Abstract
Members of the protein family called ATPases associated with various cellular activities (AAA+) play a crucial role in transforming chemical energy into biological events. AAA+ proteins are complex molecular machines and typically form ring-shaped oligomeric complexes that are crucial for ATPase activity and mechanism of action. The Escherichia coli transcription activator phage shock protein F (PspF) is an AAA+ mechanochemical enzyme that functions to sense and relay the energy derived from nucleoside triphosphate hydrolysis to catalyze transcription by the σ54-RNA polymerase. Closed promoter complexes formed by the σ54-RNA polymerase are substrates for the action of PspF. By using a protein fragmentation approach, we identify here at least one σ54-binding surface in the PspF AAA+ domain. Results suggest that ATP hydrolysis by PspF is coupled to the exposure of at least one σ54-binding surface. This nucleotide hydrolysis-dependent presentation of a substrate binding surface can explain why complexes that form between σ54 and PspF are transient and could be part of a mechanism used generally by other AAA+ proteins to regulate activity.
Numerous cellular activities rely upon ATP binding and hydrolysis by mechanochemical enzymes. Sequence analysis and structural comparisons have revealed a major protein family [ATPase associated with various cellular activities (AAA+)] that functions in diverse cellular activities in all three kingdoms of life (1). Members of the AAA+ family are typified by their ability to self-assemble into oligomeric structures and have the conserved function of converting the energy derived from ATP hydrolysis into mechanical force to remodel their target substrates. Substrates for AAA+ proteins include large protein complexes, nucleic acids, and nucleoprotein complexes (1, 2). How ATP binding and hydrolysis are coupled to function in AAA+ proteins is not well understood at a mechanistic level.
Transcription activator proteins that target the bacterial RNA polymerase (RNAP) associated with the enhancer-dependent promoter specificity factor σ54 are AAA+ proteins (3–5). In bacteria, two classes of σ factors exist, the σ70 and the σ54 class (6). Even though members of both classes bind to a common core RNAP, the two types of holoenzyme are differently regulated. Unlike the σ70 class, the σ54–RNAP forms closed promoter complexes that rarely spontaneously isomerize to form open promoter complexes from which transcription can initiate. Efficient conversion to open complexes of σ54–RNAP closed complexes strictly depends on NTP hydrolysis by σ54 AAA+ activator proteins (7).
The major issues that surround functioning of AAA+ proteins are (i) how NTP binding and hydrolysis lead to new functional states that have the discrete properties needed to interact with and remodel their targets, and (ii) where the substrate-binding surfaces are localized. Recently, we showed that stable complex formation between some AAA+ activator proteins and σ54 required the presence of ADP-AlFx, a transition state analogue of ATP at the point of hydrolysis (8). We proposed that NTP hydrolysis permits conformational changes in σ54-dependent activators that permit stable binding of the activator to σ54, an interaction important for efficient energy coupling that leads to open complex formation (5, 8). Unstable NTP-independent complexes between σ54 and the activator DctD have been demonstrated, and may occur before NTP hydrolysis (4, 9–10).
To investigate how NTP regulates the activity of σ54-dependent activators, we conducted protein-fragmentation studies on one σ54 activator protein, the Escherichia coli σ54-activator phage shock protein F (PspF) that is involved in stress response (11, 12). The objective was to obtain biochemically active partial sequences of PspF that contained σ54-binding determinants. Our results led to the identification of residues in PspF that interact directly with σ54, indicating that NTP hydrolysis exposes at least one σ54 interacting surface in PspF. Comparison with known AAA+ protein structures suggests that the σ54-binding surface in PspF is an “insertion” within the AAA+ domain that is subjected to movements upon NTP hydrolysis (5), hence linking nucleotide interactions to substrate remodeling.
Materials and Methods
Plasmids and Strains.
Plasmid pJS101 (pET28b+∷pspFΔHTH) encodes E. coli PspF residues 1–292 with expression controlled by the T7 promoter in vector pET28b+ and was the template for plasmid pPB1 encoding PspF residues 1–275. DNA encoding residues 1–275 was amplified by using the T7 (Novagen) and Fcd (5′-ACGTAAAAAGCTTCACAGTGTTGGAAGCGAGGT-3′) primers (underlined sequence indicates the restriction site for HindIII) and cloned into NdeI-HindIII-digested pET28b+ (Novagen). pPB1 encodes PspF residues 1–275 as an amino-terminal 6 His-tagged fusion protein.
pPB1 was template to amplify DNA encoding PspF fragments F1–4 by using appropriate primers for PspF F1 (residues 69–134), PspF F2 (residues 69–102), PspF F3 (residues 89–134), and PspF F4 (residues 69–93). PCR fragments were inserted into NcoI-HindIII-digested pMBP∷Parallel1 (13). Plasmids pPB2, pPB3, pPB4, and pPB5 encode PspF fragments F1-F4, respectively, in-frame with the E. coli malE gene. Mutant PspF fragments F4 were constructed as described above by using pPB1 harboring the relevant mutations Ser-75→Ala (S75A), H80A, T86A, and T86S as the template.
E. coli strain JM109 containing the σ54-dependent transcriptional pspAp-lacZ fusion carried by the tetracycline-resistant plasmid pSJ1 (S. E. Jones and M.B., unpublished work), a derivative of pMR25 (gift of M. R. K. Alley), was used to measure PspF1–275 activity in vivo. pPB1 derivatives containing the mutant forms of pspF1–275 was XbaI-HindIII-digested, and the mutant sequences were cloned into the chloramphenicol-resistant pACT3 plasmid (14) so as to express the wild-type or mutant PspF1–275 proteins under the control of a tac promoter.
β-Galactosidase Assays.
Overnight cultures of E. coli strain JM109∷pSJ1 (pspA-lacZ fusion) containing pACT3-pspF1–275 derivative plasmids were diluted 100-fold into LB with the appropriate antibiotics. After 3 h at 37°C, 1 mM isopropyl β-d-thiogalactoside (IPTG) was added and β-galactosidase activities were determined after a 2-h incubation. β-galactosidase activities were assayed as described (15) and expressed as units per OD600 (Miller units).
Proteins.
Plasmids pPB2, pPB3, pPB4, and pPB5 were transformed into E. coli strain TB1 (New England Biolabs), and MBP fusions were overexpressed. Freshly transformed TB1 cells (≈100–200 cfu) were used to inoculate 1 liter of LB media and grown at 37°C. At OD600 of 0.4–0.6, the cultures were induced with 1 mM IPTG, and the PspF fragments were overexpressed for 2 h. Harvested cells were resuspended in 10 mM Na2HPO4, pH 7.0/30 mM NaCl/0.05% (vol/vol) Tween 20/10 mM 2-mercaptoethanol/10 mM EDTA/1 mM PMSF and disrupted by using a French pressure cell; subsequently, they were lysate-centrifuged (15,000 × g for 30 min) and NaCl was added to 0.5 M. Supernatant was loaded onto 2 ml of amylose resin column (New England Biolabs), the column was washed with 10 mM Na2HPO4, pH 7.2/0.5 M NaCl/10 mM 2-mercaptoethanol/1 mM EDTA, and proteins were eluted with 10 mM maltose. Fractions containing the fusion protein were pooled, dialyzed against storage buffer (10 mM Tris⋅HCl, pH 8.0/50 mM NaCl/50% (vol/vol) glycerol/0.1 mM EDTA/1 mM DTT) and stored at −80°C.
PspF1–275 was purified as described (16). For 32P end-labeling, a heart muscle kinase-tagged Klebsiella pneumoniae σ54 was purified, as described (17). K. pneumoniae σ54 (residues 1–477), its derivative ΔR1σ54 lacking region 1 (residues 57–477), and σ54 region 1 (residues 1–56) were purified as described (18, 19). E. coli core RNAP was purchased from Epicentre Technologies (Madison, WI).
ATP Binding and Hydrolysis Assays.
ATP binding assays by UV crosslinking were performed as described (20). Briefly, 20-μl samples contained 5 μM PspF1–275 in reaction buffer A (35 mM Tris-acetate, pH 8.0/70 mM K-acetate/5 mM Mg-acetate/19 mM NH4-acetate/0.7 mM DTT) and with 0.75 μCi [γ-32P]ATP (3,000 Ci/mmol; 1 Ci = 37 GBq) ± 3 mM ATP. Samples were illuminated by a UV lamp (254 nm, UVG-54, Ultraviolet Products, San Gabriel, CA) at a 3-cm distance (15 min, 0°C) and analyzed by 15% SDS gel. The dried gel was visualized by phosphorimaging (Fuji Bas-1500).
The ATPase assay was performed in reaction buffer A with 2 μM PspF1–275, as described (11). Released Pi was separated from ATP by using thin layer chromatography as described (21) and measured by phosphorimaging.
Native Gel Mobility Shift Assays.
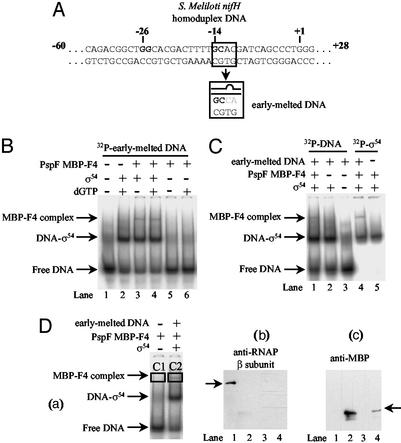
Sinorhizobium meliloti nifH homoduplex promoter probe and a heteroduplex variant that mimics the conformation of promoter DNA at position −12 (where DNA melting originates) as it exists within closed promoter complexes (by having a 2-bp mismatch next to the conserved GC-region of σ54-dependent promoters), called the early-melted probe (see Fig. 2A), were constructed as described (22, 23). Binding reactions were performed in 25 mM Tris-acetate, pH 8.0/8 mM Mg-acetate/100 mM KCl/1 mM DTT/3.5% (wt/vol) PEG-6000 with 16 nM promoter probe end-labeled on one strand with T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol). Where indicated, σ54, ΔR1σ54, or purified σ54 region 1 was present at 0.5 μM, and NTPs were present at 2 mM. For assays with PspF MBP-F4, the σ54–early-melted probe complex was formed (see Fig. 2A; ref. 22), and 4 μM PspF MBP-F4 was added for 10 min before resolving on a 4.5% native polyacrylamide gel. Native gels were run in 25 mM Tris/200 mM glycine buffer, pH 8.6. Complexes were detected by phosphorimager (Fuji BAS-1500) analysis. With 32P-end-labeled proteins, 50 ng of α-lactoalbumin was included in the binding reactions.
Figure 2.
Stable binding of PspF fragment MBP-F4 to a σ54–early-melted promoter probe complex. (A) Sequences (residues shown are −35 to +3) of the S. meliloti homoduplex and early-melted promoter probes. (B) Native gel showing the NTP-independent binding of PspF MBP-F4 to early-melted promoter probe bound σ54. The background smear migrating at the same position as the σ54–early-melted probe complex is present in all lanes, even if protein is added. (C) Native gel showing that σ54 is part of the PspF MBP-F4-dependent complex. Reactions were carried out as in B but, where indicated, contained either 16 nM 32P-end-labeled early-melted DNA (lanes 1–3) or 1 μM 32P-end-labeled σ54 (lanes 4–5). (D) Western blot analysis of the protein content of the PspF MBP-F4-dependent complex. (a) Native gel showing PspF MBP-F4 complex formation. Proteins in the boxed complexes were blotted by using antibodies against the anti-RNAP β-subunit (b) and E. coli anti-MBP protein (c). Lanes 1 and 2 contain purified E. coli core RNAP and PspF MBP-F4, respectively. Lanes 3 and 4 contain the contents of the gel-isolated complexes C1 and C2, respectively. Arrows indicate the migration position of the E. coli RNAP and purified PspF MBP-F4.
Immunoblotting.
PspF MBP-F4 complexes were separated on native-gels and visualized by autoradiography of the wet gel. Bands were excised and applied to a 12.5% SDS gel. After electrophoresis, proteins were transferred onto nitrocellulose membranes by using a semidry transblot system (Bio-Rad). Antibodies specific for maltose binding protein (MBP; New England Biolabs), the RNAP β-subunit (gift of Akira Ishihama, Tokyo), and alkaline phosphatase-conjugated anti-rabbit IgG (Promega) were used.
DNase I Footprinting.
Binding reactions were conducted as described above but with 50 nM early-melted promoter probe (template strand end-labeled). DNase I footprinting was conducted essentially as described (23, 24). Briefly, DNase I (1.75 × 103 units, Amersham Pharmacia) was added to a 10-μl binding reaction for 1 min, and the reaction was stopped by adding 10 mM EDTA. Complexes were separated on native gels, DNA was excised and eluted into 0.1 mM EDTA, pH 8.0, at 37°C overnight. Recovery of isolated DNA was determined by dry Cerenkov counting, and equal numbers of counts were loaded onto a denaturing 10% polyacrylamide gel. The dried gel was visualized by phosphorimaging.
In Vitro Transcription Assays.
The template for the in vitro transcription assays was the supercoiled plasmid pSLE1 (11), which contains the E. coli pspA promoter. Activator-dependent transcription was performed as described (25). Reactions were done at 37°C and contained 100 mM holoenzyme (1:5 ratio of core RNAP:σ54). After activation with 4 μM PspF1–275, the reactions were incubated for 20 min to allow open complex formation. To allow synthesis of transcripts, heparin (100 μg/ml−1, final concentration), a mixture of rNTPs and 12.5 μCi [α-32P]UTP (800 Ci/mmol) were added for an additional 10 min. The reactions were stopped with 4 μl of formamide loading dye, heated to 95°C for 3 min, and half the sample was run directly on a 6% denaturing sequencing gel. Dried gels were visualized by phosphorimaging.
V8 Protease Footprinting.
Reactions (20 μl) contained 10 μM PspF1–275, 50 nM of 32P-end-labeled σ54, and 150 nM early-melted DNA probe (see above). After incubation for 10 min at 37°C, 150 ng of V8 protease (Sigma) was added for 2 min, followed by the addition of 500 μM dichloroisocoumarin to stop digestion. Reactions were run on a 4.5% native gel to separate promoter-bound σ54 from free σ54. Promoter-bound σ54 complexes were isolated and analyzed on a 15% SDS gel.
Results
PspF Fragment MBP-F4 Stably Binds a Binary σ54–DNA Complex in an NTP-Independent Manner.
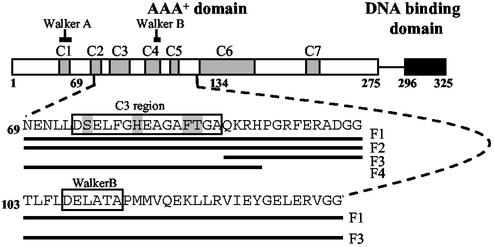
Four fragments of E. coli PspF (Fig. 1) were chosen on the basis of known defects of members of the σ54 activator proteins that identified sequences important for interacting with σ54 (10, 26, 27). These fragments variably contain the Walker B motif associated with nucleotide binding and/or the GAFTGA amino acid sequence, which is strongly implicated in productive engagement of the σ54–RNAP but which contains some residues not required for ATP hydrolysis per se (Fig. 1 and Table 1; refs. 10, 26–29). The end point of the fragments was based on structure considerations using a model of PspF based on the AAA+ protein p97 for guidance (5). Fragments with “complete” secondary structure elements were chosen. The fragments were purified as MBP fusion proteins (hereafter called PspF MBP-F1 to MBP-F4).
Figure 1.
Schematic showing the domain organization of E. coli PspF (to scale). PspF contains 325 amino acid residues and is composed of two domains. The carboxyl-terminal domain (residues 296–325, highlighted in black) contains a helix-turn-helix DNA-binding domain. (Top) The amino-terminal AAA+ domain (residues 1–275) is directly responsible for ATP hydrolysis and transcriptional activation. This domain is highly conserved among σ54 activators; the seven conserved regions (C1–C7) are indicated in gray. The Walker A and Walker B motifs involved in NTP binding and hydrolysis are indicated. The amino acid sequences of the PspF fragments F1 (residues 69–134), F2 (residues 69–102), F3 (residues 89–134), and F4 (residues 69–93) are shown. (Middle) C3 region containing the signature motif of σ54 activators, D/ESELFGH and GAFTGA, is shown in box. (Bottom) Walker B motif is boxed. Residues that were targeted for mutagenesis are highlighted (see text).
Table 1.
Properties of mutant forms of PspF1–275
| Amino acid substitution | Transcriptional activation in vivo, Miller units* | Transcriptional activation in vitro, arbitrary units† | ADP-AlFx-dependent oligomerization of PspF‡ | ADP-AlFx-dependent binding of PspF to σ54§ | ATP binding¶ | ATP hydrolysis, turnover min−1‖ |
|---|---|---|---|---|---|---|
| None | 878 | 223 | + | + | + | 13.5 |
| S75A | 12 | <0.1 | − | − | + | 0.9 |
| H80A | 12 | 1.54 | − | − | + | 0.1 |
| F85A | 12 | <0.1 | − | − | + | 1 |
| F85L | 11 | <0.1 | − | − | + | 1.5 |
| T86A | 12 | <0.1 | − | − | + | 23.2 |
| T86S | 786 | 187 | −** | + | + | 20.9 |
Expression of β-galactosidase from pspAp-lacZ fusions with mutant PspF1–275 proteins in E. coli. PspF1–275 proteins were overexpressed from pACT3-pspF1–275 derivative plasmids. β-Galactosidase values are averaged from triplicate determinations with two different isolates of each strain. Standard error was <10%. The level of β-Galactosidase activity in the absence of pACT3 derivative plasmid is 8.
pspAp-specific transcription, as described in Materials and Methods in the presence of 4 μM PspF1–275. The amount of transcript is in arbitrary units and was quantified by PhosphorImager analysis.
Indicates the ability of the PspF1–275 mutants to oligomerize in the presence of ADP-AlFx (8). +, wild-type levels of oligomerization. −, no oligomerization was detected.
Indicates the ability of σ54 to form a stable complex with the oligomerized PspF1–275 (8).
The ATP binding activities of the variant forms of PspF1–275 were measured with 5 μM purified protein, as described in Materials and Methods. +, wild-type levels of ATP binding.
The ATPase activities of the variant forms of PspF1–275 were measured with 2 μM purified protein after a 1-h reaction, as described in Materials and Methods. Hydrolysis was measured as the release of 32Pi, which was detected by TLC and quantified by PhosphorImager analysis.
The failure to see ADP-AlFx-dependent oligomerization with PspF1–275 harboring the T86S mutation is unexpected and has been explored in ref. 8.
Initially, we screened for the ability of the four PspF MBP-fragments to interact with σ54 in the presence and absence of ADP-AlFx by native PAGE. No stable σ54-PspF fragment complexes were detected (data not shown). Because promoter-bound σ54 constitutes a target for the activator, we tested the PspF fragments for binding to S. meliloti nifH-promoter DNA-bound σ54. By using these conditions, no complexes were detected with the PspF fragments (data not shown). Nucleotide hydrolysis-dependent productive interactions between PspF and σ54 do occur when σ54 is bound to the early-melted DNA, a promoter probe that mimics the state of the DNA in closed promoter complexes as formed with the holoenzyme (Fig. 2A, ref. 23, and see Materials and Methods). Strikingly, when we screened the PspF-fragments for stable binding to σ54 under conditions where σ54 was bound to the early-melted probe, we detected a slower migrating complex in reactions containing PspF fragment MBP-F4 (Fig. 2B; hereafter called the PspF MBP-F4 complex). The presence of NTPs (dGTP, GTP, or ADP-AlFx) had no obvious effect on the formation of this PspF MBP-F4-dependent complex (Fig. 2B and data not shown). Binding of PspF MBP-F4 to the σ54–early-melted probe complex was not competed out by the intact AAA+ domain of PspF (residues 1–275, hereafter called PspF1–275; Fig. 1), which is consistent with the view that at least one σ54-binding site was inaccessible before binding to ADP-AlFx or NTP hydrolysis (data not shown).
We analyzed the composition of the PspF MBP-F4 complex by immunoblotting with antibodies against MBP (to confirm the presence of PspF MBP-F4) and the RNAP β-subunit (to rule out contaminating core RNAP) and by using 32P-labeled σ54. Results shown in Fig. 2 C and D establish that the PspF MBP-F4 complex contained σ54 and the MBP-tagged PspF F4, but not core RNAP. We conclude that the σ54 that bound to the early-melted probe is in a conformational state favorable for detectable interaction with PspF MBP-F4. The basis of the failure for fragments MBP-F1, MBP-F2, and MBP-F3 to bind to σ54–early-melted probe complexes is not understood, but could be related to masking effects of adjacent sequences or simply to proper protein folding.
The early-melted promoter probe contains a segment with double-stranded and single-stranded DNA juxtaposed, the fork junction structure. Interactions between σ54 and the fork junction are crucial for regulated transcription by the σ54–RNAP (22, 30, 31). Binding assays with PspF MBP-F4 and promoter probe variants containing a fork junction at −12 but lacking downstream DNA sequences (essentially, truncated variants of the early-melted probe) resulted in PspF MBP-F4 complex formation (data not shown). This result suggests that binding of PspF MBP-F4 to σ54 does not require the presence of DNA downstream of the −12 position (with respect to the transcription start site at +1), but does critically require the fork-junction structure.
Amino-Terminal 56 Residues of σ54 Are Required for Complex Formation with PspF MBP-F4.
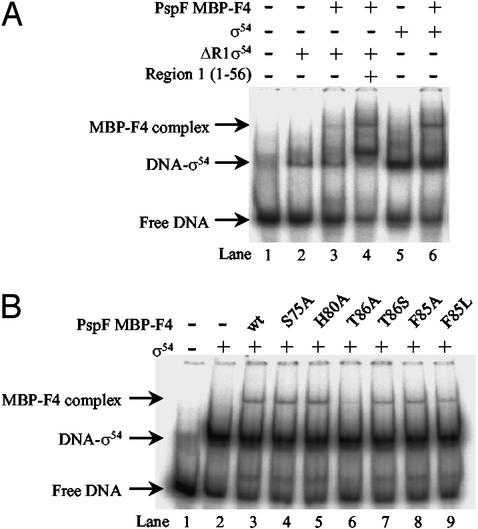
In natural σ54–RNAP closed complexes, distortion of the DNA next to the consensus promoter GC element at −12 (where DNA melting originates) depends upon region 1 of σ54 (32). Because σ54 region 1 plays a central role during regulated transcription activation (3) and binds to PspF (8), we examined the involvement of σ54 region 1 in binding PspF MBP-F4 by using an amino-terminal-truncated form of σ54 that lacks region 1 residues 1–56 (ΔR1σ54). ΔR1σ54 binds the early-melted probe far less efficiently than the wild-type σ54 (Fig. 3A, compare lanes 2 and 5). The PspF MBP-F4 complex formed far less efficiently (by more than 10-fold) with the ΔR1σ54 bound to the early-melted probe (Fig. 3A, lane 3). However, addition of region 1 sequences in trans markedly improves the binding activity of ΔR1σ54 to the early-melted probe and the amount of PspF MBP-F4 complex formation (Fig. 3A, lane 4). Region 1 alone does not bind the early-melted probe or PspF MBP-F4 (data not shown). Overall, these results indicate that formation of the PspF MBP-F4 complex requires region 1-dependent binding of σ54 to the early-melted probe and may involve contact of PspF MBP-F4 to Region 1.
Figure 3.
Region 1 of σ54 and the GAFTGA motif of PspF are required for stable formation of the PspF MBP-F4-σ54–early-melted DNA complex. (A) Native gel showing the reduced ability of PspF MBP-F4 to form a stable complex with ΔR1σ54 bound to the early-melted promoter probe. For the reaction shown in lane 4, σ54 region 1 was added in trans to the ΔR1σ54–early-melted promoter probe complex before adding PspF F4. (B) Native gel showing the ability of mutant PspF MBP-F4 fragments to form complexes with early-melted bound σ54.
The Integrity of the GAFTGA Motif Is Essential for PspF MBP-F4 Complex Formation.
It has been suggested that the highly conserved region C3, in the AAA+ domain of σ54-dependent activators (Fig. 1), plays an important role in coupling NTP hydrolysis to biological output (10, 26, 27). A six-amino acid motif (GAFTGA; residues 82–86) within the C3 region is strongly implicated in interactions with σ54 (10, 26–28). Parts of the GAFTGA motif are essential for aspects of successful energy coupling between an early-melted probe-bound σ54 and PspF, but not for NTP binding and hydrolysis per se (ref. 23 and our unpublished data). Studies on the S. meliloti C4-dicarboxylate transport protein D (DctD), a σ54-dependent activator, suggested that a second highly conserved motif within C3, known as the D/ESELFGH motif (residues 74–80), also contributed to a σ54 interaction site (5, 10).
To analyze the contribution of highly conserved C3 residues to PspF MBP-F4 activity, we constructed six mutations in the context of PspF MBP-F4: S75A, H80A, F85A, F85L, T86A, and T86S (Fig. 1). In the context of PspF1–275, none of these mutant proteins significantly activated transcription from the pspA promoter in vivo or in vitro, except the conservative T86S mutant (Table 1). Binding assays with σ54 and the early-melted probe showed that PspF MBP-F4 harboring the S75A and H80A mutations in the D/ESELFGH motif were able to form PspF MBP-F4 complexes as well as the wild-type PspF MBP-F4 (Fig. 3B, lanes 4 and 5). PspF MBP-F4 with the T86A mutation in the GAFTGA motif failed to form a detectable PspF MBP-F4 complex, contrasting the wild-type PspF MBP-F4 and PspF MBP-F4 harboring the more conserved substitution T86S (Fig. 3B, lanes 6 and 7). We confirmed by matrix-assisted laser desorption ionization-time of flight mass spectrometry that the PspF MBP-F4 protein harboring the T86A mutation was intact (data not shown). Therefore, in the context of the PspF MBP-F4, it seems that T86 contributes to an energetically favorable interaction with σ54. Further, PspF1–275 containing the T86A mutation is active for ATP binding and hydrolysis but not for forming an ADP-AlFx-dependent stable binary complex with σ54 (Table 1, ref. 8). Interestingly, the adjacent substitutions F85A and F85L in PspF MBP-F4, as well as the S75A and H80A mutations, did not diminish PspF MBP-F4 complex formation. However, because these mutations, unlike the T86A mutation in the context of PspF1–275, showed defects in ATP hydrolysis but not in ATP binding (Table 1), we conclude that conserved residues S75, H80, and F85 in the C3 region are required for full ATP hydrolysis. Overall, the results strongly suggest that (i) certain residues in the GAFTGA motif, and apparently not those of the D/ESELFGH motif, in the conserved C3 region of PspF are required for binding to σ54, and (ii) NTP hydrolysis is required to present sequences in C3 for stable binding to σ54.
Stable Binding of PspF MBP-F4 to the σ54–Early-Melted Probe Complex Changes the Interaction σ54 Makes with DNA.
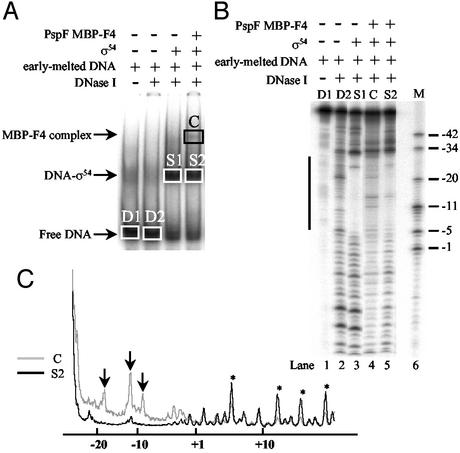
Previously, we showed by DNaseI footprinting that the stable association of σ54 with the ADP-AlFx form of the PspF1–275 leads to extended σ54–DNA interactions in the downstream direction toward the transcription start site (8). To investigate whether the stable binding of PspF MBP-F4 to σ54 changes the σ54–early-melted probe interactions, we conducted DNaseI footprinting experiments on the PspF MBP-F4 complex. As expected for the σ54–early-melted probe complex, σ54 protected the DNA between positions −5 and −33 from DNaseI cleavage (Fig. 4B, lane 3). Strikingly, analysis of the PspF MBP-F4 complex indicated that binding of PspF MBP-F4 to σ54–early-melted probe complex changes the DNA contacts made by σ54, as evidenced by (i) the hypersensitivity of the DNA to DNaseI in the −5 to −33 region, and particularly around position −12, where DNA melting originates (Fig. 4B, compare lanes 3 and 4; also, marked by arrows in Fig. 4C), and (ii) the extended protection of DNA from DNaseI cleavage in the downstream direction beyond position −5 (Fig. 4B, compare lanes 3 and 4; also, marked by stars in Fig. 4C). We also noted that the DNA-cleavage pattern upstream of position −34 was reproducibly more protected in the ternary PspF MBP-F4 complex than in the binary σ54–early-melted probe complex. However, we cannot offer an explanation for this observation. Overall, the DNA footprinting data suggest that PspF MBP-F4 exerts an NTP-independent effect on σ54–region 1 interactions with the fork-junction structure at position −12, which is consistent with (i) the action of PspF in NTP-dependent reactions (23, 33) and (ii) requirement of region 1 for ternary complex formation by PspF MBP-F4.
Figure 4.
DNase I footprint of the PspF MBP-F4 complex. (A) Native gel showing PspF F4 complex formation. Unbound probe and complexes that were isolated for analysis by denaturing PAGE are boxed. (B) Denaturing gel (10%) showing σ54 and PspF MBP-F4 complex-dependent footprints on the early-melted probe. Solid line represents the DNA segment that becomes protected upon σ54 binding and hypersensitive upon PspF MBP-F4 complex formation. (C) Scan of lanes 4 and 5 from B. *, Positions in the promoter that become protected only in the PspF MBP-F4 complex. ↓, The enhanced reactivity of the DNA to DNase I around position −12 in the PspF MBP-F4 complex.
Stable Binding of PspF to σ54 Requires NTP Interactions.
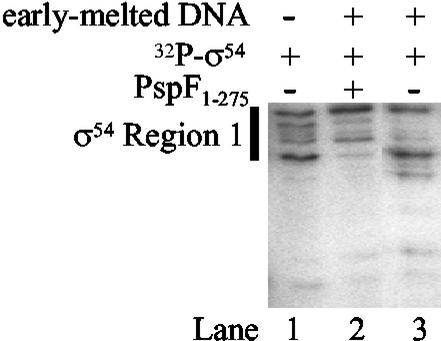
Results so far clearly indicate that (i) a sequence exists in PspF, critically including the GAFTGA motif, which makes a direct and functionally significant contact with σ54, and (ii) in the absence of NTP, this sequence is not fully available to make a stable interaction with σ54 when present in the context of intact PspF or PspF1–275 (8). Therefore, PspF may make a stable interaction with σ54 in one or a few restricted NTP-dependent functional states, as mimicked by our protein fragmentation results. However, weak interactions between σ54–RNAP and PspF as well as some other activators, such as DctD, have been reported to occur in the absence of NTP (9, 10, 26). To investigate this further, we formed the σ54–early-melted probe complex by using 32P-end-labeled σ54, exposed it to PspF1–275 in the absence of NTP, and treated the reaction with V8 protease to try to detect σ54–PspF1–275 interactions by protein footprinting. As expected, no stable ternary complex between σ54–early-melted probe complex and PspF1–275 was detected when the reactions were analyzed by native PAGE (data not shown). However, when the σ54–early-melted promoter complex that had been exposed to PspF1–275 and V8 protease was isolated from a native gel and analyzed by SDS/PAGE, we detected an altered cleavage pattern across σ54 region 1 (Fig. 5, lane 2). This cleavage pattern was not found in the absence of PspF1–275 (Fig. 5, lane 3). Because we failed to observe a stable ternary complex between PspF1–275, σ54, and early-melted probe by native PAGE, we interpret the altered V8 protease cleavage pattern as evidence that a weak σ54–PspF1–275 interaction independent of NTP can occur.
Figure 5.
Analysis of PspF–σ54 interactions by V8 protease footprinting. Lane 1, V8 protease digestion pattern of 32P-σ54; lane 3, when bound to the early-melted promoter probe; lane 2, V8 digestion of 32P-σ54 bound to the early-melted promoter probe in the presence of unlabeled PspF1–275. Region 1 of σ54 is indicated by a thick black line.
Discussion
The AAA+ Domain of PspF.
σ54-dependent activators are complex molecular machines that use the energy released by NTP hydrolysis to remodel the σ54–RNAP closed complex to initiate open complex formation (3, 5). Functioning of AAA+ proteins is believed to involve conformational changes reflecting the state of the bound NTP. Our fragmentation study demonstrates that at least one σ54-binding surface of PspF is localized within the C3 region of the AAA+ domain, and that NTP hydrolysis may be used to present one σ54-binding sequence.
PspF-σ54 Interactions.
Three types of binding between PspF and σ54 are observed: (i) NTP (ADP-AlFx)-dependent, but promoter-DNA independent (8); (ii) promoter-DNA dependent, but NTP-independent stable binding by PspF MBP-F4 (this work); and (iii) promoter-DNA dependent, but NTP-independent transient binding (this work). This result suggests that PspF must exist in at least two different states, each capable of interacting with σ54. The NTP-independent binding of PspF MBP-F4 to the σ54–early-melted promoter probe complex, and the usual requirement of ADP-AlFx for the stable binding of PspF to σ54 (8), suggest that the σ54-interacting surface in PspF responsible for stable binding to σ54 is normally masked in the absence of NTP. Because PspF MBP-F4 complex formation is strictly dependent on σ54 region 1 sequences and a fork-junction DNA structure that is universally used by RNAP en route to transcription initiation (31), we propose that the interaction σ54 region 1 makes with the fork-junction structure generates a local small structure that is recognized by PspF.
Residue T86 in the GAFTGA Motif.
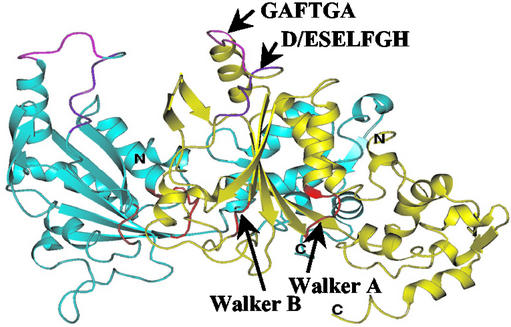
The GAFTGA motif is a signature motif of σ54-dependent activators. It makes a crucial contribution in PspF binding to σ54, and T86 seems to make an energetically significant contribution to the binding of PspF MBP-F4 to σ54, contrasting with F85. Because the PspF1–275 F85A mutant has a markedly reduced ATPase activity compared with the PspF1–275 T86A or T86S mutants (Table 1), we propose that the residue F85 is more closely associated with interactions involving nucleotide than interactions directly involving σ54. Properties of the F85A and T86A substitutions in the context of PspF1–275 indicate that the integrity of the GAFTGA motif is also required for the NTP-dependent oligomerization of PspF (Table 1), suggesting that the GAFTGA motif is part of a network of interactions that involves indirect T86 and F85 interactions with NTP. Consistent with this view, structure-based homology modeling of PspF using the AAA+ proteins p97 and HslU known structures indicate that the σ54 interacting surface is proximal to the oligomerization interface within a dimer of PspF (Fig. 6; ref. 5).
Figure 6.
Homology model of the PspF dimer based on the crystal structures of HslU and p97 (5). One promoter is colored blue and the other is colored yellow. The GAFTGA (in pink) and the D/ESELFGH (in purple) motifs are indicated. The Walker A and Walker B motifs implicated in NTP interactions are shown in red.
Notably, a single amino acid, T158 in the activating region 1 of the σ70-dependent transcription activator CRP (cAMP receptor protein) makes one of the most energetically favorable interactions with the α-subunit of the RNAP for transcription activation (34). However, in the case of PspF, the binding interaction with σ54 drives isomerization of the σ54–RNAP, whereas for CRP, the binding interaction with α simply increases promoter occupancy without remodeling the σ70–RNAP promoter complex. A critical feature of σ54-dependent transcription is that the binding interaction between σ54 and activator is coupled to NTP hydrolysis, allowing energy coupling to occur. Even though the binding energy between the activator and σ54 is modest and could otherwise be associated with a modest activation of transcription, energy coupling allows high rates of open complex formation and transcription initiation.
Mechanochemical Functioning of PspF.
It seems that (i) in the absence of NTP, PspF makes transient interactions with σ54 in the closed complex, but this interaction is not sufficient for transcription initiation; (ii) when bound to NTP and at the point of hydrolysis, PspF adopts a different functional state that allows a stable interaction with σ54 that leads to energy coupling and catalysis of open complex formation; (iii) NTP hydrolysis leads to domain movements in PspF that result in the presentation of C3 sequences in a specific structural context capable of interacting with the nucleoprotein target, the σ54–RNAP closed complex. The latter point probably explains why other PspF fragments, notably PspF MBP-F1 and PspF MBP-F2, which also contain the GAFTGA motif but within different contexts, fail to form a stable complex with the early-melted promoter probe-bound σ54.
Our fragmentation approach is a retention of function approach. It has the advantage of not being based on loss of function analysis, which suffers from potential indirect deleterious effects of mutations. Thus, the fragmentation analysis of PspF is widely applicable to explore how NTP hydrolysis conditions biological output in many AAA+ proteins.
Overview.
The changes in protein conformation we envisage in PspF and other AAA+ proteins appear to be orchestrated by the binding and subsequent hydrolysis of NTPs. The identification of a σ54-binding surface provides one of the few extant examples where NTP hydrolysis causes conformational changes that result in the unmasking and/or stable presentation of a target-binding surface within an AAA+ domain. In the E. coli RuvB protein which promotes branch migration of Holliday junctions in conjunction with RuvA, NTP hydrolysis causes the protrusion of a unique β-hairpin from the AAA+ domain, which then stably interacts with RuvA (35). Strikingly, and as predicted for PspF, in the E. coli AAA+ protease HslU (that functions together with HslV), the HslV-binding surface is located in a hydrophobic pocket at the HslU–HslU interface within the AAA+ domain. During ATP hydrolysis, the HslV-binding surfaces distend to form the functional HslUV complex (36), which suggests the shared use of target-binding site “presentation” modulated by NTP interactions. A model of PspF (derived from p97 and HslU) also suggests that the C3 region is a loop structure that could be relocated upon ATP hydrolysis (Fig. 6 and ref. 5).
Acknowledgments
We thank Professor Akira Ishihama for the antibodies against the β-subunit of E. coli RNAP and Susan Jones for comments on the manuscript. This work was supported by a Biotechnology and Biological Sciences Research Council grant to M.B.
Abbreviations
- AAA+
ATPases associated with various cellular activities
- RNAP
RNA polymerase
- PspF
phage shock protein F
- MBP
maltose binding protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Neuwald A F, Aravind L, Spouge J L, Koonin E V. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 2.Vale R D. J Cell Biol. 2000;150:13–19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck M, Gallegos M T, Studholme D J, Guo Y, Gralla J D. J Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu H, Hoover T R. Curr Opin Microbiol. 2001;4:138–144. doi: 10.1016/s1369-5274(00)00179-x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Chaney M K, Wigneshweraraj S R, Schumacher J, Bordes P, Cannon W, Buck M. Mol Microbiol. 2002;45:895–903. doi: 10.1046/j.1365-2958.2002.03065.x. [DOI] [PubMed] [Google Scholar]
- 6.Merrick M J. Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 7.Rombel I, North A, Hwang I, Wyman C, Kustu S. Cold Spring Harbor Symp Quant Biol. 1998;63:157–166. doi: 10.1101/sqb.1998.63.157. [DOI] [PubMed] [Google Scholar]
- 8.Chaney M, Grande R, Wigneshweraraj S R, Cannon W, Casaz P, Gallegos M T, Schumacher J, Jones S, Elderkin S, Dago A E, et al. Genes Dev. 2001;15:2282–2294. doi: 10.1101/gad.205501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J H, Hoover T R. Proc Natl Acad Sci USA. 1995;92:9702–9706. doi: 10.1073/pnas.92.21.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y K, Lee J H, Brewer J M, Hoover T R. Mol Microbiol. 1997;26:373–386. doi: 10.1046/j.1365-2958.1997.5851955.x. [DOI] [PubMed] [Google Scholar]
- 11.Elderkin S, Jones S, Schumacher J, Studholme D, Buck M. J Mol Biol. 2002;320:23–27. doi: 10.1016/S0022-2836(02)00404-7. [DOI] [PubMed] [Google Scholar]
- 12.Jovanovic G, Weiner L, Model P. J Bacteriol. 1996;178:1936–1945. doi: 10.1128/jb.178.7.1936-1945.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheffield P, Garrard S, Derewenda Z. Protein Expression Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 14.Dykxhoorn D M, St Pierre R, Linn T. Gene. 1996;177:133–136. doi: 10.1016/0378-1119(96)00289-2. [DOI] [PubMed] [Google Scholar]
- 15.Miller J H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 16.Jovanovic G, Rakonjac J, Model P. J Mol Biol. 1999;285:469–483. doi: 10.1006/jmbi.1998.2263. [DOI] [PubMed] [Google Scholar]
- 17.Casaz P, Buck M. Proc Natl Acad Sci USA. 1997;94:12145–12150. doi: 10.1073/pnas.94.22.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallegos M T, Buck M. J Mol Biol. 1999;288:539–553. doi: 10.1006/jmbi.1999.2704. [DOI] [PubMed] [Google Scholar]
- 19.Cannon W, Missailidis S, Smith C, Cottier A, Austin S, Moore M, Buck M. J Mol Biol. 1995;248:781–803. doi: 10.1006/jmbi.1995.0260. [DOI] [PubMed] [Google Scholar]
- 20.Sarkar G, Edery I, Sonenberg N. J Biol Chem. 1985;260:13831–13837. [PubMed] [Google Scholar]
- 21.Bochner B R, Ames B N. J Biol Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- 22.Cannon W, Gallegos M T, Casaz P, Buck M. Genes Dev. 1999;13:357–370. doi: 10.1101/gad.13.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cannon W, Gallegos M T, Buck M. Nat Struct Biol. 2000;7:594–601. doi: 10.1038/76830. [DOI] [PubMed] [Google Scholar]
- 24.Cannon W, Gallegos M T, Buck M. J Biol Chem. 2001;276:386–394. doi: 10.1074/jbc.M007779200. [DOI] [PubMed] [Google Scholar]
- 25.Chaney M, Buck M. Mol Microbiol. 1999;33:1200–1209. doi: 10.1046/j.1365-2958.1999.01566.x. [DOI] [PubMed] [Google Scholar]
- 26.Lew C M, Gralla J D. J Biol Chem. 2002;277:41517–41524. doi: 10.1074/jbc.M206912200. [DOI] [PubMed] [Google Scholar]
- 27.Rombel I, Peters-Wendisch P, Mesecar A, Thorgeirsson T, Shin Y K, Kustu S. J Bacteriol. 1999;181:4628–4638. doi: 10.1128/jb.181.15.4628-4638.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez V, Olvera L, Soberon X, Morett E. Mol Microbiol. 1998;28:55–67. doi: 10.1046/j.1365-2958.1998.00772.x. [DOI] [PubMed] [Google Scholar]
- 29.Morett E, Segovia L. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, Lew C M, Gralla J D. Genes Dev. 2000;14:2242–2255. doi: 10.1101/gad.794800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo Y, Wang L, Gralla J D. EMBO J. 1999;18:3736–3745. doi: 10.1093/emboj/18.13.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris L, Cannon W, Claverie-Martin F, Austin S, Buck M. J Biol Chem. 1994;269:11563–11571. [PubMed] [Google Scholar]
- 33.Wigneshweraraj S R, Chaney M K, Ishihama A, Buck M. J Mol Biol. 2001;306:681–701. doi: 10.1006/jmbi.2000.4393. [DOI] [PubMed] [Google Scholar]
- 34.Benoff B, Yang H, Lawson C L, Parkinson G, Liu J, Blatter E, Ebright Y W, Berman H M, Ebright R H. Science. 2002;297:1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 35.Han Y W, Iwasaki H, Miyata T, Mayanagi K, Yamada K, Morikawa K, Shinagawa H. J Biol Chem. 2001;276:35024–35028. doi: 10.1074/jbc.M103611200. [DOI] [PubMed] [Google Scholar]
- 36.Seong I S, Kang M S, Choi M K, Lee J W, Koh O J, Wang J, Eom S H, Chung C H. J Biol Chem. 2002;277:25976–25982. doi: 10.1074/jbc.M202793200. [DOI] [PubMed] [Google Scholar]