Abstract
The p53 tumor suppressor protein plays a crucial role in tumorigenesis by controlling cell-cycle progression and apoptosis. We have previously described a transcript designated tumor suppressor activated pathway-6 (TSAP6) that is up-regulated in the p53-inducible cell line, LTR6. Cloning of the murine and human full-length TSAP6 cDNA revealed that it encodes a 488-aa protein with five to six transmembrane domains. This gene is the murine and human homologue of the recently published rat pHyde. Antibodies raised against murine and human TSAP6 recognize a 50- to 55-kDa band induced by p53. Analysis of the TSAP6 promoter identified a functional p53-responsive element. Functional studies demonstrated that TSAP6 antisense cDNA diminished levels of the 50- to 55-kDa protein and decreased significantly the levels of p53-induced apoptosis. Furthermore, TSAP6 small interfering RNA inhibited apoptosis in TSAP6-overexpressing cells. Yeast two-hybrid analysis followed by GST/in vitro-transcribed/translated pull-down assays and in vivo coimmunoprecipitations revealed that TSAP6 associated with Nix, a proapoptotic Bcl-2-related protein and the Myt1 kinase, a negative regulator of the G2/M transition. Moreover, TSAP6 enhanced the susceptibility of cells to apoptosis and cooperated with Nix to exacerbate this effect. Cell-cycle studies indicated that TSAP6 could augment Myt1 activity. Overall, these data suggest that TSAP6 may act downstream to p53 to interface apoptosis and cell-cycle progression.
A series of 10 differentially expressed genes designated as either tumor suppressor activated pathway (TSAP) or tumor suppressor inhibited pathway (TSIP) have been described that were either up- or down-regulated, respectively, by p53 activation in LTR6 cells (1). LTR6 cells are derivatives of the murine myeloid M1 cell line carrying the Val-135 temperature-sensitive p53 mutant (2). After shifting to 32°C, LTR6 cells acquire wild-type p53 function and subsequently undergo massive apoptosis (2). Among the isolated genes characterized subsequently are Siah1b (TSAP3) and presenilin-1 (TSIP2) (1). Siah1b is the mammalian homologue of the Drosophila seven in absentia gene, Sina (3, 4). Presenilin-1, a predisposition gene for familial Alzheimer's disease (5), is inhibited by p53 activation and functions as an antiapoptotic molecule (6). TSAP6 represents a molecule up-regulated by p53. Recently it was reported that pHyde, the rat homologue of TSAP6, could induce apoptosis in a caspase-dependent manner in prostate cancer cells (7, 8).
The p53 tumor suppressor protein functions to maintain genomic integrity. It prevents the proliferation of cancer-prone cells primarily by enlisting two biological processes: cell-cycle arrest and apoptosis (9, 10). The proapoptotic effects of p53 are mediated by a variety of mechanisms (9, 11–13). Part of the cell-cycle regulatory function of p53 involves the induction of p21waf-1 (14, 15), an inhibitor of cyclin-dependent kinases, which inhibits cell-cycle progression at both G1 and G2 (16–18). p53 also blocks cells at the G2/M checkpoint by inhibiting the function of p34cdc2, the cyclin-dependent kinase required for entry into mitosis. The enzymatic activity of p34cdc2 is subjected to negative regulation by the Wee1 kinase, which phosphorylates p34cdc2 on Tyr-15 (19), and Myt1, a dual-specificity kinase, that phosphorylates p34cdc2 on both Thr-14 and Tyr-15 residues (20, 21).
In the present study we characterize TSAP6. TSAP6 is transcriptionally activated by p53, whereas its gene product associates with Myt1 and Nix proteins. Elevation of TSAP6 expression augments cell-cycle delay and apoptosis, suggesting that TSAP6 might play a pivotal role in tumor suppression.
Materials and Methods
TSAP6 cDNA and Promoter Cloning.
To obtain the murine TSAP6 full-length cDNA, an antisense primer (5′-GTGAGTACATATCACATGTATGGGGTGTCA-3′, GenBank accession no. U50961) was designed for 5′ rapid amplification of cDNA ends on a murine liver Marathon cDNA library (CLONTECH). Human TSAP6 full-length cDNA was cloned from a human pooled-tissue cDNA (CLONTECH). Nonoverlapping fragments covering ≈20,700 bp on chromosome 1 upstream of the first exon of mouse TSAP6 were cloned from DNA derived from mouse embryonic stem cells.
Antibodies and Cells.
Anti-TSAP6 S15N antibody was raised against a peptide derived from the sequence of murine TSAP6 SNPTEKEHLQHRQSN. Anti-TSAP6a was generated against amino acids 16–30 (DSDSSLAKVPDEAPK) of the human TSAP6 protein. The anti-Myt1 (3027) antibody (22) and anti-Nix (Abcam, Cambridge, U.K.) were used for immunoblotting. LTR6-as2 and LTR6-as4 are two polyclonal LTR6 cell lines stably transfected with pBK-RSV (Promega) containing murine TSAP6 antisense. The HeLa-39 and HeLa-51 monoclonal cell lines stably express hemagglutinin (HA)-TSAP6 and were selected with G418. HA-TSAP6-inducible HeLa cells (Invitrogen) were grown in the presence of blasticidin (5 μg/ml) and hygromycin (150 μg/ml). Doxycycline (ICN) was added at the indicated times to induce HA-TSAP6 expression.
Yeast Two-Hybrid (Y2H) Screen.
Full-length human TSAP6 was fused in frame with the LexA DNA-binding domain of pEG202. A cDNA library (≈106) derived from the murine LTR6 cell line, incubated at 32°C for 4 h, was cloned into the galactose-inducible pYESTrp2 vector (Invitrogen). Y2H screening was performed as described (23).
Protein-Interaction Assays.
GST/in vitro-transcribed/translated (IVT) and 293T cell transfection interaction analysis were performed by using standard procedures. For detection of endogenous interactions, U20S, HeLa-39, K562, and human fetal tissue-derived lysates were immunoprecipitated with anti-TSAP6 antibody or an IgY-negative control and immunoblotted with either anti-Nix or anti-Myt1 antibody.
Apoptotic and TSAP6 Small Interfering RNA (siRNA) Studies.
Cell death was assessed by either measuring the DNA content of isolated nuclei stained with propidium iodide or staining cells with annexin V-phycoerythrin (Roche, Indianapolis). For siRNA experiments, TSAP6 mRNA was knocked down with small interfering RNA molecules (24). RNA duplex with 3′-dTdT overhang directed against TSAP6 5′-AAGATCCTGGTGGATGTGAGC-3′ was synthesized (Dharmacon, Lafayette, CO). Mouse Trt siRNA duplex 5′-AACCATCACTTACAAGAAACC-3′ was used as control.
Northern Blot Analysis and Luciferase Assays.
Northern blot analysis was carried out by using 3 μg of poly(A)+ mRNA. For luciferase assays, 293T cells were transfected by using Lipofectamine (GIBCO/BRL) with 0.8 μg of the indicated TSAP6-Luc constructs together with 0.8 μg of either pCMVc5-p53 or vector control.
Cell-Synchronization Studies.
HeLa cells were synchronized at the G1/S border by double-thymidine block (DTB) as described (25).
Results
TSAP6 Is a Putative Multipass Membrane Protein.
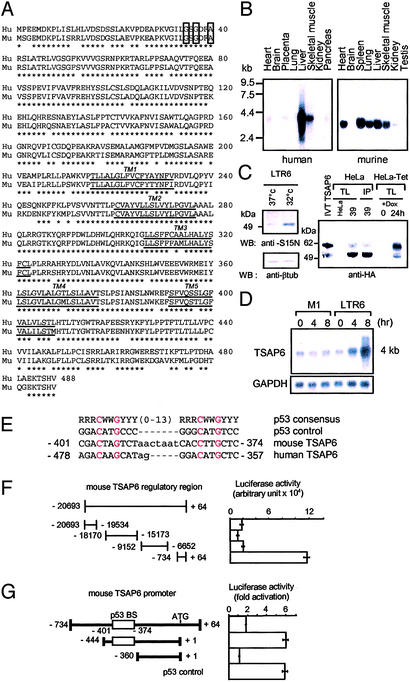
Full-length human and murine TSAP6 cDNA were cloned. The deduced amino acid sequence predicts that both proteins are of 488 aa, sharing 87% identity (Fig. 1A). TSAP6 is predicted to have five to six transmembrane-spanning regions. An iterative PSI-BLAST search revealed a GXGXXG/A motif, also known as Rossmann fold (26), within the NH2-terminal cytoplasmic portion of TSAP6, a domain prevalent in molecules containing oxidoreductase and dehydrogenase activities. BLAST search analysis revealed that TSAP6 is homologous to the previously reported pHyde (GenBank accession no. AF238864) and dudulin-2 (GenBank accession no. NM_018234). Furthermore, TSAP6 displays significant homology with six-transmembrane epithelial antigen of the prostate (STEAP), a molecule overexpressed in prostate cancer cells (27), and TIARP, tumor necrosis factor-α-induced adipose-related protein, implicated in adipocyte differentiation (28).
Figure 1.
TSAP6 encodes a putative five- to six-transmembrane protein that is transactivated by p53. (A) Amino acid sequence comparison of human (Hu) and murine (Mu) TSAP6. The five-transmembrane regions (TM1–5) are underlined. The conserved Rossmann-fold motif is boxed in bold. *, amino acid identities between human and mouse TSAP6. (B) Multitissue Northern blots probed with either human (Left) or murine (Right) TSAP6. (C) Anti-murine TSAP6 (S15N) Western blot analysis of extracts from LTR6 cells either before (lane 1) or after (lane 2) temperature shift to 32°C to induce p53 activity. Anti-HA immunoblot detects a doublet of ≈50 and 55 kDa from nonradiolabeled IVT TSAP6-HA, HeLa-39 lysates, and HeLa-TRex cells 24 h after the addition of doxycycline. Anti-TSAP6a immunoprecipitated (IP) the same species (Right, lane 4). (D) Northern blot analysis of TSAP6 expression levels in LTR6 cells compared with its parental p53 null cell line, M1, before and after temperature shift. (E) Sequences of the potential p53 response elements within the mouse and human TSAP6 promoters aligned with the consensus p53-binding site (14) (where R indicates purine, Y indicates pyrimidine, and W indicates A or T). Also shown is the sequence of a consensus p53-binding site (p53 control) used as a positive control in G. (F) The indicated fragments of the mouse TSAP6 promoter were transfected either alone or together with pCMVc50-p53 and tested in luciferase assays. Numbers represent arbitrary units. (G) TSAP6 promoter sequences containing the predicted p53-binding site (p53 BS) are transactivated by p53. Luciferase activity is shown as fold activation. Data represent the mean ± SD (n = 3).
Northern blot analysis on human (Fig. 1B Left) and murine (Fig. 1B Right) multitissue blots revealed differentially expressed TSAP6 transcripts of 4.3 and 3.8 kb, respectively. Last, fluorescence in situ hybridization localized TSAP6 to human chromosome 2q14.2 (data not shown).
The protein-expression pattern of TSAP6 was also assessed. The anti-murine TSAP6 (S15N) antibody recognized an ≈50- to 55-kDa species in cell extracts derived from LTR6 cells, the amount of which increased when wild-type p53 function was activated at 32°C (Fig. 1C Left). Overexpression of human HA-TSAP6 by IVT, stable expression in HeLa cells (HeLa-39), or induced expression after doxycycline treatment of HeLa Tet-on cells resulted in the appearance of a doublet in the 50- to 55-kDa region (Fig. 1C Right). Furthermore, anti-human TSAP6 (TSAP6a) immunoprecipitated overexpressed HA-TSAP6 (Fig. 1C Right, lane 4).
TSAP6 Transcripts Are Up-Regulated by p53.
Northern blot analysis confirmed the up-regulation of TSAP6 4–8 h after p53 activation in LTR6 cells (Fig. 1D) and in MCF7 cells after actinomycin treatment, which activates the endogenous p53 protein (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). Luciferase assays were performed in 293T cells to monitor transactivation of the TSAP6 promoter by p53. To that end, four nonoverlapping fragments covering ≈20 kb of the first exon of the murine TSAP6 promoter were cloned in front of the luciferase reporter gene (TSAP6-Luc). The proximal −734 +64 sequence contains a putative p53-binding site, the sequence of which can be aligned with a typical p53 consensus site and appears to be conserved with its human counterpart (Fig. 1E). Luciferase assays showed strong induction of −734 +64 TSAP6-Luc by p53 (Fig. 1F). Further mapping showed that fragments containing the p53-binding site retained transactivation by p53, whereas −360 +1 TSAP6-Luc, lacking this p53-binding site, was unresponsive (Fig. 1G). Overall, these data argue that TSAP6 can be transcriptionally activated by p53.
TSAP6 Antisense Dampens p53-Induced Apoptosis.
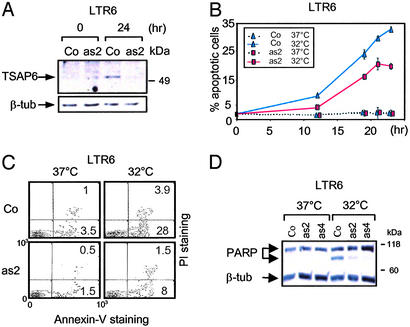
To investigate a possible role of TSAP6 in p53-mediated apoptosis, LTR6 cells carrying either vector control or TSAP6 antisense (-as2 and -as4) were generated. Immunoblot analysis confirmed that by 24 h after p53 activation, endogenous TSAP6 expression was partially compromised in LTR6-as2 compared with their parental LTR6 cells (Fig. 2A). DNA fragmentation analysis by flow cytometry revealed that by 19 and 23 h after p53 activation, LTR6-as2 exhibited a significant reduction in cell death of ≈35% and ≈41%, respectively, as compared with LTR6 control cells (Fig. 2B). A similar conclusion was reached by annexin V staining. Two-color flow-cytometry analysis revealed that by 18 h after p53 activation, 28% of LTR6 cells became annexin V+ as compared with only 8% of LTR6-as2 cells (Fig. 2C). In addition, cleavage of poly(ADP-ribose) polymerase (PARP), another indicator of apoptosis, was notably diminished 18 h after p53 activation in LTR6-as2 and -as4 cells as compared with LTR6 cells (Fig. 2D).
Figure 2.
TSAP6 antisense impairs p53-induced apoptosis. LTR6-control (Co) or LTR6-TSAP6 antisense (-as2 and -as4) cells were cultured at 32°C to activate p53. At the indicated time points, cells were harvested and assayed for apoptosis. (A) TSAP6 proteins levels in LTR6-as2 cells are diminished 24 h after p53 activation as detected by immunoblotting with the S15N anti-TSAP6 antibody. β-tub, β-tubulin. (B and C) Apoptosis was measured by flow cytometry by either determining the percentage of nuclei undergoing DNA fragmentation (B) or annexin V staining (C). The percentage of cell death represents the mean of three independent experiments. LTR6 (Co) represents LTR6 cells with or without vector. PI, propidium iodide. (D) Reduced PARP cleavage in LTR6-as2 and -as4 cells after p53 activation. Total lysates were generated and analyzed by immunoblotting with an anti-PARP antibody.
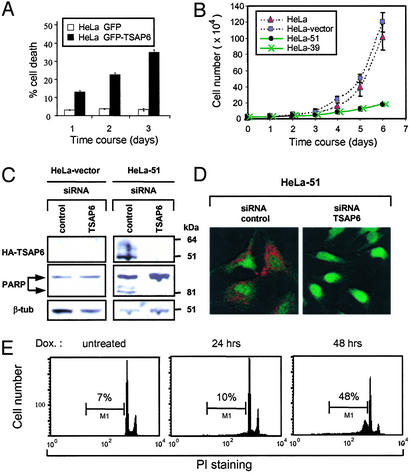
A role of TSAP6 in apoptosis was explored further in transient transfection assays. HeLa cells were transfected with either pGFP or pGFP-TSAP6 expression constructs. GFP+ cells were directly visualized and scored for apoptosis over 3 days by determining the percentage of cells that had undergone nuclear shrinkage. These data show that in HeLa cells, GFP-TSAP6 promoted a time-dependent augmentation of apoptosis (Fig. 3A). HeLa cells stably expressing HA-TSAP6 (clones 39 and 51) were isolated and assessed for their growth rate. Fig. 3B shows that compared with parental HeLa and HeLa-vector control cells, which exhibited a relatively similar rate of increase in cell number, HeLa-39 and HeLa-51 cells showed a marked reduction. Moreover, TSAP6 overexpression in HeLa-51 cells correlated with the partial cleavage of PARP (Fig. 3C Middle), suggesting the activation of cell death in these cells. Importantly, this PARP cleavage is likely due to overexpressed TSAP6, because TSAP6 depletion by siRNA lead to the disappearance of cleaved PARP (Fig. 3C). Immunoblot and immunofluorescence analysis using an anti-HA antibody confirmed the efficient depletion of overexpressed TSAP6 (Fig. 3 C and D).
Figure 3.
TSAP6 sensitizes cells toward apoptosis. (A) Transient expression of GFP-TSAP6 in HeLa cells promotes apoptosis. Plasmids expressing GFP or GFP-TSAP6 were transfected into HeLa cells. GFP+ cells were scored for apoptosis by monitoring nuclear shrinkage. (B) HeLa and HeLa-vector control cells were analyzed in parallel with HeLa-39 and -51 clones for cell growth. At the indicated time points, cells were harvested and counted. Experiments were performed in triplicate (mean ± SD). (C) HeLa-vector (Left) and HeLa-51 cells (Right) were transfected with either an siRNA control or siRNA TSAP6. Immunoblot analysis with an anti-HA antibody confirmed the efficient depletion of overexpressed TSAP6 (Top Right), which resulted in reduced PARP cleavage (Middle Right). β-tub, β-tubulin. (D) Immunofluorescence analysis using an anti-HA antibody (red) verified the depletion of overexpressed TSAP6 by siRNA. (E) Induced expression of TSAP6 promotes apoptosis. Flow cytometry was performed on propidium iodide (PI)-stained nuclei derived from HeLa-Trex-HA-TSAP6 cells before or at the indicated times after the addition of doxycycline (Dox.).
In addition, HA-TSAP6-inducible HeLa cells were also used in apoptosis assays. Immunoblotting with an anti-HA antibody showed HA-TSAP6 expression 24 h after induction (Fig. 1C Right). DNA fragmentation analysis by flow cytometry revealed that by 48 h after doxycycline treatment, 48% of the cells were apoptotic as represented by the sub-G1 population (Fig. 3E). Overall, these data imply that TSAP6 promotes apoptosis.
TSAP6 Interacts with Nix and with the Myt1 Kinase.
To characterize further the functional relevance of TSAP6, a Y2H screen was carried out by using full-length human TSAP6 as “bait” and a cDNA library derived from murine LTR6 cells cultured at 32°C for 4 h. Initial Y2H analysis identified Nix (29) as a TSAP6 binding partner. Three overlapping fragments of Nix were identified consisting of the last 62, 167, and 206 aa, respectively. In addition, a 150-aa COOH-terminal fragment of the Myt1 kinase (Myt1-150) was also identified as a TSAP6 interactor. Yeast mating assays confirmed a specific TSAP6 interaction with both Myt1 or Nix Y2H fragments as well as their respective full-length human counterparts (data not shown). Furthermore, in GST pull-down assays, radiolabeled IVT TSAP6 bound specifically with the GST-Nix and -Myt1 fusion proteins but not with the negative control GST-NKTR. A reciprocal interaction was also detected between GST-TSAP6 and IVT-labeled Myt1 and Nix, demonstrating a direct association between these two proteins and TSAP6 (see Fig. 7, which is published as supporting information on the PNAS web site).
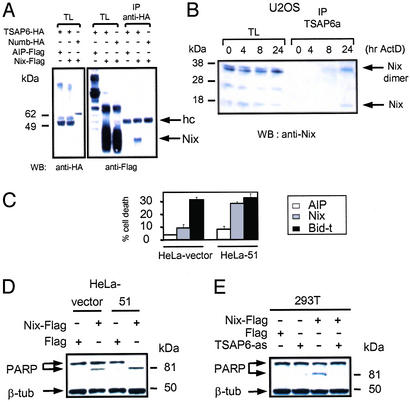
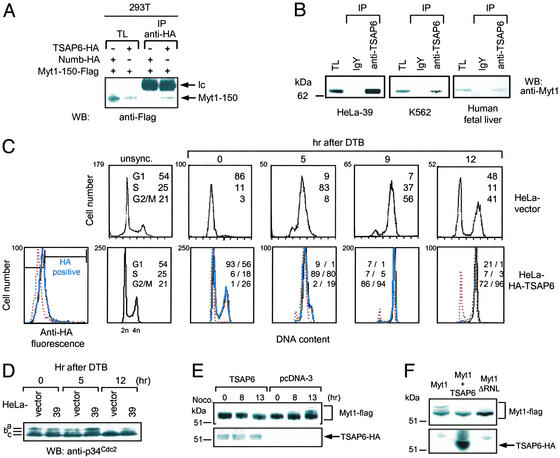
In vivo interaction analysis was performed to corroborate the above findings. 293T cells were transiently cotransfected with plasmids expressing HA-TSAP6 in combination with either Flag-Nix (Fig. 4A) or Flag-Myt1-150 (Fig. 5A). Western blot analysis of protein complexes immunoprecipitated with anti-HA antibodies revealed that Flag-Nix and Flag-Myt1-150 but not Flag-apoptosis-linked gene-2-interacting protein 1 (AIP) were coprecipitated with TSAP6.
Figure 4.
Interaction and functional analysis of TSAP6 and Nix. (A) HA-TSAP6 and Flag-Nix interact in 293T cells. 293T cells were transiently cotransfected with the indicated expression plasmids. Nix expression levels in total cell lysates (TL) and anti-HA immunoprecipitates (IP) were determined by immunoblotting with anti-Flag antibody. hc, Ig heavy chains; WB, Western blot. (B) Endogenous TSAP6 and Nix interact in U2OS cells under conditions that promote apoptosis. Cell lysates derived from U2OS cells treated with adriamycin were incubated with an anti-TSAP6 antibody coupled to IgY-agarose followed by immunoblotting with an anti-Nix antibody. The Nix dimer is detected as a 38-kDa protein. ActD, actinomycin D. (C) HeLa-51 cells overexpressing Nix are more susceptible to apoptosis. HeLa-vector or HeLa-51 cells were transiently transfected with expression plasmids encoding Flag-tagged proteins containing the AIP negative control, Nix, or COOH-terminal portion of Bid (Bid-t). Flag-positive cells were scored for apoptosis based on nuclear shrinkage. Experiments were performed in triplicate (mean ± SD). (D) Enhanced PARP cleavage in HeLa-51 cells overexpressing Nix as determined by immunoblot analysis with an anti-PARP antibody. (E) TSAP6 antisense impairs Nix-induced cell death. 293T cells were cotransfected with the indicated expression plasmids. Twenty-four hours after transfection, cell lysates were analyzed for PARP cleavage.
Figure 5.
TSAP6 interacts with Myt1 and influences cell-cycle progression. (A) Overexpressed HA-TSAP6 immunoprecipitates (IP) Flag-Myt1-150 from extracts of 293T cells. WB, Western blot; Ic, Ig light chain. (B) Endogenous TSAP6 and Myt1 interact in HeLa-39 cells (Left), K562 cells (Center), or human fetal liver tissue (Right). Lysates were incubated with either a control antibody or anti-TSAP6a antibody followed by incubation with agarose anti-IgY and Western blot analysis with anti-Myt1 antibody. TL, total cell lysate. (C) Overproduction of TSAP6 induces a G2/M delay. Polyclonal HeLa-vector and HeLa HA-TSAP6 cells were synchronized at the G1/S border by a DTB procedure and analyzed by flow cytometry after release from the block. Flow-cytometry analysis was performed separately on either gated HA-positive (solid blue line) or HA-negative (dotted red line) subpopulations (Left). (D) TSAP6 maintains p34cdc2 in an inactive state. Cell lysates derived from either synchronized HeLa vector or HeLa-39 cells were immunoblotted by using an anti-p34Cdc2 antibody. (E) TSAP6 overexpression prevents Myt1 hyperphosphorylation during mitotic entry. 293T cells overexpressing pcDNA3 or pcDNA-TSAP6 in combination with Flag-Myt1 were treated with nocodazole (Noco) for the indicated times. Western blot analysis with anti-Flag (Upper) or anti-HA (Lower) antibodies was used to reveal Myt1 and TSAP6 proteins, respectively, in total cell lysates. (F) The effect of TSAP6 on Myt1 phosphorylation is similar to that of the Myt1ΔRNL mutant alone.
Initial attempts to demonstrate a physical association between endogenous TSAP6 and Nix in nonstimulated cells were unsuccessful. We reasoned that such an association might require an initial signal such as a cell-death stimulus. We therefore took advantage of the observation that activation of endogenous p53 by adriamycin in U2OS cells promotes apoptosis (13). Total cell lysates derived from U2OS cells treated with adriamycin for 0, 4, 8, or 24 h were used to immunoprecipitate TSAP6. Immunoblot analysis with an anti-Nix antibody demonstrated that TSAP6 interacted with Nix in a time-dependent manner (Fig. 4B). Furthermore, an interaction between endogenous TSAP6 and Myt1 was demonstrated also. As illustrated in Fig. 5B, TSAP6 coprecipitated endogenous Myt1 from HeLa-39 cells, K562 cells, and human fetal liver. Thus, TSAP6 associates in vivo with both Nix and the Myt1 kinase.
TSAP6 Cooperates with Nix to Promote Apoptosis.
The finding that TSAP6 interacts with Nix, a proapoptotic protein, suggested that TSAP6 might exert cell death-related activities, in part, through Nix. We initially investigated the effects of Nix on TSAP6-overexpressing cells by transiently transfecting HeLa-vector and HeLa-51 stable clones with Flag-tagged constructs expressing a negative control (AIP), Nix, or the COOH-terminal portion of Bid (Bid-t). Bid-t is a potent mitochondrial-dependent proapoptotic protein used as a positive control. Flag-positive cells were scored for apoptosis by assessing nuclear shrinkage. Although AIP had negligible effects on cell death in either cell line, excess Nix clearly exacerbated cell death in HeLa cells stably overexpressing TSAP6 (Fig. 4C). This effect appeared to be specific for Nix, because Bid-t promoted cell death to a similar degree in both cell lines. Moreover, immunoblot analysis demonstrated enhanced PARP cleavage in HeLa-51 cells transfected with Nix (Fig. 4D). We reasoned that if TSAP6 and Nix signal through a common pathway, TSAP6 antisense should inhibit the cell-death effects of Nix. Indeed, cotransfection experiments in 293T cells demonstrated that although overexpression of Nix resulted in partial PARP cleavage, introduction of TSAP6 antisense blocked this effect (Fig. 4E). Taken together, these data suggest that through their association TSAP6 and Nix may cooperate to augment apoptosis.
TSAP6 Positively Regulates Myt1.
The observation that TSAP6 and Myt1 interact suggested that TSAP6 could also participate in cell cycle-related activities. To examine the impact of TSAP6 on cell-cycle progression, HeLa HA-TSAP6 polyclonal cells were used in cell-synchronization studies. A DTB procedure was performed to synchronize cells at the G1/S border. HA-negative and HA-positive populations were gated (Fig. 5C Lower Left) and analyzed by flow cytometry. As expected, HeLa-vector cells displayed a normal progression through S, G2, and M phases of the cell cycle (Fig. 5C Upper). Compared with HA-negative cells, the HA-positive subpopulation of the HA-TSAP6 polyclonal pool contained an additional, predominantly G2/M population (≈25%) that was initially observed before DTB release (Fig. 5C Lower). By 12 h, at a time when the HA-negative population initiated a return to G1, a majority of the HA-positive population was still in G2/M. Furthermore, this population was determined to be predominantly in G2 by analyzing the mitotic index and the phosphohistone H3+ and BrdUrd+ populations after DTB release of HeLa-39 and -51 cells (see Fig. 8, which is published as supporting information on the PNAS web site).
The hyperphosphorylation of p34cdc2 and Myt1 have been directly correlated with the down-modulation of their respective activities (22, 30). Thus, we addressed whether overexpressed TSAP6 could influence the phosphorylation status of either p34cdc2 or Myt1. First, total cell lysates derived from synchronized HeLa-control or HeLa-39 cells either before or after DTB release were analyzed by immunoblotting for p34cdc2. Compared with HeLa vector cells, HeLa-39 cells retained a comparably hyperphosphorylated form of p34cdc2 by 12 h after DTB release (Fig. 5D). This is consistent with previous reports on the effects of overexpressed Myt1 on p34cdc2 phosphorylation (25).
Next, 293T cells were cotransfected with Flag-Myt1 in combination with either HA-TSAP6 or pcDNA3 control and treated with nocodazole for different times to induce a block in mitosis. Anti-Flag immunoblot analysis on total cell lysates transfected with pcDNA3 revealed a time-dependent increase in the appearance of additional higher molecular weight species of Myt1 (Fig. 5E). By contrast, these hyperphosphorylated species of Myt1 were not detectable in lysates derived from TSAP6-overexpressing cells even by 13 h. A similar hypophosphorylated phenotype was observed by overexpressing the Myt1ΔRNL mutant alone, which lacks the cyclin B1/p34cdc2-binding motif (ref. 25; Fig. 5F). Overall, these results indicate that TSAP6 modulates the G2/M transition and regulates the level of phosphorylation of p34cdc2 and Myt1.
Discussion
TSAP6 was originally cloned from the murine LTR6 cell line as a transcript induced in response to wild-type p53 activation (1). Accordingly, we have observed that TSAP6 transcripts were also increased by endogenous activation of p53. Moreover, the identification of a p53-response element within the promoter region of TSAP6, conserved between the human and murine genes, argues in favor of the involvement of p53 in the regulation of TSAP6 expression. That TSAP6 antisense impaired p53-mediated apoptosis hinted at the possibility that TSAP6 may participate in cell death-related activities. This observation prompted us to explore its function further by identifying relevant TSAP6-interacting partners. A Y2H hunt revealed that TSAP6 physically associates with Nix and the Myt1 kinase, and this conclusion was confirmed by in vitro and in vivo studies.
Our studies point to a cooperation between TSAP6 and Nix in promoting apoptosis. This is supported by the observation that cells overexpressing TSAP6 are more sensitive toward apoptosis and that Nix exacerbates this effect. Similar to Nix, the p53-inducible Bax, Noxa, and Puma promote apoptosis at the mitochondria and associate with Bcl-2 (11, 12, 31). The observations that TSAP6 is up-regulated by p53 and interacts with Nix suggest yet another level of control by p53 on apoptosis.
Nix, a Bcl-2 family member, was originally cloned based on its high degree of homology with BNIP3 (29). Nix was shown to promote apoptosis through its targeting to the mitochondria (29, 32–34). Deletion of the Nix transmembrane domain abrogated both its ability to associate with the mitochondria and to induce apoptosis (29). Recently, a previously uncharacterized isoform of Nix was identified, Nix-short, which was shown to heterodimerize with Nix and block its proapoptotic activity (35).
The identification of Myt1 kinase as a TSAP6 binding partner indicated that TSAP6 could also participate in events related to cell-cycle control. The studies presented here provide evidence that TSAP6 promotes a delay at the G2/M transition of the cell cycle. Similarly, Liu et al. (25) observed that by 14 h after DTB release, the majority of HeLa cells overproducing Myt1 had accumulated at the G2/M border. Given the similar impact of TSAP6 and Myt1 on cell-cycle progression and the direct association between these two molecules, we postulated that TSAP6 could act as a positive regulator of Myt1. The maintenance of p34cdc2 in an inactive state by TSAP6 overexpresssion, a pattern highly reminiscent of cells overproducing Myt1 (25), supports this possibility. Finally, the observation that TSAP6 prevents the hyperphosphorylation of Myt1 during entry into mitosis reinforces this hypothesis.
The phosphorylation of Myt1 has been attributed, at least in part, to p90rsk kinase, Akt kinase, and cyclin B1/p34cdc2, all of which down-regulate the activity of Myt1 (22, 36, 37). By contrast, TSAP6 apparently counteracts these effects by promoting Myt1 activity. Interestingly, the binding domain of cyclin B1/p34cdc2 has been mapped to an RNL motif present within the COOH-terminal region of Myt1. Consistent with the present data, TSAP6 maintained Myt1 in a hypophosphorylated state, similar to the effects of Myt1ΔRNL alone, suggesting that TSAP6 could interfere with MPF recruitment to Myt1. Alternatively, through its association with Myt1, TSAP6 could either recruit Myt1-specific phosphatases or mask the accessibility of its phosphorylation sites. Future studies may aim at addressing these issues.
Supplementary Material
Acknowledgments
We thank N. Privat for technical assistance and J.-P. Roperch for his participation in the initial analysis of some of the Northern blots; W. Taylor (Cleveland Clinic Foundation, Cleveland) for helpful discussions; A. Greenberg (Manitoba Institute of Cell Biology, Winnipeg, MB, Canada) for providing the Nix constructs; and A. Nebreda (European Molecular Biology Laboratory, Heidelberg) for providing the anti-Myt1 (3027) antibody.
Abbreviations
- TSAP
tumor suppressor activated pathway
- HA
hemagglutinin
- Y2H
yeast two-hybrid
- IVT
in vitro-transcribed/translated
- siRNA
small interfering RNA
- DTB
double-thymidine block
- PARP
poly(ADP-ribose) polymerase
- AIP
apoptosis-linked gene-2-interacting protein 1
Footnotes
References
- 1.Amson R B, Nemani M, Roperch J-P, Israeli D, Bougueleret L, Le Gall I, Medhioub M, Linares-Cruz G, Lethrosne F, Pasturaud P, et al. Proc Natl Acad Sci USA. 1996;93:3953–3957. doi: 10.1073/pnas.93.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Nature. 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 3.Nemani M, Linares-Cruz G, Bruzzoni-Giovanelli H, Roperch J-P, Tuynder M, Bougueleret L, Cherif D, Medhioub M, Pasturaud P, Alvaro V, et al. Proc Natl Acad Sci USA. 1996;93:9039–9042. doi: 10.1073/pnas.93.17.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu G, Chung Y L, Glover T, Valentine V, Look A T, Fearon E R. Genomics. 1997;46:103–111. doi: 10.1006/geno.1997.4997. [DOI] [PubMed] [Google Scholar]
- 5.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature. 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 6.Roperch J-P, Alvaro V, Prieur S, Tuynder M, Nemani M, Lethrosne F, Piouffre L, Gendron M C, Israeli D, Dausset J, et al. Nat Med. 1998;4:835–838. doi: 10.1038/nm0798-835. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Steiner M S, Rinaldy A, Lu Y. Oncogene. 2001;20:5982–5990. doi: 10.1038/sj.onc.1204831. [DOI] [PubMed] [Google Scholar]
- 8.Steiner M S, Zhang X, Wang Y, Lu Y. Cancer Res. 2000;60:4419–4425. [PubMed] [Google Scholar]
- 9.Gottlieb T M, Oren M. Semin Cancer Biol. 1998;8:359–368. doi: 10.1006/scbi.1998.0098. [DOI] [PubMed] [Google Scholar]
- 10.Sionov R V, Haupt Y. Oncogene. 1999;18:6145–6157. doi: 10.1038/sj.onc.1203130. [DOI] [PubMed] [Google Scholar]
- 11.Miyashita T, Reed J C. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 12.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 13.Nakano K, Vousden K H. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 14.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 15.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 16.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal M L, Agarwal A, Taylor W R, Stark G R. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart N, Hicks G G, Paraskevas F, Mowat M. Oncogene. 1995;10:109–115. [PubMed] [Google Scholar]
- 19.Parker L L, Piwnica-Worms H. Science. 1992;257:1955–1957. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- 20.Mueller P R, Coleman T R, Kumagai A, Dunphy W G. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- 21.Liu F, Stanton J J, Wu Z, Piwnica-Worms H. Mol Cell Biol. 1997;17:571–583. doi: 10.1128/mcb.17.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer A, Gavin A C, Nebreda A R. EMBO J. 1998;17:5037–5047. doi: 10.1093/emboj/17.17.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finley R L, Brent R. Proc Natl Acad Sci USA. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 25.Liu F, Rothblum Oviatt C, Ryan C E, Piwnica-Worms H. Mol Cell Biol. 1999;19:5113–5123. doi: 10.1128/mcb.19.7.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eventoff W, Rossmann M G. CRC Crit Rev Biochem. 1975;3:111–140. doi: 10.3109/10409237509102554. [DOI] [PubMed] [Google Scholar]
- 27.Hubert R S, Vivanco I, Chen E, Rastegar S, Leong K, Mitchell S C, Madraswala R, Zhou Y, Kuo J, Raitano A B, Jakobovits A, Saffran D C, Afar D E. Proc Natl Acad Sci USA. 1999;96:14523–14528. doi: 10.1073/pnas.96.25.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moldes M, Lasnier F, Gauthereau X, Klein C, Pairault J, Feve B, Chambaut-Guerin A M. J Biol Chem. 2001;276:33938–33946. doi: 10.1074/jbc.M105726200. [DOI] [PubMed] [Google Scholar]
- 29.Chen G, Cizeau J, Vande Velde C, Park J H, Bozek G, Bolton J, Shi L, Dubik D, Greenberg A. J Biol Chem. 1999;274:7–10. doi: 10.1074/jbc.274.1.7. [DOI] [PubMed] [Google Scholar]
- 30.Booher R N, Holman P S, Fattaey A. J Biol Chem. 1997;272:22300–22306. doi: 10.1074/jbc.272.35.22300. [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Zhang L, Hwang P M, Kinzler K W, Vogelstein B. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 32.Yasuda M, Han J W, Dionne C A, Boyd J M, Chinnadurai G. Cancer Res. 1999;59:533–537. [PubMed] [Google Scholar]
- 33.Matsushima M, Fujiwara T, Takahashi E, Minaguchi T, Eguchi Y, Tsujimoto Y, Suzumori K, Nakamura Y. Genes Chromosomes Cancer. 1998;21:230–235. [PubMed] [Google Scholar]
- 34.Chen G, Ray R, Dubik D, Shi L, Cizeau J, Bleackley R C, Saxena S, Gietz R D, Greenberg A H. J Exp Med. 1997;186:1975–1983. doi: 10.1084/jem.186.12.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yussman M G, Toyokawa T, Odley A, Lynch R A, Wu G, Colbert M C, Aronow B J, Lorenz J N, Dorn G W. Nat Med. 2002;8:725–730. doi: 10.1038/nm719. [DOI] [PubMed] [Google Scholar]
- 36.Okumura E, Fukuhara T, Yoshida H, Hanada Si S, Kozutsumi R, Mori M, Tachibana K, Kishimoto T. Nat Cell Biol. 2002;4:111–116. doi: 10.1038/ncb741. [DOI] [PubMed] [Google Scholar]
- 37.Wells N J, Watanabe N, Tokusumi T, Jiang W, Verdecia M A, Hunter T. J Cell Sci. 1999;112:3361–3371. doi: 10.1242/jcs.112.19.3361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.