Abstract
Background
Analysis of DNA microarray data takes as input spot intensity measurements from scanner software and returns differential expression of genes between two conditions, together with a statistical significance assessment. This process typically consists of two steps: data normalization and identification of differentially expressed genes through statistical analysis. The Expresso microarray experiment management system implements these steps with a two-stage, log-linear ANOVA mixed model technique, tailored to individual experimental designs. The complement of tools in TM4, on the other hand, is based on a number of preset design choices that limit its flexibility. In the TM4 microarray analysis suite, normalization, filter, and analysis methods form an analysis pipeline. TM4 computes integrated intensity values (IIV) from the average intensities and spot pixel counts returned by the scanner software as input to its normalization steps. By contrast, Expresso can use either IIV data or median intensity values (MIV). Here, we compare Expresso and TM4 analysis of two experiments and assess the results against qRT-PCR data.
Results
The Expresso analysis using MIV data consistently identifies more genes as differentially expressed, when compared to Expresso analysis with IIV data. The typical TM4 normalization and filtering pipeline corrects systematic intensity-specific bias on a per microarray basis. Subsequent statistical analysis with Expresso or a TM4 t-test can effectively identify differentially expressed genes. The best agreement with qRT-PCR data is obtained through the use of Expresso analysis and MIV data.
Conclusion
The results of this research are of practical value to biologists who analyze microarray data sets. The TM4 normalization and filtering pipeline corrects microarray-specific systematic bias and complements the normalization stage in Expresso analysis. The results of Expresso using MIV data have the best agreement with qRT-PCR results. In one experiment, MIV is a better choice than IIV as input to data normalization and statistical analysis methods, as it yields as greater number of statistically significant differentially expressed genes; TM4 does not support the choice of MIV input data. Overall, the more flexible and extensive statistical models of Expresso achieve more accurate analytical results, when judged by the yardstick of qRT-PCR data, in the context of an experimental design of modest complexity.
Background
DNA microarrays are a powerful means of monitoring the expression of thousands of genes simultaneously. A variety of computational and statistical methods have been proposed to extract information from the large quantity of data generated from microarray experiments. Many methods assume, as we do here, the use of cDNA labeled with one of two fluorescent dyes to differentiate two treatments on a single microarray, implying data from two images to be analyzed. These methods include a number of data normalization techniques to reduce the effects of systematic errors and various kinds of statistical tests to identify differentially expressed genes in comparisons among different experimental conditions. There is as yet no single method that can be recommended under all circumstances for either normalization or identification of differential gene expression.
In recent years, ANOVA methods have gained popularity for identification of differential gene expression. The power of ANOVA methods derives from their flexibility in fitting and comparing different models to a given set of data [1]. One such method is the two-stage, log-linear ANOVA mixed models technique of Wolfinger, et al., [2]. Its first stage uses a normalization model designed to remove global effects across all microarrays. Its second stage uses a gene-specific model to estimate gene-treatment interactions as ratios of gene expression under control and treated conditions, along with a statistical significance. Kerr [3] notes that the global normalization model employed in this technique is conducive to combining data across genes for realistic and robust models of error, especially when random effects are included. Pan [4] compares different microarray statistical analysis methods and demonstrates that the log-linear ANOVA mixed model approach performs better than the t-test and regression approaches. The regression approach, although flexible and robust, assumes that the data is drawn from a normal distribution, while the t-test is limited due to very few degrees of freedom. Chu, et al., [5] compare two log-linear ANOVA mixed models for probe-level, oligonucleotide array data and found that both types of models capture key measurable sources of variability of oligonucleotide arrays for real and simulated data. Cui and Churchill [6] review the use of a mixed ANOVA model for analyzing a cDNA microarray experiment and conclude that such models provide a powerful way to obtain information from experiments with multiple factors or sources of variation. Rosa, et al., [7] review issues of analyzing cDNA microarrays with mixed linear models and puts such analysis in the larger context of Bayesian analysis procedures and adjustments for multiple testing.
Data normalization is the first step in analyzing microarray data; numerous data normalization methods have been proposed and investigated. While refinements of existing methods continue to appear (e.g., Futschik and Crompton [8]), naive methods, such as total intensity normalization, are still in use (e.g., Held, et al., [9]). Xie, et al., [10] did a comparative study of normalization methods and test statistics to analyze the results of a DNA-protein binding microarray experiment. Using performance and bias correction criteria, Bolstad, et al., [11] evaluate the cyclic lowess method, the contrast method, the quantile method, and baseline array scaling methods, both linear and non-linear; they demonstrate that normalization methods incorporating data from all microarrays perform better than methods employing a baseline array.
Several software tools that combine data normalization and statistical analysis are currently available. Dudoit, et al., [12] review these software tools with an emphasis on the TM4 microarray software suite, Bioconductor in R, and the BioArray Software Environment (BASE) system. Saeed, et al., [13] describe the features and capabilities of TM4, while Quackenbush [14] describes the normalization and transformation methods implemented in it. Williams, et al., [15], Zhu, et al., [16], and Khaitovich, et al., [17] have used TM4 in microarray data analysis. Another system is Expresso, an experiment management system that serves as a unifying framework to study data driven applications such as microarray experiments [18-20]. Expresso has adapted the two-stage ANOVA mixed models technique of Wolfinger, et al., [2] to the particular needs of individual microarray data sets. Our experience with numerous such data sets has demonstrated that modeling the underlying experiment carefully and completely is essential to obtaining meaningful and defensible results. Use of tools that require experiments to conform to their analysis methods are less than satisfactory.
In this paper, we compare the Expresso analysis methodology to the approach provided in the TM4 microarray analysis software suite [13]. Each is invoked to identify differentially expressed genes in two experimental data sets, each of which uses an Arabidopsis thaliana oligonucleotide array. Along the way, we demonstrate differences between the use of integrated intensity values (IIV) and median intensity values (MIV) as inputs. We report interactions between normalization and gene identification methods. We use quantitative reverse-transcriptase PCR (qRT-PCR) results to assess the consistency of genes reported by TM4 and Expresso as having significant differential expression.
Results and discussion
Here, we report a portion of the results obtained in our comparison of Expresso analysis and the TM4 pipeline (see Materials and Methods). Figure 1 illustrates the overall flow of the statistical analyses of microarray data that were done in this study. We began with microarray data in GPR format from Experiment 1 and Experiment 2. Median intensity values (MIV) from the GPR files can be analyzed by the Expresso GP and GOT models directly. ExpressConverter provides integrated intensity values (IIV) for further Expresso and TM4 analysis. The MIDAS normalization and filtering pipeline executes these steps in order: total intensity normalization (subscript T), lowess normalization (subscript L), standard deviation regularization (subscript S), and low intensity filter (subscript F). MIDAS allows tapping the output of any step in the pipeline; for example, IIVTL signifies an MEV file after total intensity normalization followed by lowess normalization. The identification of genes with significant differential expression was performed on all GPR and MEV files, using the Expresso GP and GOT models and the t-test in MEV.
Figure 1.
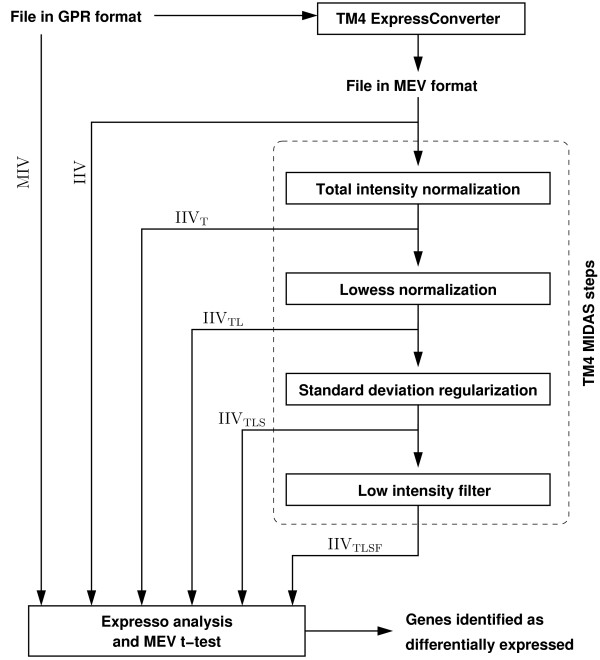
Overall flow of the statistical analyses of microarray data. Data input is in GPR format and provides the MIV for each spot. TM4 analysis requires ExpressConverter to generate MEV format containing the IIV for each spot. The normalization steps performed by MIDAS are T (total intensity normalization), L (lowess normalization), S (standard deviation regularization), and F (low intensity filter). Differential gene expression is obtained from the Expresso GP model, the Expresso GOT model (Experiment 2 only), and the t-test in MEV.
Normalization and low intensity filtering in TM4
Quackenbush [14] describes the use of ratio-intensity plots (RI-plots) to detect and normalize for any systematic intensity-dependent dye bias using lowess normalization (see Materials and Methods). We evaluated the effect of lowess normalization within the context of the flow in Figure 1 by creating RI-plots after each step for the second replicate microarray in Experiment 1, WT plant. Supplementary Figure 1 - see Additional file: 1 contains these RI-plots. The IIVTL is indeed effective, for this data set, in correcting systematic dye bias, suggesting that preprocessing by these two normalization steps in MIDAS may be a good practice in many situations.
The normalization and filtering pipeline affects the number of genes identified as differentially expressed in both the GP and GOT models. See Table 1. For example, in Experiment 1, the GP model using IIV input data identifies 567 up-expressed genes in the WT microarrays, while it identifies only 460 WT genes as up-expressed if IIVTLSF (processed by the complete MIDAS pipeline) input data is used.
Table 1.
Number of differentially expressed genes. Numbers of identified differentially expressed genes in the GP and GOT models after preprocessing by 0 or more MIDAS pipeline steps — IIV, IIVT, IIVTL, IIVTLS, or IIVTSLF. For Experiment 1, up-expression (+) and down-expression (-) numbers are given for both WT and antiPLD. For Experiment 2, + and -1 numbers are given for all 4 genotypes separately. The ALL entries correspond to the H1 hypotheses of the GOT model (see Materials and Methods).
| Genotype | IIV | IIVT | IIVTL | IIVTLS | IIVTLSF | |
| GP Model — Experiment 1 | ||||||
| WT | + | 567 | 571 | 487 | 495 | 460 |
| WT | - | 552 | 553 | 499 | 499 | 460 |
| antiPLD | + | 442 | 421 | 440 | 471 | 431 |
| antiPLD | - | 336 | 346 | 381 | 363 | 334 |
| GOT Model — Experiment 2 | ||||||
| Col-0 | + | 227 | 309 | 242 | 240 | 270 |
| Col-0 | - | 122 | 171 | 178 | 180 | 183 |
| Cvi-0 | + | 106 | 108 | 82 | 78 | 71 |
| Cvi-0 | - | 63 | 93 | 74 | 71 | 67 |
| WS | + | 234 | 346 | 279 | 269 | 150 |
| WS | - | 331 | 347 | 340 | 340 | 224 |
| Th | + | 238 | 398 | 350 | 363 | 361 |
| Th | - | 196 | 245 | 241 | 246 | 240 |
| ALL | + | 298 | 428 | 357 | 354 | 290 |
| ALL | - | 202 | 261 | 253 | 248 | 195 |
Small changes in the number of genes identified as up- or down-expressed after successive MIDAS steps may mask larger changes in the composition of sets of up- and down-expressed genes. To obtain a more precise view of the effects of MIDAS changes, we computed retention counts (RC) and retention percentages (RP) between the gene successive sets whose numbers are in Table 1. RC is the number of genes in the set before the MIDAS step that remain in the set after the step. RP is the percentage of remaining genes with respect to the number of genes in the set after the MIDAS step. Table 2 contains the RC and RP values corresponding to the counts in Table 1. For Experiment 1, there is a tremendous drop in retention during the lowess normalization that follows the total intensity normalization. There is not a drop of corresponding magnitude for Experiment 2. For both experiments, normalization has a significant effect on the sets of genes identified as differentially expressed.
Table 2.
Retention counts and percentages. Retention counts (RC) and retention percentages (RP) for the differentially expressed gene sets of Table 1. RC is the number of genes in a set before the MIDAS step that remain in the set after that step. RC is the number of genes in the set before the MIDAS step that remain in the set after that step. RP is the percentage of remaining genes with respect to the number of genes in the set after the MIDAS step. RC and RP are reported for the intersections IIV∩ IIVT, IIVT ∩ IIVTL, IIVTL ∩ IIVTLS, and IIVTLS ∩ IIVTLSF, as well as intersection IIV∩ ∩IIVTLSF, which corresponds to the effect of MIDAS steps from the start of the pipeline to the end. The ALL entries correspond to the H1 hypotheses of the GOT model (see Materials and Methods).
| IIV ∩ | IIVT∩ | IIVTL∩ | IIVTLS∩ | IIV ∩ | |||||||
| IIVT | IIVTL | IIVTLS | IIVTLSF | IIVTLSF | |||||||
| Genotype | RC | RP | RC | RP | RC | RP | RC | RP | RC | RP | |
| GP Model — Experiment 1 | |||||||||||
| WT | + | 545 | 95.45 | 74 | 15.20 | 285 | 57.58 | 456 | 99.13 | 59 | 12.83 |
| WT | - | 532 | 96.20 | 55 | 11.02 | 306 | 61.32 | 459 | 99.78 | 53 | 11.52 |
| antiPLD | + | 403 | 95.72 | 58 | 13.18 | 278 | 59.02 | 430 | 99.77 | 46 | 10.67 |
| antiPLD | - | 326 | 94.22 | 45 | 11.81 | 203 | 55.92 | 330 | 98.80 | 33 | 9.88 |
| GOT Model — Experiment 2 | |||||||||||
| Col-0 | + | 224 | 72.49 | 221 | 91.32 | 219 | 91.25 | 172 | 63.70 | 159 | 58.89 |
| Col-0 | - | 121 | 70.76 | 150 | 84.27 | 165 | 91.67 | 120 | 65.57 | 92 | 50.27 |
| Cvi-0 | + | 81 | 75.00 | 65 | 79.27 | 71 | 91.25 | 49 | 69.01 | 36 | 50.70 |
| Cvi-0 | - | 60 | 64.52 | 60 | 81.08 | 63 | 88.73 | 41 | 61.19 | 38 | 56.72 |
| ws | + | 223 | 64.45 | 230 | 82.44 | 248 | 92.19 | 108 | 72.00 | 101 | 67.33 |
| ws | - | 293 | 84.44 | 267 | 78.53 | 314 | 92.35 | 180 | 80.36 | 149 | 66.52 |
| Th | + | 236 | 59.30 | 308 | 88.00 | 330 | 90.91 | 256 | 70.91 | 201 | 55.68 |
| Th | - | 183 | 74.69 | 210 | 87.14 | 225 | 91.46 | 172 | 71.67 | 144 | 60.00 |
| ALL | + | 282 | 65.89 | 307 | 85.99 | 333 | 94.07 | 200 | 68.97 | 177 | 61.03 |
| ALL | - | 196 | 75.10 | 196 | 77.47 | 230 | 92.74 | 133 | 68.21 | 103 | 52.82 |
In Experiment 1 results, the number of genes commonly assessed by Expresso as significantly expressed when using IIV and IIVT is high. For example, there is 95.45% retention of WT genes (545 total) assessed as up-expressed when using IIVT data in Expresso compared to that when using IIV data. Retention percentage of these genes assessed as expressed however went down after doing lowess normalization. There is only 15.20% retention of WT genes (74 total) assessed as up-expressed in the results when using IIVTS data in Expresso compared to when using IIVT. While we observe increase in the retention percentage in IIVTLS (from IIVTL) and IIVTLSF (from IIVTLS), there's low retention percentage in the results using IIVTLSF from IIV data. This can be traced in the low retention percentage of IIVTL from IIVT. Hence, the normalization method that affects the results in Experiment 1 the most is lowess normalization.
The results of Expresso on Experiment 2 show that low retention percentages happen after appli-cation of total intensity normalization (lowest is 59.30%) and after application of low intensity filtering (lowest is 61.19%). The low retention percentages shown in the IIV ∩ IIVTLSF column implies that the normalization pipeline also significantly affects the results in Expresso analysis of Experiment 2.
Choice of intensity signal data
The input to statistical analysis of microarray experiments is a set of real numbers that represent the measured intensity signal for each spot in a microarray. Much statistical analysis of microarray data has traditionally used median intensity values (MIV). The alternative used in TM4 is the integrated intensity value (IIV). (See Materials and Methods.) Since IIV is intended to integrate the measured intensity across the biological sample printed at a spot, one might expect IIV to be a more accurate assessment of the biological measurement than MIV data. For example, a spot having 100 pixels and a median intensity of 5,000 has the same IIV as a spot having 50 pixels and a median intensity of 10,000.
This study provides the opportunity to observe the difference that choosing MIV or IIV makes on the sets of genes ultimately identified as differentially expressed. We used the GP model to analyze unnormalized MIV and IIV data from Experiment 1, and we used the GOT model to analyze unnormalized MIV and IIV data from Experiment 2. Table 3 reports a summary of the results. In Experiment 1, 725 WT genes are assessed as up-expressed and 774 WT genes are down-expressed when MIV data are used in Expresso. These numbers decreased to 567 up-expressed genes and 552 down-expressed genes when IIV data are used instead. A similar trend is observed in Experiment 2 results when using MIV and IIV data. These results suggest that employing IIV input data with Expresso analysis leads to more conservative results than employing MIV input data.
Table 3.
Comparison of MIV and IIV. We compare MIV and IIV input data with respect to the sets of genes ultimately identified as differentially expressed. The genotype and set labelings are the same as those in Table 1. The GP model was used to analyze the unnormalized MIV and IIV data from Experiment 1. The GOT model was used to analyze the unnormalized MIV and IIV data from Experiment 2. Up-expressed and down-expressed counts are reported for both experiments and all genotypes. The count of genes in the intersections is found in column Common. For convenience, percentages of the intersection with respect to the MIV and IIV sets are tabulated in the last two columns. The ALL entries correspond to the H1 hypotheses of the GOT model (see Materials and Methods).
| Genotype | MIV | IIV | Common | % of common in MIV | % of common in IIV | |
| GP Model — Experiment 1 | ||||||
| WT | + | 725 | 567 | 501 | 69.10% | 88.36% |
| WT | - | 774 | 552 | 515 | 66.54% | 93.30% |
| antiPLD | + | 518 | 422 | 357 | 68.92% | 84.60% |
| antiPLD | - | 505 | 336 | 313 | 61.98% | 93.15% |
| GOT Model — Experiment 2 | ||||||
| Col-0 | + | 311 | 227 | 198 | 63.67% | 87.22% |
| Col-0 | - | 190 | 122 | 110 | 57.89% | 90.16% |
| Cvi-0 | + | 185 | 106 | 89 | 48.11% | 83.96% |
| Cvi-0 | - | 97 | 63 | 59 | 60.82% | 93.65% |
| ws | + | 326 | 234 | 181 | 55.52% | 77.35% |
| ws | - | 482 | 331 | 273 | 56.64% | 82.48% |
| Th | + | 363 | 238 | 219 | 60.33% | 92.02% |
| Th | - | 251 | 196 | 166 | 66.14% | 84.69% |
| ALL | + | 448 | 298 | 249 | 55.58% | 83.56% |
| ALL | - | 294 | 202 | 159 | 54.08% | 78.71% |
Comparison of statistical methods
We compared the performance of the GP model, the GOT model, and the t-test of MEV in identifying differentially expressed genes in Experiment 2. We used the IIVTLSF data of Experiment 2 as input to these methods. We also contrast these results with MIV data analyzed in GP. Table 4 reports counts for these analyses.
Table 4.
Comparison of Expresso analysis and MEV t test. We compare the number of genes identified as differentially expressed by Expresso analysis and the MEV t-test. Counts for both up-expressed and down-expressed genes, as well as all four genotypes of Experiment 2, are reported. The first three analyses — the GP model, the GOT model, and the MEV t-test — take the IIVTLSF data as input. For point of comparison, the last analysis uses the GP model on MIV input.
| GP Model | GOT Model | MEV t-test | GP Model | |||||
| IIVTLSF | IIVTLSF | IIVTLSF | MIV | |||||
| Genotype | + | - | + | - | + | - | + | - |
| Col-0 | 1837 | 1563 | 270 | 183 | 2125 | 2007 | 1761 | 1212 |
| Cvi-0 | 817 | 1248 | 71 | 67 | 478 | 823 | 669 | 1403 |
| WS | 2606 | 2002 | 150 | 224 | 535 | 388 | 2422 | 1184 |
| Th | 2016 | 1314 | 361 | 240 | 1144 | 827 | 2395 | 1457 |
| Totals | 7276 | 6127 | 852 | 714 | 4282 | 4045 | 7247 | 5256 |
The plot in Figure 2a demonstrates that the estimates of Iog2(fold change) are the same in GP and GOT. As Figure 2b shows, the p values by GOT are smaller compared to the p values calculated by GP. Use of the MEV t-test resulted in fewer genes assessed as significantly expressed when compared to the numbers for the GP model. The results obtained when MIV data was used as input to GP, is closest to the results when using IIVTLSF.
Figure 2.
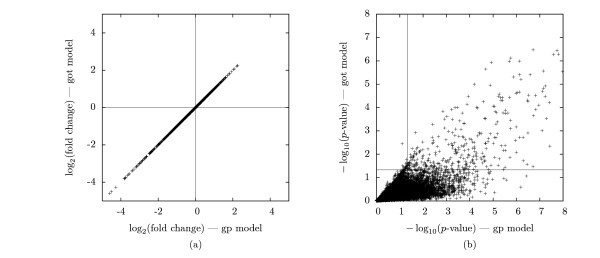
Comparison of GP and GOT models. Comparison of estimated log2(fold change) and the corresponding p-value estimates for the GP and GOT model results of the WS ecotype values in Table 4 (a) This is a scatter plot of the estimated log2(fold change) values from the GP and GOT models; these values are essentially identical, (b) This is a scatter plot of the - log10 (p-value), again for the GP and GOT models. The dotted lines correspond to - log10(0.05), as our significance cutoff is 0.05. For the GP model, it is the points to the right of the dotted line that are significant. For the GOT model, it is the points above the dotted line that are significant.
To compare the effectiveness of Expresso and TM4 in identifying gene differential expression, we compared the identified direction of differential expression of a select set of genes per genotype in Experiment 2 with results obtained by qRT-PCR. See Table 5. The lowest overall percentage (71.9%) of agreement is between the qRT-PCR results and the MEV t-test results using IIVTLSF. The log(fold change) estimates of the GP model has 77.1% percentage agreement with the qRT-PCR results, which is slightly higher than the percentage for the MEV t-test. The results of the GP model using MIV data demonstrated the greatest agreement, 90.1%, with the qRT-PCR results.
Table 5.
Comparison of qRT-PCR with Expresso and TM4. We compare the qRT-PCR results with identified up-expressed and down-expressed genes in Experiment 2 using Expresso and TM4. Results of qRT-PCR are available for each of the four ecotypes in the numbers n in the second column. Numbers of agreement or non-agreement are shown in the S, D, and F columns. The S (same) column tabulates the number of genes for which the sign of the log(fold change) for statistically significant differential expression matches the direction of change in the corresponding qRT-PCR result. The D (differing) column tabulates the number of genes for which the sign of the log(fold change) for statistically significant differential expression is in the opposite direction of the change in the corresponding qRT-PCR result. The F (filtered) column tabulates the number of genes for which there is a qRT-PCR result, but for which either the gene was filtered by MIDAS low intensity filtering or the analysis method did not assess the change in expression as statistically significant. The MEV t-test (first grouping) results are for the typical TM4 process, which involves IIV input data followed by the four MIDAS steps, which we denote IIVTLSF The GP model (second grouping) gives the same numbers and uses the same IIVTLSFinput data, the GP model (third grouping) results use MIV input data and has no filtered genes.
| GP Model | GP Model | MEV t-test | |||||||
| qRT-PCR | MIV | IIVTLSF | IIVTLSF | ||||||
| Genotype | n | S | D | S | D | F | S | D | F |
| Col-0 | 55 | 50 | 5 | 43 | 8 | 4 | 40 | 9 | 4 |
| Cvi-0 | 52 | 46 | 6 | 38 | 9 | 5 | 36 | 7 | 9 |
| ws | 59 | 54 | 5 | 46 | 10 | 3 | 41 | 9 | 9 |
| Th | 26 | 23 | 3 | 21 | 3 | 2 | 19 | 3 | 4 |
| Total | 192 | 173 | 19 | 148 | 30 | 14 | 138 | 28 | 26 |
| Percentage | 90.1% | 9.9% | 77.1% | 15.6% | 7.3% | 71.9% | 14.6% | 13.5% | |
Figures 3, 4, and 5 present the actual assessed Iog2(fold change) values for 50 selected Col-0 genes in Experiment 2, along with their qRT-PCR values. These are the 50 Col-0 genes, among the 55 with qRT-PCR values, for which we have expression values for all methods. For each gene, a histogram of the Iog2(fold change) estimates of qRT-PCR, the GP model using MIV, the GP model using IIVTLSF, and the MEV t-test is given. The 50 histograms are spread over three figures to enhance readability and are in increasing order by qRT-PCR estimated change. In general, the log2(fold change) estimates of the GP model and of the MEV t-test, all IIVTLSF input data, are approximately the same, while being slightly different from estimates of the GP model using MIV input data. As might be expected, disagreement between qRT-PCR and microarray results are more prevalent for small estimated log2(fold change) values. The histograms for genes AT4G09020 (Figure 3), AT1G35580 (Figure 4), and AT3G29360 (Figure 4) show that the direction of log(fold change) estimate of qRT-PCR matches the direction of the GP model using MIV input data, while differing from the direction of the estimates of the GP model and the MEV t-test using IIVTLSF input data.
Figure 3.
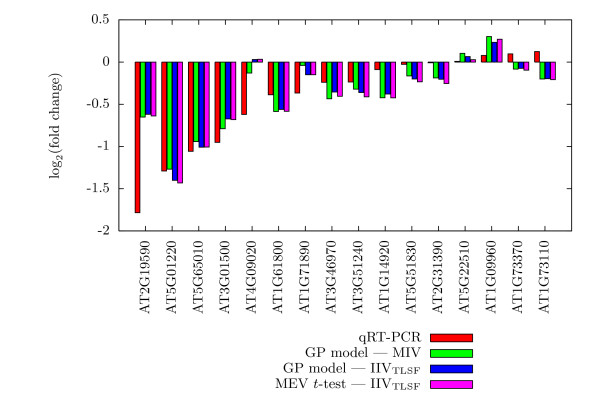
Comparison of qRT-PCR results, Part I. Comparison of qRT-PCR results to Expresso and MEV t-test results for first 16 of 50 selected genes of the Col-0 genotype of Experiment 2.
Figure 4.
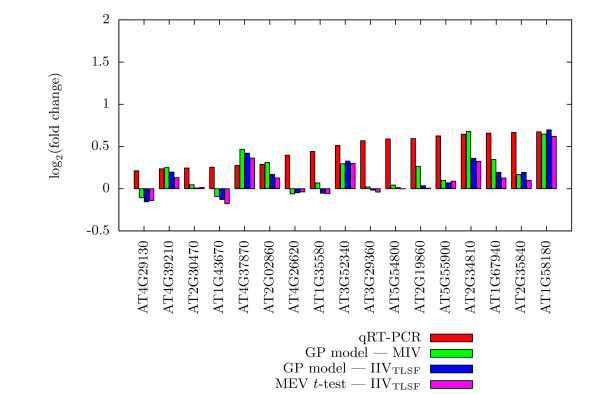
Comparison of qRT-PCR results, Part II. Comparison of qRT-PCR results to Expresso and MEV t-test results for the middle 17 of 50 selected genes of the Col-0 genotype of Experiment 2.
Figure 5.
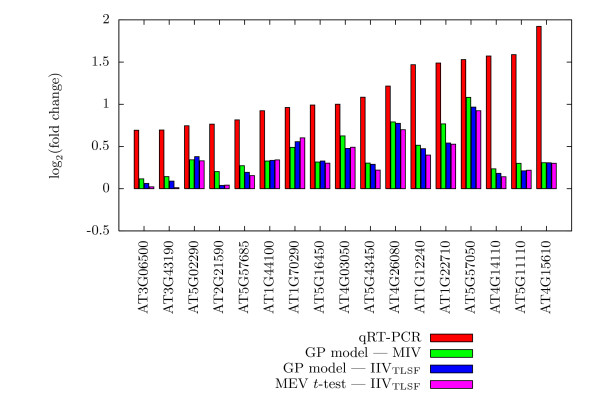
Comparison of qRT-PCR results, Part III. Comparison of qRT-PCR results to Expresso and MEV t-test results for the last 17 of 50 selected genes of the Col-0 genotype of Experiment 2.
Table 6 summarizes genotype-specific correlation results, which demonstrate that the GP model us-ing MIV input data has the highest correlation with qRT-PCR compared to the GP model and the MEV t-test using IIVTLSF input data. The highest correlation of 0.85 is for the Col-0 results of qRT-PCR versus the GP model using MIV input. The corresponding correlations for Cvi-0, WS, and Th are 0.73, 0.66, and 0.84, respectively, which are all best among the analysis methods.
Table 6.
Correlation of qRT-PCR with Expresso and TM4. We calculate the correlation of qRT-PCR results from the Expresso GP model and the MEV t-test. For each comparison, the analytical result for each gene for which there is a qRT-PCR result for a particular genotype is assembled into a result vector. We used SAS to compute a Pearson correlation of each result vector with the corresponding vector of qRT-PCR results. The computed correlations are as reported above.
| Genotype-Specific Correlation | ||||
| Comparison | Col-0 | Cvi-0 | WS | Th |
| qRT-PCR versus GP model with MIV input | 0.85 | 0.73 | 0.66 | 0.84 |
| qRT-PCR versus GP model with IIVTLSF input | 0.83 | 0.69 | 0.61 | 0.78 |
| qRT-PCR versus MEV t-test with IIVTLSF input | 0.83 | 0.65 | 0.64 | 0.77 |
Conclusion
Our integration and comparison of Expresso analysis and the capabilities of TM4 has highlighted successes in microarray analysis, some similarities, and some differences. The success of microarray analysis is demonstrated by considerable agreement between qRT-PCR results and the results of all the examined microarray analysis methods. The greatest agreement was found when median intensity value (MIV) inputs were analyzed with the Expresso GP analysis model. We also found that the use of integrated intensity value (IIV) inputs for Expresso analysis consistently resulted in fewer genes identified as differentially expressed when compared to results from MIV inputs. This suggests that the use of IIV inputs is more conservative than the use of MIV inputs, while MIV inputs may give greater agreement to qRT-PCR results than IIV inputs.
Our results demonstrate that the MIDAS normalization and filtering pipeline corrects systematic intensity-dependent dye bias on a per microarray basis. The normalization stage in Expresso analysis removes global effects across all microarrays and complements the per microarray normalization methods of MIDAS. The generally better agreement of Expresso analysis with qRT-PCR results when compared to the MEV t-test suggests that it would be desirable for MEV to have an ANOVA test that has the greater flexibility of the Expresso gene model.
Methods
Median and integrated intensity values
This research considers two ways of measuring spot intensity, one or both of which are reported by typical microarray image processing software. The median intensity value (MIV) of a spot is the median value of all the pixels identified as part of the spot. The integrated intensity value (IIV) of a spot is the total value of all the pixels identified as part of the spot. In this research, both are background-corrected values. If the IIV data is unavailable, but the radius of the bounding circle of the spot and its average intensity value are available, then the IIV data can be estimated as the product of the average intensity and the number of pixels in the circle. This is the estimate used by ExpressConverter (below) when it converts GPR format data to MEV format data.
Microarray data sets
We used data sets from two experiments that used the Arabidopsis Oligonucleotide Microarrays [21], which include 25,712 elements, each a gene-specific 70-mer (Qiagen/Operon, Valencia, CA) for a known or putative open reading frames in Arabidopsis thaliana. There are 48 blocks per microarray, 25 rows by 24 columns (600 spots) per block, and 28,800 spots per microarray, including spots for the 25,712 gene-specific 70-mers and 302 control elements. The remaining 2,786 spots are blank.
Experiment 1
Experiment 1 compares the responses of Arabidopsis thaliana wild type, ecotype Columbia (henceforth, WT), and of an antisense plant for phospholipase D α (antiPLD) in Columbia background to drought stress [22]. Plants were harvested at a single time point, and two biological replicate hybridizations were done for each of WT and antiPLD. Scan Array Express (PerkinElmer Life and Analytical Sciences, Inc., Boston, MA USA) was used to quantitate the four microarrays. By default, ScanArray Express performs a global lowess normalization of median intensities per microarray. ScanArray Express output its results to four files in GenePix (GPR) format, which constitute the Experiment 1 data set.
Experiment 2
Li, et al., [23] compare the responses to elevated CO2 of a wild Arabidopsis thaliana relative (Thellungiella halophila, ecotype Shandong; Th) and of three Arabidopsis thaliana ecotypes: Wassilewskija (WS), Columbia (Col-0), and Cape Verde Islands (Cvi-0). Three biological replicate hybridizations were done for each genotype. GenePix (Axon Instruments, Union City, CA USA) was used to quantitate the twelve microarrays. GenePix also performs by default a global lowess normalization of median intensities per microarray. The output of GenePix is twelve GPR files, which constitute the Experiment 2 data set.
Real-time quantitative RT-PCR
For verification of microarray results in Experiment 2, Li, et al., [23] performed real-time quantitative reverse-transcriptase PCR (qRT-PCR) for selected genes — 55 in Col-0; 52 in Cvi-0; 59 in WS; 26 in Th. Supplementary Table 1 - see Additional file: 2 contains the annotation of the selected genes. In brief, primer pairs were selected to represent unique sequences in the Arabidopsis thaliana genome and in the Thellungiella sequences deposited in NCBI. Thellungiella actin (CX129618) cDNA primers and Arabidopsis thaliana. Ubiquitin-10 cDNA primers were used as internal controls in the qRT-PCR analyses. RT-PCR products were detected using the fluorescent dye SYBR-green (Applied Biosystems, Foster City, CA USA) and the ABI PRISM/Taqman 7900 Sequence Detection System (Applied Biosystems, Foster City, CA USA). Dissociation curves were generated for each reaction to ensure specific amplification. Three repeats were done for each gene. The averaged threshold cycle numbers were used to estimate original mRNA levels.
The microarray results of Experiment 2 suggested that exposure to elevated CO2 resulted in changes in expression of many genes associated with carbon metabolism, and those associated with pho-tosynthetic carbon metabolism in particular. This included genes encoding proteins that transport pho-tosynthate out of the chloroplast, where carbon fixation takes place, for export to other parts of the cell, and also genes encoding transport proteins that export carbon skeletons out of the cell to other tissues where growth is taking place. Because of this finding, it was important to validate the results obtained for gene expression associated with carbon metabolism with qRT-PCR. Hence, a number of the genes in Supplementary Table 1 - see Additional file: 2 are related to carbon metabolism.
Expresso analysis
Expresso analysis employs a general and flexible method to identify differentially expressed genes that is adapted from the two-stage analysis method of Wolfinger, et al., [2]. In general, Expresso analysis consists of two log-linear ANOVA mixed models, called the normalization model and gene model. The first estimates and removes the experiment-wise systematic errors, while the second estimates and removes the gene specific errors. The residual that remains is the log-ratio estimate for each gene. In particular, the Tukey-Kramer multiple comparison of treatment effects on each gene is performed to estimate its expression level and the significance of (confidence in) that expression level. Expresso analysis is implemented for and executed on SAS (SAS/STAT version 8.2, SAS Institute Inc., Gary, NC USA).
The original model of Wolfinger, et al., [2] includes the treatment and the array as the main effects. In previous Expresso analysis, we have extended that model to experiment-appropriate models that include additional fixed and random effects. Here, the design of the two-dye oligonucleotide microarray used in Experiment 1 and Experiment 2 includes various controls strategically positioned in different blocks of the microarray. This makes it possible to estimate the random block effect in each microarray. Furthermore, the dye effect is included in the normalization model to estimate and remove the global dye bias.
For this research, we developed two Expresso models, one whose gene model assesses the gene-(plant sample) effect (the GP model) and the other whose gene model assesses the gene-genotype-treatment effect (the GOT model). The GP model is much like previous Expresso models and is applicable to both Experiment 1 and Experiment 2. However, the GOT model is specific to analyzing Experiment 2. In both experiments, we used the GP model to estimate the differences in response of individual genotypes to treatment (drought stress versus control or ozone stress versus ambient ozone). However, the GOT model was used to estimate the effects of treatment (ozone stress), aside from the effect of individual genotypes.
Expresso GP model
The normalization model is
yspdab = μ + Pp + Dd + Aa + (P × A)pa + Bba + rspdab.
Each yspdab value is the log2-transformed intensity of spot s within the dye d image in block b of array a. (A spot may represent a gene, a control, or a blank.) The global mean of the yspdab values, over all microarrays, is μ. The fixed effects in the model are the plant sample effect Pp, where p indexes the various distinct plant samples from which mRNA was obtained, and the dye effect Dd, where d has two values for the two dyes. The random effects in the model are the array effect Aa, where a indexes the microarrays, the interaction effect (P × A)pa of plant sample p with microarray a, and the block effect Bba, where b identifies the block within microarray a. The model residual is rspdab. This differs from the normalization model in Wolfinger, et al., [2], in that it incorporates dye and block effects. It is a refinement of the Expresso normalization model in Watkinson, et al., [18], in that it has no printing pin effect, which is specific to the analysis in the earlier paper, and includes the block effect.
The second stage of the analysis uses the residual values rspdab computed in the first stage to estimate the interaction between an individual gene g and each plant sample p at a significance level ≤ α = 0.05. Index g is added to the residual values rspdab resulting to rgspdab. The value of g is determined using the mapping of s index values to g index values. The gene model is
rgspdab = Gg + (G × P)gp + (G × D)gd + (G × A)ga + λgspdab.
Here, g is a spot that represents a gene (not a blank or control) within the dye d image in block b of array a. The value Gg is the mean of residual values for all spots that represent gene g in all images. The interactions (G × P)gp of gene g with plant sample p and of (G × D)gd of gene g with dye d are the fixed effects. The interaction (G × A)ga of gene g with mi-croarray a is a random effect. The λgspdab values are stochastic errors. This differs from the gene model in Wolfinger, et al., [2], in that it incorporates interactions between gene and dye and between gene and array. It refines the Expresso gene model in Watkinson, et al., [18] to include the interaction between gene and array.
The estimate of the expression level of each gene in each treatment comparison is done by computing the pair-wise least square mean differences of gene-treatment effects. The Tukey-Kramer multiple comparison of gene-(plant sample) effects on each gene is made to estimate the p values associated with each calculated expression level. If there are ρ plant samples, then there are possible pairwise comparisons. If we index the plant samples from 1 to ρ, then the null hypothesis for gene g and comparison i, j, where 1 ≤ i <j ≤ ρ, is
Ho: (G × P)gi = (G × P)gi.
The difference (G × P)gi - (G × P)gj is the estimate of the log2(fold change) of gene g in the experimental comparison Pi versus Pj. The analysis also yields a p-value for the statistical confidence in each difference.
The above GP model was used to analyze both Experiment 1 and Experiment 2. In both experiments, there are 48 blocks per array. In Experiment 1, there are four arrays and four plant samples, namely, WT-control, WT-stressed, antiPLD-control, and antiPLD-stressed. In Experiment 2, there are 12 arrays and eight plant samples, namely, Col-0-test, Col-0-control, Cvi-0-test, Cvi-0-control, WS-test, WS-control, Th-test, and Th-control.
Expresso GOT model
We wanted to estimate the gene-treatment effects separately from gene-genotype-treatment interaction effects using just one model. To do this, we designed the gene-genotype-treatment model, an alternative set of log-linear ANOVA mixed models, for the elevated CO2 experiment where we unfold the genotype information from the plant sample factor in the GP model. This resulted in a normalization model that includes the genotype effect (Oo) with 4 levels (Col-0, Cvi-0, WS, and Th) and basic treatment effect (Tt) with 2 levels (test and control). The random array (Aa) effect however needs to be removed from the model since it confounds the genotype effect.
The normalization model is
ysotdab = μ + Oo + TT + Dd + (O × T)ot + Bba + rsotdab.
Each ysotdab value is the log2-transformed intensity of spot s for genotype o and treatment t within the dye d image in block b of array a. We have that μ is as in the GP model. The fixed effects in the model are the genotype effect Oo, where o indexes the genotype (organism), the treatment effect Tt, where t is the treatment, and the dye effect Dd, where d has two values for the two dyes. The random effects in the model are the interaction effect (O × T)ot of genotype o with microarray a, and the block effect Bba, where b identifies the block within microarray a. The model residual is rsotdab. This differs from the normalization model in Wolfinger, et al., [2], in that it incorporates genotype (organism), dye, and block effects. It is a refinement of the Expresso normalization model in Watkinson, et al., [18], in that it has no printing pin effect, and includes the genotype and block effects.
The second stage of the analysis uses the residual values rsotdab computed in the first stage to estimate the interaction among an individual gene g, each genotype o, and each treatment t, at a significance level ≤ α = 0.05. Index g is added to the residual values rsotdab resulting to rgsotdab. The value of g is determined using the mapping of s index values to g index values. The gene model is
rgsotdab = Gg + (G × O)go + (G × T)gt + (G × O × T)got + (G × D)gd + λgsotdab
Here, Gg are as for the GP model. The interactions (G × O)go of gene g with genotype o, (G × T)gt of gene g with treatment t, (G × O × T)got of gene g with genotype o and treatment t, and (G × D)gd of gene g with dye d are fixed effects. The λgsotdab values are stochastic errors. This differs from the gene model in Wolfinger, et al., [2], in that it incorporates interactions between gene and genotype, between gene and genotype with treatment, and between gene and array. It refines the Expresso gene model in Watkinson, et al., [18] to include interactions between gene and genotype and between gene and genotype with treatment.
We do pairwise comparisons as in the GP model, but there are two plausible classes of null hypotheses to test within the GOT gene model. If τ is the number of treatments, then the class 1 null hypothesis for gene g and comparison i,j, where 1 ≤ i <j ≤ τ, is
H1: (G × T)gi = (G × T)gj.
The difference (G × T)gi - (G × T)gj is the estimate of the log2(fold change) of gene g in the Ti versus Tjcomparison. This particular comparison looks for gene-treatment effects that are independent of genotype.
We can still estimate the expression level of each gene with respect to a specific genotype level by computing the pair-wise least square differences of gene-genotype-treatment interaction effects. The class 1 null hypothesis for gene g and comparison i, j, where 1 ≤ i <j ≤ τ, is
H2: (G × O × T)goi = (G × O × T)goj.
The difference (G × O × T)goi - (G × O × T)goj is the estimate of the log2 (fold change) of gene g of a specific genotype o for the Ti versus Tj comparison. The analysis also yields a p-value for the statistical confidence in each difference.
The GOT model applies to Experiment 2 in a straightforward way. There are 12 arrays, two treatments, and four genotypes, namely, Col-0, Cvi-0, WS, and Th.
TM4 microarray software suite
The TM4 microarray software suite consists of several components freely available from the TM4 web site [24]. In this research, we employed these components: ExpressConverter, Microarray Data Analysis Software (MIDAS), and Microarray Experiment Viewer (MEV). ExpressConverter converts microarray data from various data formats, such as the GenePix Results (GPR) format, to the MEV format, which is used by MIDAS and MEV. MEV format includes only integrated intensity values (IIV), which is the kind of intensity values expected of all TM4 components.
MIDAS data normalization methods and filters
Low intensity and saturated spots are marked by quantitation programs. These spots are filtered out from the data before doing any further normalization or statistical analysis. Data normalization methods proceed from the assumption that only a relatively small proportion of the genes change significantly in expression level between the two hybridized mRNA samples. This assumption is reasonable for these data sets since the hybridizations and subsequent analysis address nearly all Arabidopsis thaliana genes. The MIDAS component of TM4 provides a number of data normalization methods and filters and supports applying them in a pipelined fashion [13,14].
Total intensity normalization
While our assumption implies that the average measured intensities of the two channels of a cDNA or oligonucleotide microarray should be almost the same, these averages are often significantly different, due to differences in the inherent fluorescence of the two dyes. The total intensity normalization step in TM4 is a straightforward means to eliminate this global dye bias. For each spot i, where 1 ≤ i ≤ n, let Ri and Gi be the measured intensities of the spot in the two channels. The normalized intensity data for spot i is = κGi and = Ri, where κ is the normalization factor Quackenbush [14] discusses this normalization in the context of sev-eral variations that are possible to address differing channel intensities.
Lowess normalization
Beyond the global dye bias, there is dye bias that is dependent on the measured spot intensities [25,26]. TM4 constructs a scatter plot, called an RI-plot, of the points (xi, yi), where 1 ≤ i ≤ n, given by xi = log10(RiGi) and yi = log2(Ri/Gi). Under our assumption, the RI-plot should be very nearly symmetric with respect to the line y = 0. In lowess normalization, TM4 applies the lowess method of Cleveland [27] to fit a locally weighted regression curve to the RI-plot; TM4 then adjusts spot intensities to eliminate any systematic intensity-dependent bias. Additional details on correcting intensity-dependent bias is found in [14].
Standard deviation regularization
After total intensity and lowess normalizations eliminate dye bias on a global (per microarray) scale, TM4 employs standard deviation regularization to ensure that the per-block variances of log(Ri/Gi) values are the same [25,28]. Quackenbush [14] provides the formulas for this normalization step.
Low intensity filtering
Since the relative error in the log(Ri/Gi) values increases if Ri or Gi is close to background levels, spots with low intensities are filtered out. Quackenbush [14] provides additional details, which essentially require that both Ri and Gi intensities be above two standard deviations of the respective backgrounds.
The MIDAS pipeline
We applied a MIDAS pipeline consisting of total intensity normalization, lowess normalization, standard deviation regularization, and low intensity filtering to both microarray data sets. MIDAS default parameters were used throughout; the default low intensity filter cut-off is RiGi < 10,000.
TM4 MEV analysis
The Multi Experiment Viewer (MEV) component of TM4 provides a number of statistical analyses and clustering algorithms to identify differentially expressed genes. We report results from the one-class t-test analysis applied to output of the MIDAS pipeline. This test assumes that the paired distribution of treated and control groups is normally distributed. Since the intensities measured from the same spot are correlated, we can apply the one-class t-test for the two-group comparison.
Authors' contributions
AAS performed the data preprocessing, Expresso analysis of all data sets, developed the database of statistical results, and drafted the manuscript. SPM performed the drought stress experiment (experiment 1), while PL performed the elevated CO2 experiment (experiment 2). PL performed the qRT-PCR of 192 genes from experiment 2. WS and PL performed TM4 analysis of experiments 1 and 2 respectively. LSH supervised the data analysis. LSH, HJB, and RG conceived of the study and coordinated the work. All authors read and approved the manuscript.
Supplementary Material
Intensity-dependent dye bias in Rl-plots. Supplementary Figure 1 is a PDF file that contains six RI-plots that illustrate specific intensity-dependent dye bias as the microarray data is processed through the normalization steps.
Genes subjected to qRT-PCR. Supplementary Table 1 is a PDF file that contains the annotation of genes subjected to qRT-PCR and the functional categories represented. For verification of microarray results in Experiment 2, Li, et al., [23] performed real-time quantitative reverse-transcriptase PCR (qRT-PCR) for selected genes — 55 in Col-0; 52 in Cvi-0; 59 in WS; 26 in Th. The AT numbers of those genes, their annotation, and categorizations into biological functions are in the table.
Acknowledgments
Acknowledgements
The work has been supported by the National Science Foundation (DBI-0223905 and BIO/IBN 0219322) and Virginia Tech and UIUC institutional funds.
Contributor Information
Allan A Sioson, Email: asioson@vt.edu.
Shrinivasrao P Mane, Email: smane@vt.edu.
Pinghua Li, Email: pinghli@life.uiuc.edu.
Wei Sha, Email: wsha@vt.edu.
Lenwood S Heath, Email: heath@vt.edu.
Hans J Bohnert, Email: bohnerth@life.uiuc.edu.
Ruth Grene, Email: grene@vt.edu.
References
- Churchill GA. Using ANOVA to Analyze Microarray Data. Bio Techniques. 2004;37:173–177. doi: 10.2144/04372TE01. [DOI] [PubMed] [Google Scholar]
- Wolfinger R, Gibson G, Wolfinger E, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules R. Assessing Gene Significance from cDNA Microarray Expression Data via Mixed Models. Journal of Computational Biology. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- Kerr MK. Linear Models for Microarray Data Analysis: Hidden Similarities and Differences. Journal of Computational Biology. 2003;10:891–901. doi: 10.1089/106652703322756131. [DOI] [PubMed] [Google Scholar]
- Pan W. A Comparative Review of Statistical Methods for Discovering Differentially Expressed Genes in Replicated Microarray Experiments. Bioinformatics. 2002;18:546–554. doi: 10.1093/bioinformatics/18.4.546. [DOI] [PubMed] [Google Scholar]
- Chu TM, Weir B, Wolfinger RD. Comparison of Li-Wong and Loglinear Mixed Models for the Statistical Analysis of Oligonucleotide Arrays. Bioinformatics. 2004;20:500–506. doi: 10.1093/bioinformatics/btg435. [DOI] [PubMed] [Google Scholar]
- Cui X, Churchill GA. Statistical Tests for Differential Expression in cDNA Microarray Experiments. Genome Biology. 2003;4 doi: 10.1186/gb-2003-4-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa GJ, Steibel JP, Tempelman RJ. Reassessing Design and Analysis of Two-colour Microarray Experiments using Mixed Effects Models. Comparative and Functional Genomics. 2005;6:123–131. doi: 10.1002/cfg.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futschik M, Crompton T. Model Selection and Efficiency Testing for Normalization of cDNA Microarray Data. Genome Biology. 2004;5 doi: 10.1186/gb-2004-5-8-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held M, Gase K, Baldwin IT. Microarray in Ecological Research: A Case Study of a cDNA Microarray for Plant-Herbivore Interactions. BMS Ecology. 2004;4 doi: 10.1186/1472-6785-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Jeong KS, Pan W, Khodursky A, Carlin BP. A Case Study on Choosing Normalization Methods and Test Statistics for Two-Channel Microarray Data. Comparative and Functional Genomics. 2004;5:432–444. doi: 10.1002/cfg.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B, Irizarry R, Astrand M, Speed T. A Comparison of Normalization Methods for High Density Oligonucleotide Array Data Based on Variance and Bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Gentleman RC, Quackenbush J. Open Source Software for the Analysis of Microarray Data. BioTechniques. 2003;34:s45–s51. [PubMed] [Google Scholar]
- Saeed A, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: A Free, Open-Source System for Microarray Data Management and Analysis. Bio Techniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray Data Normalization and Transformation. Nature Genetics Supplement. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Williams RD, King SN, Greer BT, Whiteford CC, Wei JS, Natrajan R, Kelsey A, Rogers S, Campbell C, Pritchard-Jones K, Khan J. Prognostic Classification of Relapsing Favorable Histology Wilms Tumor using cDNA Microarray Expression Profiling and Support Vector Machines. Genes, Chromosomes and Cancer. 2004;41:65–79. doi: 10.1002/gcc.20060. [DOI] [PubMed] [Google Scholar]
- Zhu X, Hart R, Chang MS, Kim JW, Lee SY, Cao YA, Mock D, Ke E, Saunders B, Alexander A, Grossoehme J, Lin KM, Yan Z, Hsueh R, Lee J, Scheuermann RH, Fruman DA, Seaman W, Subramaniam S, Sternweis P, Simon MI, Choi S. Analysis of the Major Patterns of B Cell Gene Expression Changes in Response to Short-Term Stimulation with 33 Single Ligands. The Journal of Immunology. 2004;173:7141–7149. doi: 10.4049/jimmunol.173.12.7141. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Weiss G, Lachmann M, Hellmann I, Enard W, Muetzel B, Wirkner U, Ansorge W, Paabo S. A Neutral Model of Transcriptome Evolution. PLoS Biology. 2004;2:682–689. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkinson JI, Sioson AA, Vasquez-Robinet C, Shukla M, Kumar D, Ellis M, Heath LS, Ramakrishnan N, Chevone B, Watson LT, van Zyl L, Egertsdotter U, Sederoff RR, Grene R. Photosynthetic Acclimation is Reflected in Specific Patterns of Gene Expression in Drought-Stressed Loblolly Pine. Plant Physiology. 2003;133:1702–1716. doi: 10.1104/pp.103.026914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioson AA, Watkinson JI, Vasquez-Robinet C, Ellis M, Shukla M, Kumar D, Ramakrishnan N, Heath LS, Grene R, Chevone BI, Kadafar K, Watson LT. Proceedings of the Next Generation Software Systems Workshop, 17th International Parallel and Distributed Processing Symposium (IPDPS '03) Nice, France; 2003. Expresso and Chips: Creating a Next Generation Microarray Experiment Management System; p. 209b. [Google Scholar]
- Heath LS, Ramakrishnan N, Sederoff RR, Whetten RW, Chevone BI, Struble CA, Jouenne VY, Chen D, van Zyl LM, Grene R. Studying the Functional Genomics of Stress Responses in Loblolly Pine using the Expresso Microarray Management System. Comparative and Functional Genomics. 2002;3:226–243. doi: 10.1002/cfg.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith D. Arabidopsis Oligonucleotide Microarrays http://www.ag.arizona.edu/microarray/
- Mane SP, Vasquez-Robinet C, Sioson AA, Heath LS, Grene R. Phospholipase D alpha is Involved in Drought Stress Signaling in Arabidopsis. Poster presented at the International Conference on Plant Lipid-Mediated Signaling, Raleigh, NC. 2005.
- Li P, Sioson AA, Mane SP, Ulanov A, Grothaus G, Heath LS, Murali TM, Bohnert HJ, Grene R. Response Diversity of Arabidopsis thaliana Ecotypes and Thellungiella halophila in elevated CO2 in the field. Manuscript submitted. 2005. [DOI] [PubMed]
- TM4 http://www.tm4.org/
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA Microarray Data: A Robust Composite Method Addressing Single and Multiple Slide Systematic Variation. Nucleic Acids Research. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I, Chen E, Hasseman J, Liang W, Frank B, Wang S, Sharov V, Saeed A, White J, Li J, Lee N, Yeatman T, Quackenbush J. Within the Fold: Assessing Differential Expression Measures and Reproducibility in Microarray Assays. Genome Biology. 2002;3:1–12. doi: 10.1186/gb-2002-3-11-research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland W. Robust Locally Weighted Regression and Smoothing Scatterplots. J Amer Stat Assoc. 1979;74:829–836. doi: 10.2307/2286407. [DOI] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance Stabilization Applied to Microarray Data Calibration and to the Quantification of Differential Expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Intensity-dependent dye bias in Rl-plots. Supplementary Figure 1 is a PDF file that contains six RI-plots that illustrate specific intensity-dependent dye bias as the microarray data is processed through the normalization steps.
Genes subjected to qRT-PCR. Supplementary Table 1 is a PDF file that contains the annotation of genes subjected to qRT-PCR and the functional categories represented. For verification of microarray results in Experiment 2, Li, et al., [23] performed real-time quantitative reverse-transcriptase PCR (qRT-PCR) for selected genes — 55 in Col-0; 52 in Cvi-0; 59 in WS; 26 in Th. The AT numbers of those genes, their annotation, and categorizations into biological functions are in the table.


