Abstract
We have cloned two genes, FIS1 and FIS2, that control both fertilization independent seed development and postpollination embryo development in Arabidopsis. These genes confer female gametophytic phenotypes. FIS2 encodes a protein with a C2H2 zinc-finger motif and three putative nuclear localization signals, indicating that it is likely to be a transcription factor. FIS1 encodes a protein with homology to the Drosophila Polycomb group gene Enhancer-of-zeste and is identical to the recently described Arabidopsis gene MEDEA. FIS1 is a protein with a number of putative functional domains, including the SET domain present in Enhancer-of-zeste-related proteins. Comparison of the position of the lesions in the fis1 and medea mutant alleles indicates that fis1 is a null allele producing a truncated polypeptide lacking all the protein domains whereas the deduced protein from medea lacks only the SET domain. We present a model of the role of FIS1 and FIS2 gene products in seed development.
Plants have two alternating life phases, a gametophytic phase giving rise to male and female gametes and, after gamete fusion during fertilization, a sporophytic phase initiating with the zygote that develops into the mature plant (1). In most diploid, sexually reproducing plants, seed development begins when the haploid egg and the homodiploid central cells of the female gametophyte are fertilized by two sperm cells, producing a diploid zygote and triploid endosperm, respectively. In apomictic plants, unreduced egg cells or diploid nucellar cells develop parthenogenetically to produce the zygote, and in some cases endosperm development is autonomous. To isolate genes that might control components of apomixis, we have isolated mutants of Arabidopsis in which some stages of seed development are initiated without pollination (2, 3). In fis1 and fis2, autonomous diploid endosperm development progresses to the cellularized stage, and occasionally zygote-like and early embryo-like structures form. In the fis3 mutant, endosperm development stops at the free-nuclear stage and autonomous embryo-like structures have not been seen (3). Another mutant, fie, also showing endosperm development without fertilization and mapping to chromosome 3, is likely to be an allele of fis3 (4). After pollination in fis1, fis2, and fis3 mutants, most embryos are arrested at the globular to torpedo stages. We suggested that the FIS genes define a complex that suppresses the development of the seed, including the embryo and endosperm, in the absence of fertilization, and after fertilization, plays a role in promoting embryo development (3). To further define the roles of these gene products in seed development we have cloned the FIS1 and FIS2 genes.
In this paper we describe the FIS1 and FIS2 genes, their deduced products, and a model of their role in triggering fertilization-independent seed development and postpollination embryo development. Altered regulation of these genes and their homologs might play a role in the development of apomictic seeds.
MATERIALS AND METHODS
Genetic Mapping of fis1 and fis2.
Two separate mapping populations were obtained by crossing line E12 (CS8116, ABRC, Ohio State University (Columbus, OH) (Nossen background), and short integument 1–2 (sin1–2) (Ws background) obtained from Aminesh Ray (Rochester University, Rochester, NY) to fis1/fis1 (L.er background). In E12 a Ds element (DsG) with the reporter gene GUS and NPTII conferring kanamycin resistance (KanR) was located south of angustifolia (an). In the sin1–2 mutant, the SIN1 gene is disrupted by a T-DNA insertion carrying KanR. The resulting F1 hybrids were back crossed to Landsberg erecta (L.er) and the F2 progeny were scored for recombinants between KanR contained in the T-DNA or the DsG and fis1. fis1/KanR recombinants were analyzed with simple sequence length polymorphism markers AthACS, nF21B7, and nT1G11 (http://genome.bio.upenn.edu/sslp_info/sslp_html), and two simple sequence length polymorphism markers found by our group from bacterial artificial chromosome (BAC) T7I23. Plant genomic DNA preparation and Southern blot analyses were performed as previously described (5). BAC IGF 11O10 was obtained from the Resource Center of the German Human Genome Project (Max-Planck-Institut, Berlin).
Plants used for fis2 Restriction Fragment Length Polymorphism analyses were recovered from two F2 populations generated by crosses with morphological markers erecta (er) and asymmetric leaves (as). In cross #1, fis2 er (L.er) plants were crossed to FIS2 ER (Columbia). In cross #2, fis2 AS (L.er) plants were crossed to FIS2 as (Columbia) plants. Recombinant FIS2/fis2 er/er and FIS2/fis2 as/as were analyzed with restriction fragment length polymorphism markers mi277 and m323. YUP9D3 ends were cloned according to published procedures (6). Cosmid 18H1 was analyzed by restriction mapping by using enzymes EcoRI and BamHI (Fig. 1B). The YUP library and the TAMU BAC library were obtained from Joe Ecker (University of Pennsylvania, Philadelphia) and from Rod Wing (Clemson University, Clemson, SC), respectively. The pOCA18 cosmid library was obtained from Neil Olszewski (University of Minnesota, St. Paul, MN).
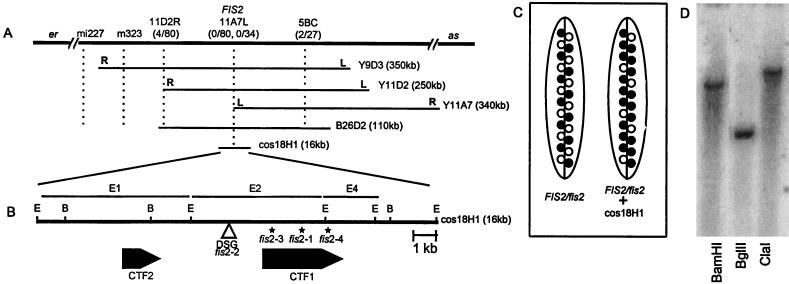
Figure 1.
(A) Position of the FIS2 locus on chromosome II. The relative position of the FIS2 locus and restriction fragment length polymorphism markers m323, mi227, YUP9D3, YUP11D2, YUP11A7 left (L) and right (R) ends, and the BAC 26D2 fragment 5BC are shown. (B) The position of the DsG insertion and the two cDNAs are shown. Asterisks (∗) mark the locations of the mutations in the three ethyl methanesulfonate-induced fis2 alleles. E, EcoRI; B, BamHI. (C) Complementation of fis2 with the cosmid 18H1. The silique on the left is from a FIS2/fis2 plant showing a ratio of 50:50 of normal and fis seeds. On the right is a silique from a FIS2/fis2 plant with an introduced 18H1 FIS2 segment. The ratio of normal to fis seeds is now 75:25 as expected for the complementation of a gametophytic mutation (see data in Table 1). ○, fis seeds; •, FIS seeds. (D) Southern blot of Arabidopsis L.er genomic DNA digested with either BamHI, BglII, or ClaI and hybridized with the 2.38 kb FIS2 cDNA insert.
Tagging of FIS2.
DsG1 and Ac1 lines were obtained from V. Sundaresan (Institute of Molecular Agrobiology, Singapore) and have been described (7). F1 plants of the DsG/Ac1 cross were screened for chimeric plants containing sectors that showed both the female gametophytic mutant phenotype and the fertilization-independent seed phenotype. One out of the 20 plants tested contained a presumptive FIS2/fis2–2 sector. A library was made from DNA of a FIS2/fis2–2 plant and screened with the DNA fragment E2 from cosmid 18H1 and the DsG 5′ end probe.
Complementation of fis2.
Cosmid pOCA18H1 was mobilized in Agrobacterium tumefaciens AGL1, and the T-DNA introduced into Arabidopsis ecotype C24 using the root explant transformation protocol (8). Four independent T1 KanR plants were crossed as females to fis2/fis2. Siliques from KanR and kanamycin-sensitive (KanS) F1 plants were scored for female gametophytic lethality.
cDNA Library Construction and Screening.
Polysomal poly(A) mRNA was extracted from Arabidopsis L.er siliques ranging from heart/torpedo to maturing seed stages using standard procedures (9). A cDNA library was made using the Choice cDNA Synthesis System (GIBCO/BRL, catalog no. 18090) according to the manufacturers protocol except that the size fractionated cDNA was cloned into Lambda Zap II vector (Stratagene). DNA probes derived from the EcoRI fragments E1 and E2 (Fig. 1B) were used to screen 200,000 clones from the cDNA library. Prehybridization and hybridization were performed in 10% PEG6000, 7% SDS, 0.25 M NaCl, 0.05 M NaPO4 (pH 7.2), 1% BSA, 1 mM EDTA at 65°C for 2 hr and 16 hr, respectively. The filters were washed at room temperature (once in 2× standard saline citrate, 1% SDS, once in 1× standard saline citrate, 1% SDS for 30 min each). Lambda phagemid DNA containing the cDNA insert was excised from positive plaques according to the Stratagene protocol.
DNA and Protein Sequence Analysis.
An Applied Biosystems Model 370A DNA Sequencer with fluorescent dye-labeled dideoxy terminators was used for sequencing. FIS2 sequence (exons and introns) from the homozygous fis2 mutants was obtained by PCR amplification of a 3,754-bp region using seven different primer pairs. Sequence of the MEDEA gene from the homozygous fis1 mutant was obtained by PCR amplification of a 4,155-bp segment using five different primer pairs designed from the MEDEA cDNA sequence (10). The sequence data were analyzed by using the GCG software (11). Sequence comparisons were performed using blast searches (12) and multiple sequence alignments were performed and analyzed with the computer programs clustalw (13), genedoc (http://www.cris.com/∼Ketchup/genedoc.shtml), and philip (14).
Southern Hybridization Analysis.
Genomic DNA from Arabidopsis seedlings was prepared by the hexadecyltrimethylammonium bromide protocol (15). Five micrograms of genomic DNA was digested with restriction enzymes before 1% agarose gel electrophoresis. The DNA was then transferred to a Hybond N membrane, prehybridized for 1 hr, hybridized, and the filters washed according to Church and Gilbert (16). The membrane containing the digested genomic DNA was probed with the radiolabeled 2.38-kb FIS2 cDNA insert.
RESULTS
Localization of FIS2.
Genetic mapping localized the FIS2 gene between the markers er and as on chromosome 2 (Fig. 1A). A yeast artificial chromosome clone, YUP9D3, that mapped in this region was used to link the genetic location of FIS2 to the molecular map. Analyses of recombinants at the fis2-er and fis2-as interval indicated that the left end of 9D3 mapped to the as side of the FIS2 gene. By using YUP9D3 as a probe, two additional yeast artificial chromosomes, 11D2 and 11A7 were isolated from the YUP library. The right end of 11D2 mapped to the er side of FIS2 and the left end of 11A7 showed no recombination with FIS2 indicating close proximity of this marker to FIS2 (Fig. 1A).
To locate FIS2 on a DNA fragment smaller than a yeast artificial chromosome, the left end of 11A7 was used as a probe to isolate a BAC clone, 26D2, and the cosmid 18H1. A physical map of BAC 26D2 was constructed with the enzymes BamHI and ClaI, and the fragments were tested on the fis2-er and fis2-as recombinant population to delineate the position of the FIS2 gene. The results of these analyses suggested that the FIS2 gene is contained in fragment E2 of BAC 26D2 (Fig. 1 A and B).
Genetic Complementation of FIS2.
The cosmid 18H1 containing the E2 fragment was introduced into the Arabidopsis ecotype C24 and the transgenic plant was pollinated by a fis2/fis2 plant. KanR F1 plants containing the introduced cosmid with the FIS2 gene were selected and their siliques scored for normal and arrested seeds (Table 1). As fis2 is a female gametophytic mutant, the expected ratio of wild-type to arrested seed in a FIS2/fis2 plant is 50:50. An introduced single copy FIS2 transgene will segregate 1:1 in the gametes so that 50% of the fis2 seeds will contain the transgene and have a FIS2 phenotype. This will shift the ratio to 75:25 in favor of normal seed. In F1 plants derived from four independent transgenic plants containing the E2 fragment, the ratio of arrested to wild-type seeds fitted the 75:25 segregation ratio (Fig. 1C and Table 1). In contrast, a 50:50 ratio was observed in the KanS plants of the same cross. These data show that the cosmid 18H1 does complement the mutant phenotype of fis2 (Table 1) and also suggest that the fis2 mutation is a gametophytic loss-of-function allele.
Table 1.
Ratio of normal:arrested seen in FIS/fis2 plants segregating for the cosmid 18H1
| Line | KanR
progeny (+18H1)
|
KanS progeny
(−18H1)
|
||
|---|---|---|---|---|
| Ratio | X2 | Ratio | X2 | |
| 1 | 179:57 | 0.051 | Not tested | – |
| 6 | 153:55 | 0.23 | Not tested | – |
| 7 | 139:47 | 0.01 | 131:119 | 0.576 |
| 10 | 210:73 | 0.1 | 54:61 | 0.43 |
| Control C24 | NA | – | 211:216 | 0.058 |
KanR plants contain the cosmic and KanS do not. Each line is derived from an independent transformation event. Expected ratios for KanR and KanS plants are 75:25 and 50:50, respectively, if the 18H1 cosmid complements the fis2 mutation. Control cross with C24 shows the segregation of fis2 without complementation. NA, not available.
Isolation of FIS2 cDNA.
Analyses of DNA from FIS2/fis2–2 indicated that the DsG transposon was inserted in the E2 segment of the genomic DNA, further delineating the position of FIS2. The E2 segment, used to probe a late silique cDNA library, hybridized to two cDNA clones of 1.43 kb and 2.38 kb. The 1.43-kb insert was identical to the 3′ end of the 2.38-kb insert indicating that they came from the same gene, CTF1 (Close To FIS #1). The 5′ end of the CTF1 cDNA was 750-bp downstream of the DsG insertion (Fig. 1B). In a second screen, a 1-kb BamHI–EcoRI DNA fragment from E1 identified another cDNA, CTF2, that mapped 2.5 kb upstream of the DsG insertion point (Fig. 1B). Thus, the tagged fis2 allele did not define which of the two cDNAs encodes FIS2.
To ascertain which of the two cDNAs does encode FIS2, both CTF1 and CTF2 sequences of three ethyl methanesulfonate-induced mutant alleles of fis2 were compared with the wild-type sequences. CTF2 sequences in these mutant alleles of fis2 were identical to the wild-type sequence, eliminating this gene as a contender for being the FIS2 gene. Each mutant allele of fis2 contained a different alteration in the CTF1 sequence. In the fis2–1 allele, there is a single nucleotide deletion at position 1,472 of the CTF1 cDNA that induces a frameshift resulting in a stop codon four amino acids downstream of the deletion. A G to A base conversion in the fis2–3 mutant occurs at the 3′ splice junction of intron four of CTF1, altering the consensus splice acceptor sequence AG to AA and probably resulting in missplicing of the FIS2 gene. In fis2–4, a G to A base substitution occurs at position 2,017 of the CTF1 cDNA creating a stop codon. The DsG tagged allele, fis2–2, did not show any nucleotide changes in either the CTF1 or CTF2 sequences. Therefore, DsG could have disrupted a critical regulatory sequence in the upstream region of the FIS2 gene or, the Ds insert may have altered chromatin structure modifying the normal expression of FIS2. These results confirm that CTF1 corresponds to the FIS2 gene. Southern hybridization with the FIS2 probe showed a single band at high and low stringency (Fig. 1D), indicating that FIS2 is encoded by a single copy gene.
Characteristics of FIS2.
Comparison of the FIS2 cDNA and genomic sequences from the Arabidopsis ecotype L.er defined 12 exons and 11 introns. The largest exon (#7) is 1,365-bp long and contains a 66-bp sequence that is repeated 12 times (repeat A) and a 51-bp sequence that is repeated seven times (repeat B). The FIS2 gene from the Arabidopsis ecotype Columbia (Col) contains a 180-bp deletion in exon #7 from nucleotide 1,434 to 1,613 of the L.er FIS2 cDNA spanning one of the B repeats and two of the A repeats. Subsequent PCR analysis of this region in ecotypes L.er, Col, WS, and C24 showed that the deletion was present only in Col (data not shown). Col also contains a 26-bp deletion in intron #6. These deletions do not appear to have any effect on FIS2 function because Col plants do not exhibit a fis phenotype and the cosmid 18H1 that complemented the fis2 mutation in the L.er ecotype originated from the Col ecotype.
FIS2 expression was not detected in shoots, leaves, bolting stem, flower buds or siliques after Northern blot analysis using 20 μg of total RNA for each tissue sample. Similarly, RNAse protection assays did not generate a protected fragment, suggesting that FIS2 is expressed at low levels. The only confirmed expression of the gene is in late silique RNA where a FIS2 cDNA was isolated from the library at a frequency of 1:100,000.
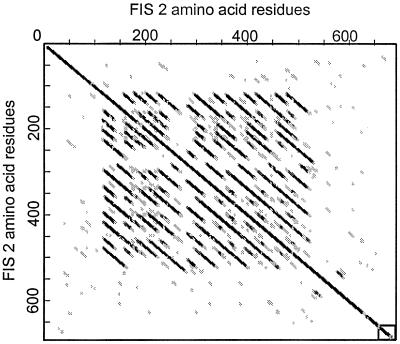
The predicted polypeptide sequence of FIS2 shows a putative C2H2 zinc-finger motif within the first 35 residues of the protein and 3 putative bipartite nuclear localization signals distributed between residues 267–540 (Fig. 2). The presence of the zinc finger motif and nuclear localization signal suggest that FIS2 might be a transcription factor. Like other zinc finger proteins (17), FIS2 has a high serine content (12.9%). The A and B repeats produce a distinctive signature when the FIS2 protein product is compared with itself by dot matrix analysis (Fig. 3). The consensus sequences for A and B repeats are H-V-N-D-D-N-V-S-S-P-P-[R/K]-A-H-S-S-K-K and L-T-T-T-Q-P-A-I-A-E-S-S-E-P-K-V. Following the consensus sequence described above, half of the A repeats have a T-S-D-I sequence and the other half have N-E-S-T. The central part of the A repeat has a consensus SPP[R/K] with similarity to the motif [T/S]PXX (X is usually a basic amino acid) that has been shown to bind in the minor groove of DNA (18).
Figure 2.
Deduced protein sequence of FIS2. The amino acid sequence corresponding to the C2H2 zinc finger motif is double underlined. The three putative nuclear localization signals are underlined with thick lines. The 12 A repeats are shown in filled boxes and the 7 B repeats are shown in open boxes. The [T/S]PXX motifs within the A repeats are underlined with thin lines. The position of the fis2–1 (deletion of T) and the fis2–4 (G → A) mutations are shown with the resulting modifications of the encoded peptide. The location of the fis2–3 mutation (G → A) at the junction of intron 5 and exon 6 is indicated by an arrow (↓). Stop codons are indicated with asterisks (∗).
Figure 3.
Bi-dimensional plot of the FIS2 predicted protein sequence showing the tandem repeats between residues 120 and 520. Within the matrix, the dark dots indicate high homology and open dots indicate no significant homology. The dot matrix was obtained using the software antherprot 3.2 (35) with a window size = 19 and a similarity threshold = 10. The principle of the method is described in Staden (1982) Nucleic Acid Res. 10, 2951–2961.
FIS1 Is Allelic to MEDEA.
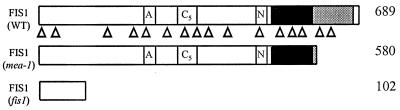
We localized the FIS1 gene to a short region on top of chromosome 1, linked to an (3). Using further genetic and physical mapping, we identified yeast artificial chromosome and BAC contigs that were likely to span the FIS1 gene. While this work was in progress, the medea mutant was described exhibiting embryo arrest in 50% of seeds after fertilization (10). MEDEA maps to the same region as fis1 (U. Grossniklaus, personal communication). MEDEA encodes a SET domain protein (Suppressor of variagation 3-9, Enhancer of zeste, Trithorax) similar to Enhancer-of-zeste (Ez) from Drosophila, a member of the Polycomb group of proteins. We investigated whether medea and fis1 were allelic mutations by sequencing the MEDEA gene from wild-type and homozygous fis1 plants (Fig. 4). In medea, the DsG element is inserted at position 1,755 of the cDNA (10). In fis1 the same cDNA shows a C to T substitution at position 320 creating a stop codon (Fig. 4), indicating that medea is an allele of FIS1.
Figure 4.
Schematic representation of the FIS1 protein. The SET domain is shown as ░⃞. The cysteine-rich domain CXC is shown as ■. Boxes labeled A, N, and C5 represent an acidic domain, a putative nuclear localization signal, and a second conserved cysteine-rich domain. The truncated FIS1 peptides resulting from the mea-1 and fis1 mutations are shown beneath the wild-type FIS1 protein. Numbers on the right indicate the number of amino acid residues in the corresponding FIS1 protein products. Triangles (▵) under the schematic representation of FIS1 protein indicate the position of the 16 introns in the corresponding DNA sequence.
We sequenced the FIS1 genomic region and compared it with the cDNA sequence (10). FIS1 has 17 exons and 16 introns with the introns ranging from 75 bp to 566 bp in length (Fig. 4). There are conserved TATAAT and CCAAT boxes located at positions −123 and −143 relative to the initiating ATG codon. In addition to its similarity to Ez (37% similarity), the FIS1 predicted polypeptide shares homology with two Arabidopsis proteins, CURLY LEAF (CLF) (39% similarity), a regulator of floral homeotic gene expression (19), and EZA1 (49% similarity), a recently identified gene of unknown function (GenBank accession no. AF100163). The C-terminal regions of FIS1 and Ez that contain the conserved SET domain and a cysteine-rich domain (CXC) show 65% sequence similarity.
DISCUSSION
FIS2, a C2H2 Zinc Finger Transcriptional Regulator.
We isolated the FIS2 gene using positional cloning and tagging. The presence of a C2H2 zinc finger signature, and three putative nuclear localization signal motifs suggests that FIS2 encodes a transcriptional regulator. Other than these motifs the FIS2 sequence does not have extensive homology to any sequence in the database. Several members of the C2H2 zinc-finger protein family, also known as the TFIIIA-like zinc finger protein gene family, play important roles in growth and development in Drosophila (20, 21) and in plants (22). Unlike animal C2H2 zinc finger proteins that contain multiple zinc finger motifs, present as tandem arrays, the plant C2H2 zinc finger proteins contain only one to four fingers (22). The region in the center of the FIS2 protein containing A and B type repeats (Fig. 2), with most of the A-type repeats containing a [T/S]PXX motif, could form a 3-dimensional structure involved in protein-protein interactions. Alternatively, it could form fingers where the [T/S]PXX motifs positioned at the tip of each finger could directly bind A+T-rich DNA sequences. Histones H1, H2A, and H2B are essential for chromatin condensation and each of them contains several [T/S]PXX motifs (23, 24).
FIS1 Is Related to the Polycomb Group Protein Ez of Drosophila.
We determined that the FIS1 gene is the same as MEDEA, a recently described gene related to the Polycomb-group gene Ez from Drosophila (10). Mutants of Ez exhibit defects in oogenesis, maternal-effect lethality, and zygotic lethality (25). Several lines of evidence indicate that Ez-related proteins play a central role in chromosome architecture and the nucleation of repressive protein complexes in chromatin (26). Comparison of the sequences between fis1 and medea indicates that whereas all of the characteristic domains of Ez-related proteins are eliminated by the N-terminal stop codon in the fis1 allele, all domains except the SET domain remain intact in the C-terminal disruption of the medea mutant allele (Fig. 4). fis1 exhibits an autonomous endosperm phenotype in 50% of seeds and an arrested embryo phenotype after pollination (3); medea has been reported to have the arrested embryo phenotype (10) but also shows a weak autonomous endosperm phenotype (27).
Do Caenorhabditis and Arabidopsis Use Similar Genes to Maintain their Germlines?
In Caenorhabditis elegans, an Ez homolog (MES2) appears to play a central role in maintaining a repressed chromatin state in the germline (28). Active MES genes are essential for normal germline development (29). It has been suggested that MES proteins act in a complex that is required to maintain a germline specific organization of chromatin from one generation to the next and that this chromatin state is essential to initiate the correct pattern of gene expression in the germline. Tandem arrays of transgenes are efficiently expressed in somatic cells but are specifically silenced in the germline of wild-type worms (30). In mes mutant lines, these tandem transgenes are activated in the germline, supporting the concept that the MES proteins repress gene expression through an influence on chromatin state. The similarities between FIS1 and MES2 in being Ez homologs and in repressing germline functions suggest that FIS1 may also act by maintaining a repressed chromatin state. Another FIS gene, FIS3, may be a homolog of another MES gene such as MES6 (a homolog of Extra sex combs in Drosophila) (28). Mutation in all three fis loci leads to autonomous partial seed development so perhaps they too, like the MES proteins, may act in a complex with cooperative roles in maintaining a repressed seed development program, normally relieved by pollination.
In C. elegans, the PIE-1 protein keeps the germline blastomeres transcriptionally quiescent whereas the surrounding somatic cells become transcriptionally active (31). When PIE-1 decays, the primordial germ cells become transcriptionally active and the MES genes are required to repress certain genes. PIE-1 is a zinc finger protein, as is FIS2. FIS2 may play a role analogous to that of PIE-1.
Model of Action of the FIS Genes.
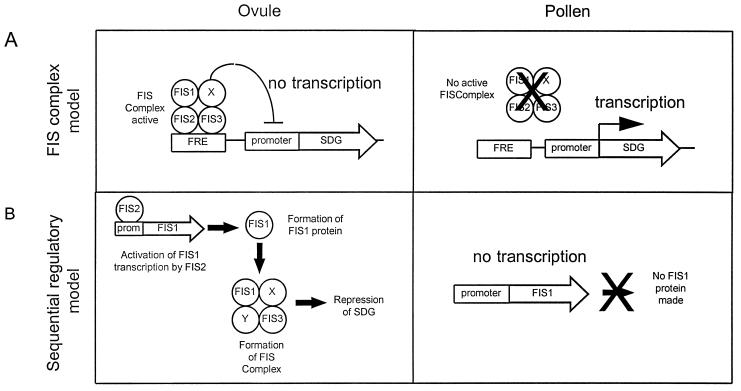
Formerly, we presented a model in which the FIS gene products act in a complex to repress genes required for seed development (3). We can now modify this model to incorporate our new data that suggest that FIS2 is a transcription factor and FIS1 a homolog of the Polycomb group protein Ez (Fig. 5A). In the revised model, FIS1 represses the activity of one or more seed development genes (SDG) and FIS2, together with FIS1 and FIS3, maintains the repressed state of the SDG. A mutation to a nonfunctional allele in any of FIS1, FIS2, or FIS3 would result in derepression of the seed development genes. SDG expression may trigger heterochronic developmental steps, such as cell division in the central or egg cells that are normally quiescent in the absence of pollination. These steps may initiate a cascade of developmental events leading to partial seed development.
Figure 5.
Model for the role of FIS protein products in seed development. The cartoon represent possible roles of FIS protein in maintaining Seed Development Gene (SDG) repression. Molecular events taking place in ovule and pollen are described. X and Y represent additional proteins that could be part of the FIS complex. (A) The “FIS complex” model. FIS1, 2, and 3 bind as a complex (FIS complex) to specific sites in SDG regulatory regions termed FRE for FIS Response Element. The FIS complex represses by interfering with the basal transcription machinery. (B) The “Sequential Regulatory” model. This model proposes that FIS2 transcriptionally regulates FIS1 and/or other FIS gene(s) and subsequently forms an active FIS complex, repressing SDG. In both models, FIS genes are not active in pollen so that SDG can be active at the appropriate stage of development.
During normal seed development, fertilization may introduce derepressed SDG from pollen to initiate seed development. The derepression of SDG in pollen could be a result of epigenetic silencing of one or more of the FIS genes in the male gamete. These processes resemble genomic imprinting that has been shown to occur in plant endosperm (32, 33).
In a second model (Fig. 5B), the FIS2 product positively regulates the FIS1 gene that represses SDG as part of a silencing mechanism. In fis2 mutants, FIS1 would not be expressed and thus FIS1 mediated silencing of SDG does not occur. FIS3 may also be under FIS2 control and/or may be in a functional complex with FIS1. This sequential model is based on the indication that FIS2 is a transcription factor and that FIS1 may play a role in gene repression. This model predicts that FIS1 gene expression would be down-regulated in a fis2 mutant.
The FIS genes also play a positive role in embryo development after pollination. In fis mutants, after pollination, embryo development is generally incomplete, with embryos arrested at the globular to torpedo stages; occasional embryos develop to maturity (3). In fis mutants, the auto-activation of genes in the female gametophyte might initiate an abnormal development pattern leading to embryo arrest even in fertilized ovules. The occasional escapees that develop to mature seeds presumably result from the leakiness of the mutation. The Polycomb group protein Ez of Drosophila has been associated with a gene activating function during particular developmental stages and acting through a specific promoter complex (34). FIS1 may have an activating role, again interacting in some way with FIS2 (and FIS3).
Apomixis requires many components of the reproductive processes to be integrated to produce autonomous viable seeds. Among these components are autonomous endosperm development and the initiation of zygotic development in the absence of fertilization. Both of these are seen to varying degrees in the fis mutants suggesting that the FIS genes may play a role in apomictic seed development and that apomixis may involve altered regulation of these genes.
Acknowledgments
We thank Janice Norman and Bjorg Sherman for technical support, Chris Helliwell for helping with the cloning the FIS2 gene, and Ueli Grossniklaus for disclosing the map location of MEDEA. This research has been supported by the Rockefeller Foundation and by the Australian Center for International Agricultural Research.
ABBREVIATIONS
- BAC
bacterial artificial chromosome
- KanR
kanamycin resistance
Footnotes
References
- 1.Reiser L, Fischer R L. Plant Cell. 1993;5:1291–1301. doi: 10.1105/tpc.5.10.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peacock W J, Luo M, Craig S, Dennis E S, Chaudhury A. Induced Mutations and Molecular Techniques for Crop Improvement. Vienna, Austria: IAEA; 1995. pp. 117–125. [Google Scholar]
- 3.Chaudhury A M, Ming L, Miller C, Craig S, Dennis E S, Peacock W J. Proc Natl Acad Sci USA. 1997;94:4223–4228. doi: 10.1073/pnas.94.8.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohad N, Margossian L, Hsu Y C, Williams C, Repetti P, Fischer R L. Proc Natl Acad Sci USA. 1996;93:5319–5324. doi: 10.1073/pnas.93.11.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapple R M, Chaudhury A M, Blomer K C, Farrell L B, Dennis E S. Aust J Plant Physiol. 1996;23:453–465. [Google Scholar]
- 6.Hermanson G G, Hoekstra M F, McElligott D L, Evans G A. Nucleic Acids Res. 1991;19:4943–4948. doi: 10.1093/nar/19.18.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sundaresan V, Springer P, Volpe T, Haward S, Jones J D G, Dean C, Ma H, Martienssen R. Genes Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- 8.Valvekens D, Van Montagu V, Van Lijsebettens M. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox K H, Goldberg R B. In: Plant Molecular Biology; A Practical Approach. Shaw C H, editor. Oxford: IRL; 1988. pp. 1–34. [Google Scholar]
- 10.Grossniklaus U, Viellecalzada J P, Hoeppner M A, Gagliano W B. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- 11.Devereux J, Haeberli P, Smithies O. Nucleic Acid Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
- 15.Taylor B, Powell A. Focus. 1982;4:4–6. [Google Scholar]
- 16.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;83:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tague B W, Goodman H M. Plant Mol Biol. 1995;28:267–279. doi: 10.1007/BF00020246. [DOI] [PubMed] [Google Scholar]
- 18.Churchill M E A, Suzuki M. EMBO J. 1989;8:4189–4195. doi: 10.1002/j.1460-2075.1989.tb08604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz E M, Coupland G. Nature (London) 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- 20.Stanojevic D, Hoey T, Levine M. Nature (London) 1989;341:331–335. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- 21.Treisman J, Desplan C. Nature (London) 1989;341:335–337. doi: 10.1038/341335a0. [DOI] [PubMed] [Google Scholar]
- 22.Takatsuji H. Cell Mol Life Sci. 1998;54:582–596. doi: 10.1007/s000180050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koning A J, Tanimoto E Y, Kiehne K, Rost T, Comai L. Plant Cell. 1991;3:657–665. doi: 10.1105/tpc.3.7.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki M. EMBO J. 1989;8:797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones R S, Gelbart W M. Genetics. 1990;126:185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenuwein T, Laible G, Dorn R, Reuter G. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grossniklaus U, Viellecalzada J P. Trends Plant Sci. 1998;3:328–328. [Google Scholar]
- 28.Holdeman R, Nehrt S, Strome S. Development (Cambridge, UK) 1998;125:2457–2467. doi: 10.1242/dev.125.13.2457. [DOI] [PubMed] [Google Scholar]
- 29.Garvin C, Holdeman R, Strome S. Genetics. 1998;148:167–185. doi: 10.1093/genetics/148.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly W G, Fire A. Development (Cambridge, UK) 1998;125:2451–2456. doi: 10.1242/dev.125.13.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seydoux G, Mello C C, Pettitt J, Wood W B, Priess J R, Fire A. Nature (London) 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- 32.Haig D, Westoby M. Philos Trans R Soc London B. 1991;333:1–13. [Google Scholar]
- 33.Kermicle J L, Alleman M. Development (Cambridge, U.K.) 1990. pp. 9–14. [PubMed] [Google Scholar]
- 34.Lajeunesse D, Shearn A. Development (Cambridge, UK) 1996;122:2189–2197. doi: 10.1242/dev.122.7.2189. [DOI] [PubMed] [Google Scholar]
- 35.Geourjon C, Deleage G. J Mol Graph. 1995;13:209–212. doi: 10.1016/0263-7855(95)00035-5. [DOI] [PubMed] [Google Scholar]