between the derivatized glycerol chain and Trp132, and hydrogen-bonding between the C8 hydroxy group and the backbone nitrogen of Asn133 (Figure 4A). The hydrogen-bond formed between the C9 hydroxy group of sialic acid and the Asn133 carbonyl is directly replaced by the nitrogen of the oxamido group. An additional hydrogen-bond is formed between C11 oxygen and the backbone nitrogen of Lys131. Superposition of oxamido-Neu5Ac with sialoadhesin structures suggests that the weaker binding of this compound could be due to a steric clash of the side-chain of Val109 with the terminal amide of the oxamido group (Figure 4D).
Figure 4. Binding of oxamido-Neu5Ac by siglec-7.
Siglec-7 is shown in complex with oxamido-Neu5Ac (A) with unbiased |Fo|-|Fc|,Φcalc density contoured at 2.25 σ. The methyl-α-glycoside of 9-amino-9-deoxy-Neu5Ac was derivatized with an oxamido group at the C9 position (B). A surface representation of siglec-7 (C) and sialoadhesin (D) shows that both possess a hydrophobic gate, formed by Lys135 and Tyr136, and Leu107 and Val109 respectively. However, these channels lie at divergent angles. (D) The sialoadhesin9–BIP(biphenyl)-Neu5Ac complex (BIP is shown with cream carbon atoms) is shown in order to highlight the orientation of the hydrophobic gate. Additionally, residues which participate in BIP binding are indicated. Superimposition of oxamido-Neu5Ac (cyan carbons) on to sialoadhesin (D), shows that Val109 could sterically hinder binding.
Derivatization of Neu5Ac at the C9 position by hydrophobic moieties has yielded increased binding to other siglecs, namely human and murine CD22 and CD33, and sialoadhesin [9,10]. Crystal structures of sialoadhesin in complex with sialiosides demonstrated that this was predominantly due to the presence of a ‘hydrophobic gate’ formed by Val109 and Leu107 [9], which sandwiches the hydrophobic substituents. By sequence alignment and modelling of members of the siglec family, Zaccai et al. [9] proposed that variations in the residues which create this cleft (which are not well conserved) might contribute to the differences in binding affinity. In siglec-7 the walls of this hydrophobic groove are formed by Lys135 and Tyr136 (Figure 4C), and differs significantly from that of sialoadhesin (Figure 4D). The hydrophobic gate of siglec-7 is deeper, with much steeper sides, and the axis of the cleft is skewed by approx. 45° compared with that of sialoadhesin.
Such differences provide promising leads for the design of siglec-specific sialosides: of the human siglecs, siglec-7 is unique in having tryptophan and lysine residues forming the hydrophobic gate. In the siglec-7–oxamido-Neu5Ac structure, the oxamido group is anchored by a hydrogen-bond with its terminal amide, aligning it with the axis of the channel. Therefore the addition of aromatic moieties to the C11 nitrogen of oxamido-Neu5Ac may enhance binding and specificity. This could be complemented by the use of Neu5Gc as the template for derivatization.
Conclusions
In the present study, we have described the structure of siglec-7 in complex with a ganglioside analogue of DSLc4. This structure has allowed detailed analysis of the primary sialic acid binding site of siglec-7. Furthermore, a sialic acid analogue carrying an oxamido group at C9 was studied in complex with siglec-7. On the basis of this structure, the 3–4-fold increase in relative inhibitory potency when compared with methyl-α-Neu5Ac has been explained in molecular detail. This compound will serve as a guide for further substitutions, with the aim of creating a ligand of high affinity and high selectivity towards siglec-7.
Acknowledgments
This work was supported by BBSRC grant B14010 awarded to P. R. C., and D. M. F. vA. S. K. and S. W. were supported by Deutsche Forschungsgemeinschaft (grant DFG Ke428.3-3). D. M. F. vA. is supported by a Wellcome Trust Senior Research Fellowship.
References
- 1.Crocker P. R., Varki A. Siglecs in the immune system. Immunology. 2001;103:137–145. doi: 10.1046/j.0019-2805.2001.01241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crocker P. R. Siglecs in innate immunity. Curr. Opin. Pharmacol. 2005;4:431–437. doi: 10.1016/j.coph.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Crocker P. R. Siglecs: sialic-acid-binding immunoglobulin-like lectins in cell-cell interactions and signalling. Curr. Opin. Struct. Biol. 2002;12:609–615. doi: 10.1016/s0959-440x(02)00375-5. [DOI] [PubMed] [Google Scholar]
- 4.Crocker P. R., Varki A. Siglecs, sialic acids and innate immunity. Trends Immunol. 2001;22:337–342. doi: 10.1016/s1471-4906(01)01930-5. [DOI] [PubMed] [Google Scholar]
- 4a.Staub E., Rosenthal A., Hinzmann B. Systematic identification of immunoreceptor tyrosine-based inhibitory motifs in the human proteome. Cell Signal. 2004;4:435–456. doi: 10.1016/j.cellsig.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Yusa S., Campbell K. S. Src homology region 2-containing protein tyrosine phosphatase-2 (SHP-2) can play a direct role in the inhibitory function of killer cell Ig- like receptors in human NK cells. J. Immunol. 2003;170:4539–4547. doi: 10.4049/jimmunol.170.9.4539. [DOI] [PubMed] [Google Scholar]
- 6.Connolly N. P., Jones M., Watt S. M. Human siglec-5: tissue distribution, novel isoforms and domain specificities for sialic acid-dependent ligand interactions. Br. J. Haematol. 2002;119:221–238. doi: 10.1046/j.1365-2141.2002.03808.x. [DOI] [PubMed] [Google Scholar]
- 7.Razi N., Varki A. Masking and unmasking of the sialic acid-binding lectin activity of CD22 (Siglec-2) on B lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7469–7474. doi: 10.1073/pnas.95.13.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Razi N., Varki A. Cryptic sialic acid binding lectins on human blood leukocytes can be unmasked by sialidase treatment or cellular activation. Glycobiology. 1999;11:1225–1234. doi: 10.1093/glycob/9.11.1225. [DOI] [PubMed] [Google Scholar]
- 8.Nicoll G., Arvil T., Lock K., Furukawa K., Bovin N., Crocker P. R. Ganglioside GD3 expression on target cells can modulate NK cell cytotoxicity via siglec-7-dependent and independent mechanisms. Eur. J. Immunol. 2003;33:1642–1648. doi: 10.1002/eji.200323693. [DOI] [PubMed] [Google Scholar]
- 9.Zaccai N. R., Maenaka K., Maenaka T., Crocker P. R., Brossmer R., Kelm S., Jones E. Y. Structure-guided design of sialic acid-based siglec inhibitors and crystallographic analysis in complex with sialoadhesin. Structure. 2003;11:557–567. doi: 10.1016/s0969-2126(03)00073-x. [DOI] [PubMed] [Google Scholar]
- 10.Kelm S., Gerlach J., Brossmer R., Danzer C. P., Nitschke L. The ligand-binding domain of CD22 is needed for inhibition of the B cell receptor signal, as demonstrated by a novel human CD22-specific inhibitor compound. J. Exp. Med. 2002;195:1207–1213. doi: 10.1084/jem.20011783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alphey M. S., Attrill H., Crocker P. R., van Aalten D. M. F. High resolution crystal structures of siglec-7. Insights into ligand specificity in the Siglec family. J. Biol. Chem. 2003;278:3372–3377. doi: 10.1074/jbc.M210602200. [DOI] [PubMed] [Google Scholar]
- 12.Yamaji T., Teranishi T., Alphey M. S., Crocker P. R., Hashimoto Y. A small region of the natural killer cell receptor, Siglec-7, is responsible for its preferred binding to α 2,8-disialyl and branched α 2,6-sialyl residues. A comparison with siglec-9. J. Biol. Chem. 2002;277:6324–6332. doi: 10.1074/jbc.M110146200. [DOI] [PubMed] [Google Scholar]
- 13.Soulieres D., Rousseau A., Deschenes J., Tremblay M., Tardif M., Pelletier G. Characterization of gangliosides in human uveal melanoma cells. Int. J.Cancer. 1991;49:498–503. doi: 10.1002/ijc.2910490404. [DOI] [PubMed] [Google Scholar]
- 14.Ando T., Ishida H., Kiso M. First total synthesis of α-(2–>3)/α-(2–>6)-disialyl lactotetraosyl ceramide and disialyl Lewis A ganglioside as cancer-associated carbohydrate antigens. Carbohydr. Res. 2003;38:503–514. doi: 10.1016/s0008-6215(02)00465-2. [DOI] [PubMed] [Google Scholar]
- 15.Ito A., Handa K., Withers D. A., Satoh M., Hakomori S. Binding specificity of siglec7 to disialogangliosides of renal cell carcinoma: possible role of disialogangliosides in tumor progression. FEBS Lett. 2001;504:82–86. doi: 10.1016/s0014-5793(01)02734-x. [DOI] [PubMed] [Google Scholar]
- 16.Miyazaki K., Ohmori K., Izawa M., Koike T., Kumamoto K., Furukawa K., Ando T., Kiso M., Yamaji T., Hashimoto Y., et al. Loss of disialyl Lewis(a), the ligand for lymphocyte inhibitory receptor sialic acid-binding immunoglobulin-like lectin-7 (siglec-7) associated with increased sialyl Lewis(a) expression on human colon cancers. Cancer Res. 2004;64:4498–4505. doi: 10.1158/0008-5472.CAN-03-3614. [DOI] [PubMed] [Google Scholar]
- 17.Angata T., Varki A. Siglec-7: a sialic acid-binding lectin of the immunoglobulin superfamily. Glycobiology. 2000;10:431–438. doi: 10.1093/glycob/10.4.431. [DOI] [PubMed] [Google Scholar]
- 18.Crocker P. R., Vinson M., Kelm S., Drickamer K. Molecular analysis of sialoside binding to sialoadhesin by NMR and site-directed mutagenesis. Biochem. J. 1999;341:355–361. [PMC free article] [PubMed] [Google Scholar]
- 19.May A. P., Robinson R. C., Vinson M., Crocker P. R., Jones E. Y. Crystal structure of the N-terminal domain of sialoadhesin in complex with 3′ sialyllactose at 1.85 Å resolution. Mol. Cell. 1998;1:719–728. doi: 10.1016/s1097-2765(00)80071-4. [DOI] [PubMed] [Google Scholar]
- 20.Stanley P., Chaney W. Control of carbohydrate processing: the lec1A CHO mutation results in partial loss of N-acetylglucosaminyltransferase I activity. Mol. Cell Biol. 1985;5:1204–1211. doi: 10.1128/mcb.5.6.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. Sect. A Found. Crystallogr. 1994;50:157–163. [Google Scholar]
- 23.Perrakis A., Morris R., Lamzin V. S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 24.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography and NMR system: a new software system for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A Found. Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 27.Nicoll G., Ni J., Liu D., Klenerman P., Munday J., Dubock S., Mattei M. G., Crocker P. R. Identification and characterization of a novel siglec, siglec-7, expressed by human natural killer cells and monocytes. J. Biol. Chem. 1999;274:34089–34095. doi: 10.1074/jbc.274.48.34089. [DOI] [PubMed] [Google Scholar]
- 28.Kelm S., Pelz A., Schauer R., Filbin M. T., Tang S., de Bellard M. E., Schnaar R. L., Mahoney J. A., Hartnell A., Bradfield P., et al. Sialoadhesin, myelin-associated glycoprotein and CD22 define a new family of sialic acid-dependent adhesion molecules of the immunoglobulin superfamily. Curr. Biol. 1994;4:965–972. doi: 10.1016/s0960-9822(00)00220-7. [DOI] [PubMed] [Google Scholar]
- 29.Lutteke T., Frank M., von der Lieth C. W. Carbohydrate structure suite (CSS): analysis of carbohydrate 3D structures derived from the PDB. Nucleic Acids Res. 2005;33:242–246. doi: 10.1093/nar/gki013. [DOI] [PMC free article] [PubMed] [Google Scholar]