Abstract
In the G protein-coupled receptor rhodopsin, the conserved NPxxY(x)5,6F motif connects the transmembrane helix VII and the cytoplasmic helix 8. The less geometrically constrained retinal analogue 9-demethyl-retinal prevents efficient transformation of rhodopsin to signaling metarhodopsin (Meta) II after retinal photoisomerization. Here, we demonstrate that Ala replacement mutations within the NPxxY(x)5,6F domain, which eliminate an interaction between aromatic residues Y306 and F313, allow formation of Meta II despite the presence of 9-demethyl-retinal. Also a disulfide bond linking residues 306 and 313 in the 9-demethyl-retinal-reconstituted mutant Y306C/F313C/C316S prevented Meta II formation, whereas the reduced form of the mutant readily transformed to Meta II after illumination. These observations suggest that the interaction between residues 306 and 313 is disrupted during the Meta I/Meta II transition. However, this enhancement in Meta II formation is not reflected in the G protein activation, which is dramatically reduced for these mutants, suggesting that changes in the Y306–F313 interaction also lead to a proper realigning of helix 8 after photoisomerization. The E134Q mutation, located in the second conserved motif, D(E)RY, rescues activity in 9-demethyl-retinal-reconstituted mutants to different degrees, depending on the position of the Ala replacement in the NPxxY(x)5,6F motif, thus revealing distinct roles for the NP and Y(x)5,6F portions. Our studies underscore the importance of the NPxxY(x)5,6F and D(E)RY motifs in providing structural constraints in rhodopsin that rearrange in response to photoisomerization during formation of the G protein-activating Meta II. The dual control of the structural rearrangements secures reliable transformation of quiescent rhodopsin to activating Meta II.
Keywords: GPCR‖NPxxY motif
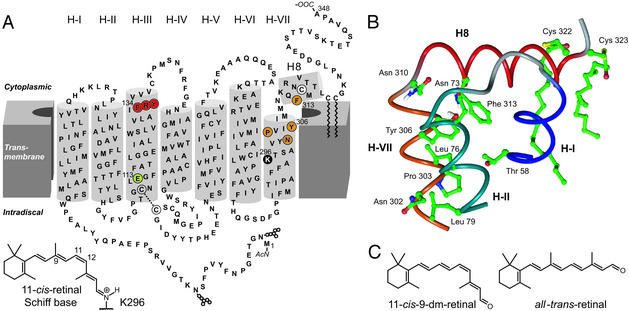
Rhodopsin is a prototypical receptor from the largest subfamily A of G protein-coupled receptors (GPCRs), which are thought to operate through similar signaling mechanisms (1, 2). The only known crystal structure of a GPCR, that of rhodopsin (3), confirmed the presence of seven transmembrane helices (H). The structure describes the receptor in its inactive ground state with the bound inverse agonist, 11-cis-retinal. The ɛ-amino group of K296 in H-VII tethers the chromophore 11-cis-retinal to opsin via a covalent Schiff base (SB) bond (Fig. 1). A salt-bridge between the protonated SB and the counterion, the conserved Glu-113 in H-III, contributes largely to constrain the receptor in the inactive conformation. Additional constraints involve hydrogen bond networks and hydrophobic interactions linking transmembrane helices, sequestration of the Arg residue of the D(E)RY motif in the hydrophobic milieu, and intramolecular interactions within the NPxxY(x)5,6F region (3, 4). Absorption of light energy isomerizes the 11-cis-retinylidene chromophore, generating the agonist all-trans-retinylidene in situ. Subsequent thermal relaxation of the retinal–protein complex leads within milliseconds to an equilibrium of metarhodopsin (Meta) states that show analogies to the low- and high-affinity states of other GPCRs activated by diffusible ligands (2). This mechanism ensures the fast response of rhodopsin to light and an increase in its activity toward the heterotrimeric retinal G protein, transducin (Gt), by a factor of 1012 (5).
Figure 1.
The conserved D(E)RY and NPxxY(x)5,6F motifs in rhodopsin. (A) Secondary structure model of bovine rhodopsin with the conserved motifs D(E)RY (red) and NPxxY(x)5,6F (orange) in transmembrane helices III (H-III) and VII (H-VII), respectively. In the crystal structure (3), an amphipathic helix following H-VII was observed. This helix, H8, is terminated by palmitoylated C322 and C323, attaching it to the lipid membranes. The chromophore 11-cis-retinal is attached to K296 in H-VII of the opsin apoprotein via a protonated SB, which is stabilized by the counterion E113 (green) in H-III. N2 and N15 at the N terminus carry carbohydrate chains. (B) Three-dimensional model of the NPxxY(x)5,6F region and neighboring helices (H-I, H-II, and H8) based on the crystal structure of rhodopsin (3, 4). The side chain of Y306 shows an interaction with F313 in H8. (C) Chemical structures of the retinal analogue 9-dm-retinal and all-trans-retinal.
The equilibrium between the active Meta II and its inactive predecessor, Meta I, can be measured by monitoring changes in the absorption of the chromophore. The spectral absorption shift is due to deprotonation of the SB and protonation of the counterion E113 (ref. 6; Meta I, λmax = 480 nm; Meta II, λmax = 380 nm). Besides protonation of E113 in Meta II, an additional proton uptake, most likely by E134 in the highly conserved D(E)RY motif at the cytoplasmic end of H-III (Fig. 1A), is necessary to reach the active conformation (7, 8). The light-induced conformational changes of the cytoplasmic domain that allow catalytic nucleotide exchange in the G protein occur in parallel with the protonation changes and a relative movement of the cytoplasmic ends of H-III and H-VI (9, 10). Besides D(E)RY, there is another highly conserved motif in GPCRs connecting H-VII and cytoplasmic helix 8 (H8). H8 is anchored to the membranes by palmitoylated residues C322 and C323 in rhodopsin. This region is important for the interaction of light-activated rhodopsin with Gt (11, 12). The crystal structure of rhodopsin indicates a hydrophobic interaction between Y306 in H-VII and F313 in the amphipathic helix (H8; Fig. 1B). A sequence alignment of GPCRs of the rhodopsin subfamily reveals that the majority of GPCRs contain an NPxxY(x)5,6F sequence. Mutations in the NPxxY(x)5,6F motif affect receptor expression, ligand affinity, receptor sequestration, heterotrimeric G protein coupling, and association with the small G proteins ARF and RhoA (for example, see refs. 13–16).
The function of the NPxxY(x)5,6F motif in the transition from the ground state to the active forms of GPCRs is not well understood. Therefore, we set out to elucidate the role of this region in rhodopsin by using a combination of structural modeling, mutagenesis, and chemical modification of the chromophore, together with enzymatic and spectroscopic assays. We found that the NPxxY(x)5,6F and D(E)RY motifs provide a dual control of the activating structural changes in this prototypical GPCR.
Materials and Methods
Materials.
The materials and proteins used were as described (17, 18). Purified antirhodopsin mAb rho-1D4 was obtained from the National Culture Center, Minneapolis. The rhodopsin mutant genes were prepared by restriction fragment replacement of the synthetic gene (19) in a modified pLITMUS cloning vector (New England Biolabs). Codon alterations were introduced by replacement of the AatII/BspEI or AatII/BstEII restriction fragment with the corresponding synthetic oligonucleotide duplex. For expression, PstI/SalI restriction fragments of the mutant genes were used to replace the WT PstI/SalI restriction fragment in the synthetic rhodopsin gene cloned into a eukaryotic expression vector (20, 21). To generate mutant genes with the additional E134Q mutation, the PstI/SalI fragments were cloned into the PstI/SalI-digested E134Q mutant expression plasmid (21), which was kindly provided by T. Sakmar (The Rockefeller University, New York). COS cell membranes were isolated essentially as described by Han and Sakmar (22). 11-cis-9-demethyl-retinal (9-dm-retinal) was prepared as described (17).
Preparation of Rhodopsins.
Rhodopsins were expressed in COS-1 cells, reconstituted with retinal, and purified as described with 0.03% dodecyl-β-maltoside (DM) in elution buffer (17). The mutant proteins exhibited the same degree of glycosylation as WT (according to SDS/PAGE), although they were expressed at slightly reduced levels. The mutant Y306C/F313C/C316S, however, was expressed at five-fold reduced levels compared with WT (data not shown). Y306C/F313C/C316S was eluted in 2 mM sodium phosphate, pH 6.0, 0.03% (wt/vol) DM containing 100 μM of an 18-mer peptide corresponding to the C-terminal rhodopsin sequence (17). A fraction of the purified pigment was stirred at 4°C for 2–3 days in the dark to allow formation of a disulfide bond by oxidation with ambient oxygen. Formation of a disulfide bond was confirmed by titration of Cys residues with 4,4′-dithiodipyridine (PDS), essentially as described (23).
UV-Visible Spectroscopy.
Spectra were taken at 20°C with a Varian Cary 50 UV-visible spectrometer with a resolution of 2 nm. Samples were illuminated for 15 sec by using a 150-W fiber optic light source equipped with a >480 nm longpass (GG495 Schott, Mainz, Germany) and a heat protection filter. Buffers used were 50 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane (BTP; pH 7.5 or pH 8.5), 0.03% DM for Ala mutants; and 50 mM BTP (pH 7.5), 130 mM NaCl, 1 mM MgCl2, 0.03% DM for Y306C/F313C/C316S. The concentration of rhodopsin pigments was determined spectroscopically by using ɛ500 = 42,700 M−1·cm−1 for all pigments, because deviation of ɛ500 between mutant and WT pigments was <3%.
Gt-Activation Assay.
Fluorescence measurements (Fluorolog 2, Spex Industries, Metuchen, NJ) were performed as described with modifications (17). Protein concentrations in the samples were 2 nM pigment (or 20 nM for Y306C/F313C/C316S pigments) and 250 nM Gt in 20 mM BTP (pH 7.5), 130 mM NaCl, 1 mM MgCl2, and 0.01% DM in a final volume of 650 μl. Bovine Gt (250 nM) and guanosine 5′-[γ-thio]triphosphate (GTP[γS]; 5 μM) were incubated in a stirring cuvette for 2 min at 20°C. Then, initial fluorescence intensity was recorded for 20 s, followed by closure of the excitation beam shutter and addition of rhodopsin (2 nM final concentration). After 5 min of incubation with receptor, the beam shutter was reopened, and 20 s later the receptor was activated by continuous illumination with orange light by using the same equipment as for UV-visible spectroscopy. For determination of the rates of Gt activation, traces were normalized to the fluorescence intensity before illumination, and initial slopes of the first 30–60 s of data after illumination were fitted by linear regression. Evaluation of the fluorescence intensity before and after the 5-min incubation period and comparison with WT rhodopsin, which is inactive in the dark, allowed detection of possible activity of the pigments in the dark.
For Gt-activation measurements with transfected COS cell membranes, 65 μl of membrane suspension was mixed in a total volume of 650 μl with buffer [20 mM BTP (pH 6.8)/130 mM NaCl/1 mM MgCl2] and Gt (500 nM final concentration). GTP[γS] uptake by Gt was initiated by addition of GTP[γS] (10 μM final concentration). For reference, the membrane suspension was incubated with 200 μM 11-cis-retinal for at least 1 h before measurements and illuminated with orange light before addition of GTP[γS].
Structural Modeling.
Simulations and graphics were done with insightii version 2000 (Accelrys, San Diego) by using Protein Data Bank ID code 1HZX as the starting structure. Molecular dynamics simulations (Discover module in insightii) were performed at 300 K for 100 ps in vacuo, followed by energy minimization. Only the cytoplasmic half of rhodopsin was allowed to move. All simulations were carried out by using ESFF force field and charges generated in this module. The charges were determined by minimizing the electrostatic energy with respect to the charges under the constraint that the sum of the charges is equal to the net charge on the molecule. Before and after each molecular dynamics run, optimization of the whole structure was done (maintaining frozen parts when necessary).
Results
Formation of the Active Receptor Conformation Is Facilitated in Rhodopsin Mutants Y306A and F313A but Not N302A or P303A.
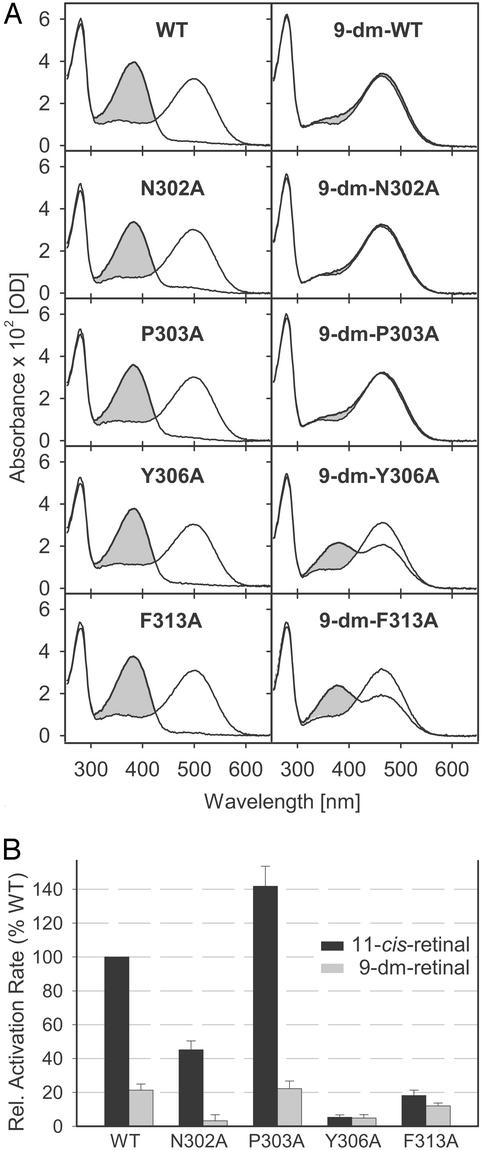
Single Ala replacement mutants, N302A, P303A, Y306A, and F313A, were generated to study the function of the NPxxY(x)5,6F motif. Mutant opsins were expressed in COS cells, reconstituted with either the native chromophore 11-cis-retinal or its derivative 9-dm-retinal lacking the methyl at C-9 on the polyene chain (Fig. 1), and purified by immunoaffinity chromatography as stable pigments. All pigments reconstituted with native chromophore showed absorption spectra similar to WT, both in the dark and after illumination (Fig. 2A Left). The same λmax values of 500 (dark ground state and 11-cis-retinal as the chromophore) and 380 nm (active Meta II, formed after illumination) suggest that the chromophore-binding site was not perturbed as a result of the mutations. Illuminated pigments showed only very low amounts of the inactive Meta I conformation (λmax = 478 nm). This result is due to the strong agonistic properties of all-trans-retinal and solubilization of rhodopsin with DM, which favors formation of active Meta II. To detect a possible effect of the mutations on the equilibrium between inactive and active receptor conformations, we investigated pigments containing 9-dm-retinal, which has only weak agonistic properties. The 9-dm-WT pigment has a maximal absorption at ≈464 nm in the dark (Fig. 2A Top Right). Photon absorption by the pigment and isomerization of 9-dm-retinal do not provide enough energy to break all inactivating constraints in the receptor, resulting in a low fraction of the 9-dm-Meta II conformation (λmax = 380 nm; refs. 17 and 24). Active 9-dm-Meta II is in equilibrium with the inactive 9-dm-Meta I conformation, contributing to the absorption peak at ≈466 nm (24). The spectra of 9-dm-Ala mutants obtained in the dark were comparable to the spectrum of 9-dm-WT. However, after illumination, differences between WT and some of the mutants were observed. 9-dm-N302A and 9-dm-P303A showed behaviors similar to 9-dm-WT, whereas 9-dm-Y306A and 9-dm-F313A showed an increased fraction of the 9-dm-Meta II photoproduct (Fig. 2A Right).
Figure 2.
Spectra of purified recombinant pigments and Gt-activation rates. (A) WT or Ala replacement opsins were reconstituted with 11-cis-retinal to form pigments, and 9-dm-pigments were obtained with 9-dm-retinal. Spectra were measured before (solid lines) and after (shaded gray) illumination with orange light. (B) Light-dependent Gt-activation rates of rhodopsins containing 11-cis-retinal (black) or 9-dm-retinal (gray) were determined from GTP[γS] uptake by Gt and are presented relative to the rate of WT reconstituted with 11-cis-retinal. Values from left to right are 100.0%, 21.3%, 45.1%, 3.2%, 141.8%, 22.2%, 5.3%, 4.8%, 18.1%, and 12.0%.
Effect of Ala Replacements on the Activation of Gt.
Despite the facilitated formation of Meta II in mutants Y306A and F313A, this did not translate into an enhanced ability to catalyze nucleotide exchange in Gt. The photoactivated Ala replacement mutants displayed profoundly reduced Gt activation, relative to WT, when reconstituted with either native (Y306A, 5.3%; F313A, 18.1%) or 9-dm-retinal chromophores (Y306A, 4.8%; F313A, 12.0%) (Fig. 2B).
In the case of mutants N302A and P303A containing the native chromophore, the light-induced activity was modestly reduced for N302A and increased for P303A. In the 9-dm-pigments, which like 9-dm-WT did not form Meta II efficiently (Fig. 2A), a large reduction in Gt activation was observed: WT [100% (11-cis-retinal) → 21.3% (9-dm-retinal)], N302A (45.1% → 3.2%), and P303A (141.8% → 22.2%). None of the mutations caused constitutive activity because none of the four Ala replacement pigments showed significant catalysis of nucleotide exchange in Gt, neither in the dark nor without chromophore (data not shown).
Residues Y306 and F313 Interact in a State-Dependent Way.
To investigate whether Y306 and F313 are in close vicinity in the active receptor conformation, the triple mutant Y306C/F313C/C316S was constructed. By oxidation, a disulfide bridge was formed reversibly between positions 306 and 313. The pigment containing 11-cis-retinal showed full light-induced Meta II formation in both reduced and oxidized states, as judged by UV-visible absorption spectra (data not shown). The oxidized 9-dm-Y306C/F313C/C316S pigment showed UV-visible spectra similar to 9-dm-WT, and poorly formed 9-dm-Meta II after illumination (Fig. 3). However, when the sulfhydryl groups at positions 306 and 313 were reduced, the UV-visible spectra resembled the spectra of 9-dm-Y306A and 9-dm-F313A, yielding a large fraction of 9-dm-Meta II after illumination. The origin of the absorption around 466 nm is complex and includes different species like 9-dm-Meta I, nonisomerized 9-dm-pigment, and, at low pH, an SB-reprotonated 9-dm-Meta II form (17, 24). A likely explanation for the spectrum of the illuminated oxidized 9-dm-Y306C/F313C/C316S pigment is the presence of an increased amount of 9-dm-Meta I compared with the reduced pigment. The activity of the Y306C/F313C/C316S pigment with and without disulfide bridges was almost abolished (2.6% of WT activity) and no significant difference between reduced and oxidized states was observed.
Figure 3.
UV-visible spectra of the Y306C/F313C/C316S mutant reconstituted with 9-demethyl-retinal. The purified pigment was measured in the dark (solid lines) and after illumination with orange light (broken lines) before (green) and after (blue) 3 days of oxidation with ambient oxygen. Reduction of the disulfide bridge between positions 306 and 313 in the oxidized pigment by incubation with 1 mM DTT for 3 h yielded the spectra shown in black.
Mutations Involving Both NPxxY(x)5,6F and D(E)RY Regions.
We have found that 9-dm-rhodopsin displayed reduced Meta II formation and G protein activity after illumination (ref. 17; also see Fig. 2A Top Right). The action of a weaker agonist than 11-cis-retinal can be fully rescued by the E134Q mutation in the conserved D(E)RY region (ref. 17; also see Fig. 4A Top Right). Here, we used UV-visible spectroscopy and Gt activation assays to investigate the equilibrium between inactive and active Meta conformations of Ala replacement mutants containing the E134Q mutation. We found that for all four double mutants, the E134Q mutation led to almost complete formation of the Meta II conformation after illumination, both for pigments containing the native chromophore 11-cis-retinal or its analogue 9-dm-retinal (Fig. 4A). Formation of 9-dm-Meta II in the E134Q/N302A mutant is increased further at higher pH. This result suggests that the N302A mutation, close to the chromophore attachment site, has an effect on SB reprotonation of 9-dm-Meta II. The activity levels of all four 9-dm double-mutant pigments were comparable to the respective pigments containing 11-cis-retinal. However, the activity was rescued to different degrees depending on the position of the Ala replacement. The rescue effect due to the E134Q mutation was significant for Ala replacements of N302 and P303 and minor for Y306 and F313 (compare Figs. 2B and 4B). Y306A and F313A have an intrinsically stable Meta II state in the presence of 9-dm-retinal and, thus, the E134Q mutation has little additional effect.
Figure 4.
Effect of the E134Q mutation on Ala replacement mutants. (A) Spectra of purified recombinant pigments at pH 7.5 obtained by regeneration of E134Q or Ala replacement/E134Q double mutants with 11-cis-retinal or 9-dm-retinal before (solid lines) and after (shaded gray) illumination with orange light. Broken line shows spectrum of 9-dm E134Q/N302A at pH 8.5 after illumination. (B) Light-dependent Gt-activation rates of rhodopsins containing 11-cis-retinal (black) or 9-dm-retinal (gray) were determined from GTP[γS] uptake by Gt and are presented relative to the rate of WT reconstituted with 11-cis-retinal. Values from left to right are 97.3%, 93.9%, 37.7%, 41.0%, 122.7%, 135.3%, 2.0%, 6.2%, 11.0%, and 17.6%.
Discussion
Formation of Meta II Is Controlled by Conserved D(E)RY and NPxxY(x)5,6F Regions.
The crystal structure of the inactive rhodopsin ground state revealed stabilizing hydrogen bond networks, which include conserved residues from the D(E)RY and NPxxY(x)5,6F regions (3, 4). The helix–turn–helix fold of the NPxxY(x)5,6F region appears to be stabilized by a hydrophobic interaction between aromatic side chains of Y306 and F313 (Fig. 1). Reducing constraints in either region by the E134Q mutation or Ala or Cys replacement of Y306 or F313 facilitated formation of Meta II, as seen in 9-dm-retinal-reconstituted mutant rhodopsins (Figs. 2–4). We conclude that the release of constraints in both the D(E)RY and NPxxY(x)5,6F regions plays a role in the formation of Meta II. Whether the transition to Meta II is complete or only partial depends on the chemical properties of the ligand. These properties determine whether the energy provided by cis/trans-isomerization of retinal (or in case of other rhodopsin-like GPCRs, binding of diffusible agonists) is sufficient to break inactivating structural constraints and to stabilize this conformation. We also observed an increased formation of Meta II in mutant pigments E134Q, Y306A, and F313A reconstituted with an “open-ring” acyclic retinal derivative, another weak agonist (O.P.E., K.P., and K.P.H., unpublished observations). No increase of 9-dm-Meta II was seen for N302A and P303A, suggesting different roles for the NP and Y(x)5,6F portions of NPxxY(x)5,6F.
The Cytoplasmic Part of the NPxxY(x)5,6F Region Contributes to a Binding Site for the G Protein.
To our surprise, the facilitated formation of Meta II in mutants lacking aromatic side chains at positions 306 and/or 313 was not reflected in an increased activity toward Gt. Rather, the light-induced activity of these pigments was dramatically reduced, arguing for the participation of the Y(x)5,6F portion of NPxxY(x)5,6F in the formation of the binding site for Gt, in agreement with earlier observations (11, 12, 25). Although yet unidentified structural perturbations in the mutants studied can affect coupling to Gt, our conclusion was supported by the rescue effect of E134Q in double-mutant studies. The E134Q mutation strongly increased the agonism of 9-dm-retinal but failed to restore WT activity in Y306A and F313A mutants. It is noteworthy that Ala replacements one helix turn apart from Y306 only modestly reduced Gt activation (N302A) or even resulted in hyperactivity (P303A).
Conformational Changes in the NPxxY(x)5,6F Region After Receptor Activation.
Several lines of evidence suggest a structural rearrangement of H8 after receptor activation (15, 26–32). Here, we showed that the release of the constraint on the NPxxY(x)5,6F region imposed by the Y306–F313 interaction occurs during the Meta I/Meta II transition as unveiled by UV-visible spectroscopy. Consistently, a state-dependent interaction between residues 306 and 313 was suggested by the mutant Y306C/F313C/C316S, which was prepared with and without a disulfide bridge. No increase of Gt activity was found when a persistently stable interlink between residues 306 and 313 was provided by a disulfide bridge. This finding supports the conclusion that Y306 and F313 are critical for proper light-induced conformational changes in the NPxxY(x)5,6F region.
Interplay Between the Chromophore and the NPxxY(x)5,6F and D(E)RY Regions.
How can retinal isomerization be transmitted to the Y(x)5,6F portion of NPxxY(x)5,6F? The key observation is that the mutant P303A forms normal Meta II and shows hyperactivity after illumination. In contrast to the Ala replacement, P303 with its imide group cannot form a hydrogen bond to the carbonyl group of the nearby amino acid, thus allowing in the middle of H-VII formation of a short 310-helical segment, which contains the SB of the chromophore. We hypothesize that retinal isomerization leads to reorganization of this region of H-VII, which is favored by Ala at position 303, probably by its propensity to form an α-helix. In the course of this structural rearrangement, the salt-bridge between the protonated SB and the counterion E113 may be broken during the Meta I/Meta II transition (reviewed in refs. 2 and 5) and the constraints on the Y(x)5,6F and D(E)RY regions may be affected. The role of a water-mediated hydrogen bond network involving N302 (3, 4, 33) remains to be elucidated.
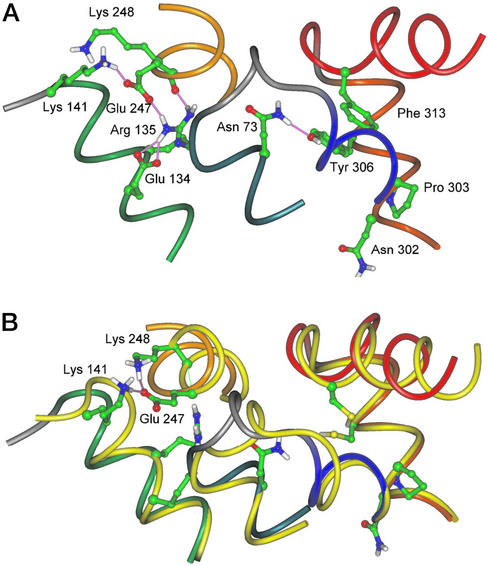
Structural changes within the D(E)RY region are crucial determinants of the rhodopsin/Gt interaction and are proposed to occur by a two-step activation mechanism that involves Meta II isoforms Meta IIa and Meta IIb, the latter formed sequentially by proton uptake (7, 8, 34). Based on the present results, we can now conclude that signal transmission from the ligand-binding domain to both the D(E)RY and the NPxxY(x)5,6F regions is essential for receptor activation. An interplay of these regions not only with the ligand but also with each other is a conclusion from the data obtained with Ala replacement/E134Q mutants: We found that E134Q can efficiently restore the activity of Ala replacement mutants in the NPxxY(x)5,6F motif containing 9-dm-retinal to the activity of the parent pigment containing 11-cis-retinal. Even in the ground state, the E134Q mutation leads to a partially active conformation in the distant fourth loop (H8) (35). We performed molecular dynamics simulations on the Y306C/F313C/C316S mutant with and without a disulfide bridge, which give an idea of how crosstalk between these two regions may occur (Fig. 5).
Figure 5.
(A) Hydrogen bonds within the D(E)RY region and between Tyr-306 (H-VII) and Asn-73 (H-II) observed after 100-ps molecular dynamics simulation of the WT ground state. In the cytoplasmic part of the NPxxY(x)5,6F region, a hydrogen bond between Asn-73 in H-II and Tyr-306 in H-VII aligns the side chain of Tyr-306 for interaction with Phe-313 (H8). In the D(E)RY region, a salt-bridge between side chains of Glu-134 and Arg-135 is seen, and Glu-247 in H-VI is linked to H-III by hydrogen bonds to both Arg-135 and Lys-141. (B) Superposition of 100-ps molecular dynamics simulation of WT and Y306C/F313C/C316S with hydrogen bonds in the D(E)RY region (mutant shown in yellow with colored side chains). In Y306C/F313C/C316S, where the interaction between aromatic side chains of Tyr-306 and Phe-313 is abolished, Lys-248 (H-VI) appears to compete for hydrogen bond interaction with the adjacent Glu-247. As a result, an intrahelical hydrogen bond between side chains of Glu-247 and Lys-248 can form, thereby facilitating dissociation of H-III from H-VI. These changes did not occur when simulations were performed with the mutant containing a disulfide bridge.
In this study, we performed experiments that were aimed at the retinal binding site and these two regions simultaneously by studying double mutants reconstituted with a chromophore analogue. We have provided evidence that the NP and Y(x)5,6F modules, in concert with the D(E)RY region and the chromophore binding site, enable retinal to functionalize the seven-helix structure as a receptor.
Acknowledgments
We thank C. Koch, R. Hauer, H. Seibel, and K. Engel for excellent technical assistance. This research was supported by the Deutsche Forschungsgemeinschaft (Sfb 449 to O.P.E. and K.P.H.), National Institutes of Health Grant EY09339, a grant from Research to Prevent Blindness (RPB) to the Department of Ophthalmology at the University of Washington, and grants from the Foundation Fighting Blindness and the E. K. Bishop Foundation (to K.P.). K.P. is an RPB Senior Investigator and a recipient of the Humboldt Research Award for Senior U.S. Scientists.
Abbreviations
- GPCR
G protein-coupled receptor
- 9-dm-retinal
11-cis-9-demethyl-retinal
- Meta
metarhodopsin
- Gt
G protein of the rod cell, transducin
- DM
dodecyl-β-maltoside
- H
helices
- SB
Schiff base
- H8
helix 8
- GTP[γS]
guanosine 5′-[γ-thio]triphosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Menon S T, Han M, Sakmar T P. Physiol Rev. 2001;81:1659–1688. doi: 10.1152/physrev.2001.81.4.1659. [DOI] [PubMed] [Google Scholar]
- 2.Okada T, Ernst O P, Palczewski K, Hofmann K P. Trends Biochem Sci. 2001;26:318–324. doi: 10.1016/s0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- 3.Palczewski K, Kumasaka T, Hori T, Behnke C A, Motoshima H, Fox B A, Le Trong I, Teller D C, Okada T, Stenkamp R E, et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 4.Teller D C, Okada T, Behnke C A, Palczewski K, Stenkamp R E. Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofmann K P. In: Handbook of Biological Physics. Hoff A J, editor. Vol. 3. Amsterdam: Elsevier; 2000. pp. 91–142. [Google Scholar]
- 6.Jäger F, Fahmy K, Sakmar T P, Siebert F. Biochemistry. 1994;33:10878–10882. doi: 10.1021/bi00202a005. [DOI] [PubMed] [Google Scholar]
- 7.Arnis S, Hofmann K P. Biochemistry. 1995;34:9333–9340. doi: 10.1021/bi00029a008. [DOI] [PubMed] [Google Scholar]
- 8.Fahmy K, Sakmar T P, Siebert F. Biochemistry. 2000;39:10607–10612. doi: 10.1021/bi000912d. [DOI] [PubMed] [Google Scholar]
- 9.Farrens D L, Altenbach C, Yang K, Hubbell W L, Khorana H G. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 10.Sheikh S P, Zvyaga T A, Lichtarge O, Sakmar T P, Bourne H R. Nature. 1996;383:347–350. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 11.Cai K, Klein-Seetharaman J, Farrens D, Zhang C, Altenbach C, Hubbell W L, Khorana H G. Biochemistry. 1999;38:7925–7930. doi: 10.1021/bi9900119. [DOI] [PubMed] [Google Scholar]
- 12.Ernst O P, Meyer C K, Marin E P, Henklein P, Fu W Y, Sakmar T P, Hofmann K P. J Biol Chem. 2000;275:1937–1943. doi: 10.1074/jbc.275.3.1937. [DOI] [PubMed] [Google Scholar]
- 13.Barak L S, Menard L, Ferguson S S, Colapietro A M, Caron M G. Biochemistry. 1995;34:15407–15414. doi: 10.1021/bi00047a003. [DOI] [PubMed] [Google Scholar]
- 14.Wess J, Nanavati S, Vogel Z, Maggio R. EMBO J. 1993;12:331–338. doi: 10.1002/j.1460-2075.1993.tb05661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prioleau C, Visiers I, Ebersole B J, Weinstein H, Sealfon S C. J Biol Chem. 2002;277:36577–36584. doi: 10.1074/jbc.M206223200. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell R, McCulloch D, Lutz E, Johnson M, MacKenzie C, Fennell M, Fink G, Zhou W, Sealfon S C. Nature. 1998;392:411–414. doi: 10.1038/32937. [DOI] [PubMed] [Google Scholar]
- 17.Meyer C K, Böhme M, Ockenfels A, Gärtner W, Hofmann K P, Ernst O P. J Biol Chem. 2000;275:19713–19718. doi: 10.1074/jbc.M000603200. [DOI] [PubMed] [Google Scholar]
- 18.Heck M, Hofmann K P. Biochemistry. 1993;32:8220–8227. doi: 10.1021/bi00083a024. [DOI] [PubMed] [Google Scholar]
- 19.Ferretti L, Karnik S S, Khorana H G, Nassal M, Oprian D D. Proc Natl Acad Sci USA. 1986;83:599–603. doi: 10.1073/pnas.83.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franke R R, Sakmar T P, Oprian D D, Khorana H G. J Biol Chem. 1988;263:2119–2122. [PubMed] [Google Scholar]
- 21.Sakmar T P, Franke R R, Khorana H G. Proc Natl Acad Sci USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han M, Sakmar T P. Methods Enzymol. 2000;315:251–267. doi: 10.1016/s0076-6879(00)15848-3. [DOI] [PubMed] [Google Scholar]
- 23.Cai K, Langen R, Hubbell W L, Khorana H G. Proc Natl Acad Sci USA. 1997;94:14267–14272. doi: 10.1073/pnas.94.26.14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel R, Fan G B, Sheves M, Siebert F. Biochemistry. 2000;39:8895–8908. doi: 10.1021/bi000852b. [DOI] [PubMed] [Google Scholar]
- 25.Marin E P, Krishna A G, Zvyaga T A, Isele J, Siebert F, Sakmar T P. J Biol Chem. 2000;275:1930–1936. doi: 10.1074/jbc.275.3.1930. [DOI] [PubMed] [Google Scholar]
- 26.Klein-Seetharaman J, Getmanova E V, Loewen M C, Reeves P J, Khorana H G. Proc Natl Acad Sci USA. 1999;96:13744–13749. doi: 10.1073/pnas.96.24.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altenbach C, Cai K, Khorana H G, Hubbell W L. Biochemistry. 1999;38:7931–7937. doi: 10.1021/bi9900121. [DOI] [PubMed] [Google Scholar]
- 28.Altenbach C, Klein-Seetharaman J, Cai K, Khorana H G, Hubbell W L. Biochemistry. 2001;40:15493–15500. doi: 10.1021/bi011545o. [DOI] [PubMed] [Google Scholar]
- 29.Imamoto Y, Kataoka M, Tokunaga F, Palczewski K. Biochemistry. 2000;39:15225–15233. doi: 10.1021/bi0018685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishna A G, Menon S T, Terry T J, Sakmar T P. Biochemistry. 2002;41:8298–8309. doi: 10.1021/bi025534m. [DOI] [PubMed] [Google Scholar]
- 31.Mielke T, Alexiev U, Glasel M, Otto H, Heyn M P. Biochemistry. 2002;41:7875–7884. doi: 10.1021/bi011862v. [DOI] [PubMed] [Google Scholar]
- 32.Abdulaev N G, Ridge K D. Proc Natl Acad Sci USA. 1998;95:12854–12859. doi: 10.1073/pnas.95.22.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau E M, Shichida Y. Proc Natl Acad Sci USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnis S, Hofmann K P. Proc Natl Acad Sci USA. 1993;90:7849–7853. doi: 10.1073/pnas.90.16.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J M, Altenbach C, Thurmond R L, Khorana H G, Hubbell W L. Proc Natl Acad Sci USA. 1997;94:14273–14278. doi: 10.1073/pnas.94.26.14273. [DOI] [PMC free article] [PubMed] [Google Scholar]