Abstract
The Cro protein from bacteriophage λ has a dimeric α+β fold that evolved from an ancestral all-α monomer. The sequence mutations responsible for this dramatic structural evolution are unknown. Here we use analysis of sequence alignments to show that Ala-33, a small side chain in the hydrophobic “ball-and-socket” dimer interface of λ Cro, was a much larger tryptophan side chain at a previous point in evolution. The retroevolutionary λ Cro-A33W mutant shows a 10-fold reduction in dimerization affinity relative to the wild type as well as a large increase in monomer thermal stability (ΔTm > 10°C), apparently due to partial filling of the hydrophobic socket from within the same monomer. An additional mutation in the dimer interface, F58D, almost completely abolishes detectable dimerization while maintaining the high monomer stability. The secondary structure content of the monomerized versions of λ Cro is similar to that of the wild-type protein, and the tertiary structure of the monomer appears relatively well defined. These results (i) support a model in which the ball-and-socket dimer interface of λ Cro was created by altered volume mutations within a limited branch of the Cro lineage and (ii) suggest the possibility that the evolution of the α+β dimer from an all-α monomer proceeded through an α+β monomer intermediate.
Protein folds can undergo a variety of dramatic evolutionary changes involving the insertion, deletion, rearrangement, or substitution of one or more secondary structure elements (1, 2). Such structural shifts could be triggered by a variety of types of mutations in the amino acid sequence. Substitution or insertion/deletion mutagenesis experiments in well studied model systems have been found to produce large structural changes in a small number of cases, and these results have been interpreted to suggest that relatively simple mutational processes could lead to the evolution of new protein folds (3–5). The mutational mechanisms underlying known naturally occurring cases of structural divergence, however, have received little experimental or theoretical attention.
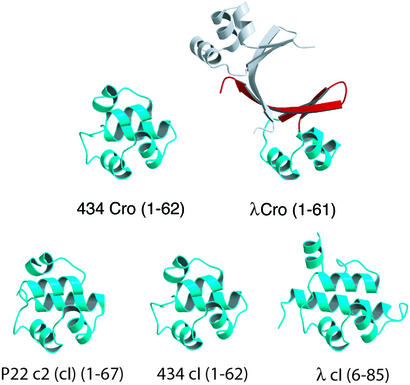
The Cro/cI superfamily contains one of the most remarkable examples of fold change in the evolution of protein structure. Cro and the N-terminal domain of cI are bacteriophage transcription factors that competitively bind to a common set of DNA operators by using a common type of helix–turn–helix motif. The closely related functions indicate that these DNA-binding domains are almost certainly paralogous, i.e. related by an ancient gene-duplication event, and thus belong to the same homologous superfamily. Of the five known Cro/cI structures, including two Cro proteins and three cI N-terminal domains, four have a fold consisting of five α-helices (6–12) (Fig. 1). In the structure of λ Cro, however, the fourth and fifth helices are replaced by a β-sheet, whereas the first three helices (including the helix–turn–helix motif) are largely retained (13, 14).
Figure 1.
Gallery of MOLSCRIPT (33) drawings of Cro and cI N-terminal domain structures showing that the fold of λ Cro is distinct both in secondary structure content and quaternary structure. The second monomer of λ Cro is shown in gray.
The oligomerization properties of λ Cro are also unique in the superfamily. Cro and cI proteins generally bind to pseudopalindromic operators as dimers involving intersubunit contacts between the C-terminal portions of the DNA-binding domains (15–19). In the all-helical family members these contacts involve side chains in or near the fourth and fifth helices, whereas in λ Cro an intersubunit β-sheet is formed from the β-sheet structures that replace these helices (see Fig. 1). The cI proteins also have a regulatory C-terminal domain that has its own dimerization interface and confers much of the actual dimerization affinity. Of the DNA-binding domains shown in Fig. 1, λ Cro is the only one that strongly self-associates in solution in the absence of DNA. The dissociation constant for the λ Cro dimer has been reported as ≈0.3–7 μM (21, 22), whereas other Cro proteins or cI N-terminal domains† have dissociation constants in at least the millimolar range (6, 7, 9, 11). The strong oligomerization of λ Cro is also partly coupled to its folding stability. The monomeric form of λ Cro is thermodynamically unstable toward unfolding, with a Tm no higher than ≈40°C (23), whereas monomeric helical Cro and cI N-terminal domains have reported Tm values of 57–70°C (24–26). These differences in secondary structure and oligomerization properties constitute a case of radical structural evolution.
The presence of the all-α fold among both Cro and cI proteins suggests that their common ancestor had such a structure, and that the dimeric α+β fold of λ Cro evolved from a monomeric all-α ancestor within a specific branch of the Cro lineage. Further support for this model comes from the recent finding that the Cro protein from bacteriophage P22 also has an all-α monomer fold (M.H.J.C., unpublished data). The question of interest, then, is how the α+β dimer evolved from an all-α monomer. In the present study, we focus on the origin of dimerization and weak monomer stability in λ Cro and show that this problem is separable from that of the mutational origin of the β-sheet secondary structure.
Materials and Methods
Sequence Mining and Multiple Alignment.
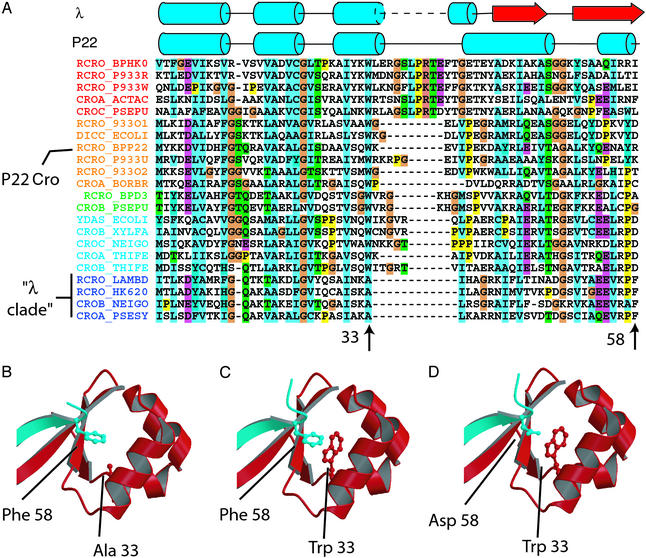
Eleven known Cro proteins (from the phages rac, D3, N15, P22, HK022, φ-80, H-19B, L, 434, λ, and HK620) were used as query sequences in standard protein (BLASTP) and six-frame translated microbial genome (TBLASTN) BLAST searches by using the NCBI server. These searches yielded a large number of hits (cutoff E < 0.001) from proteobacterial genomes, which contain a significant amount of DNA of bacteriophage origin. To generate an alignment of the highest quality possible, the resulting sequences were filtered by eliminating (i) query sequences that did not yield any BLAST hits with E < 0.001 (H-19B, L, N15, and φ-80), (ii) hits that were not part of an intact immunity operon (with an adjacent, divergently transcribed cognate cI ORF), and (iii) any sequences that showed significant similarity to their cognate cI. This last criterion was applied because it is apparent than some Cros such as 434 Cro are recent duplicates of their cognate cI proteins and do not rightly belong in the Cro lineage. The remaining sequences were linkage-clustered (in cluster sizes ranging from 2 to 6) according to BLAST similarity (E < 0.001), and profiles were generated for each cluster by using multiple alignment with CLUSTALX. Profile–profile alignment with CLUSTALX then yielded the final alignment shown in Fig. 2A. For sequence names, existing SWISS-PROT IDs were used where available. Otherwise, sequences were assigned SWISS-PROT-style names consisting of the name RCRO followed by a five-letter organism code according to the phage, prophage, or bacterial species to which they belonged. If multiple hits were found for a prophage or bacterial species, Cros were designated as CROA, CROB, etc., followed by a prophage or bacterial species code.
Figure 2.
(A) Alignment of Cro sequences generated from sequence similarity searches of microbial genomes using previously annotated Cro sequences. Alignment corresponds to residues 1–58 of P22 Cro and 5–58 of λ Cro. Sequences are named as described in Materials and Methods. RCRO_LAMBD, λ Cro; RCRO_BPP22, P22 Cro. (B) Portion of λ Cro showing ball-and-socket dimer interface and Ala-33 and Phe-58 side chains. Different monomers are shown in blue and red. (C) Model of λ Cro-A33W showing Trp-33 occupying a portion of the hydrophobic socket and clashing sterically with the Phe-58 ball. (D) Model of λ Cro-A33W/F58D.
Mutagenesis and Protein Purification.
A synthetic Cro gene in plasmid pAP119 (27) was amplified by PCR with introduction of NdeI and XhoI sites. After restriction digestion, this fragment was introduced into pET21b (Novagen), which had been digested with NdeI and XhoI. The resulting construct, pMC140, contains a Cro ORF with the additional C-terminal sequence −LEHHHHHH, which permits nickel-affinity purification. The A33W and F58D mutations were introduced sequentially by using the QuikChange site-directed mutagenesis kit (Stratagene). All Cro variants were overexpressed from the Escherichia coli strain BL21(λDE3) and purified to >95% homogeneity by chromatography on nickel-affinity and SP-Sephadex columns essentially as described (28) except that 10 mM imidazole was included in all load and wash steps for the nickel-affinity column. Also, a different protocol for the SP-Sephadex wash steps was used for A33W/F58D. In this case, a potassium-chloride gradient was applied because of the lower net positive charge of the protein. Protein concentrations were determined by using molar extinction coefficients of 3,591 M−1⋅cm−1 (wild type) or 9,150 M−1⋅cm−1 (A33W and A33W/F58D). Unless noted otherwise, all biophysical experiments were performed in 50 mM Tris, pH 7.5/250 mM KCl/0.2 mM EDTA.
Analytical Ultracentrifugation.
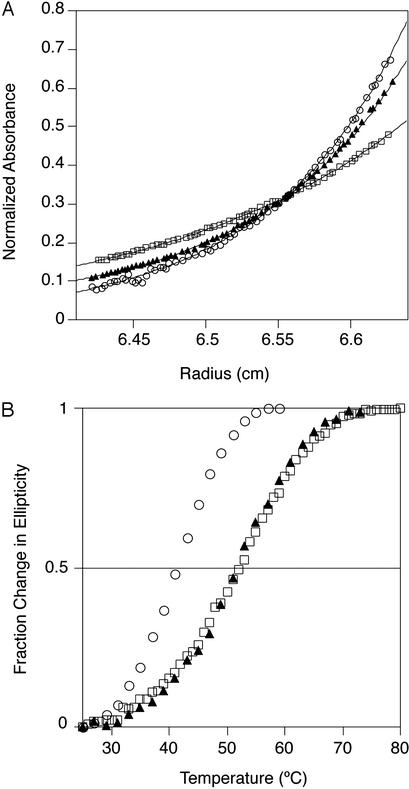
Sedimentation equilibrium experiments were performed on a Beckman Optima XL-I analytical ultracentrifuge. All proteins were dialyzed extensively before centrifugation and blanked directly against the dialysis buffer. All experiments were performed at 20°C and a rotor speed of 30,000 rpm. Data were measured as an average of 10–25 replicate scans with a radial spacing of 0.001 cm. The comparative scans in Fig. 3A were obtained at 280 nm on 30 μM samples of wild type, A33W, and A33W/F58D. These scans were successfully fit to dimer, monomer–dimer, and monomer models, respectively, by using KALEIDAGRAPH (Synergy Software, Reading, PA). Relevant parameters including molecular masses, solvent densities, and partial specific volumes were calculated from amino acid or buffer composition by using the program SEDNTERP (John Philo, Amgen, Thousand Oaks, CA, and RASMB). For all fits, the monomer molecular mass was constrained to be equal to the theoretical value computed by SEDNTERP. For measurement of dissociation constants, radial scans were performed on A33W samples (10, 20, 30, and 50 μM) at 280 nm, on wild-type Cro samples (5 and 10 μM) at 221 and 230 nm, and on A33W/F58D samples (283 μM) at 295 nm. Dissociation constants were extracted by fitting to monomer–dimer association models, and values obtained at different concentrations then were averaged. For samples of A33W/F58D at 30 and 100 μM, good fits were obtained with a single-species (monomer) model.
Figure 3.
Oligomerization and stability of ball-and-socket mutants. (A) Sedimentation equilibrium of wild-type λ Cro (○), A33W (▴), and A33W/F58D (□) at 30,000 rpm and 20°C (30 μM protein/50 mM Tris, pH 7.5/250 mM KCl/0.2 mM EDTA), monitored at 280 nm. Because of the differences in extinction coefficients, absorbances were normalized to constant integral area under the curve. Curve fits to monomer (wild type), monomer–dimer (A33W), and dimer (A33W/F58D) models are shown by solid lines. (B) Thermal melts of wild-type λ Cro (○), A33W (▴), and A33W/F58D (□) monitored by CD at 222 nm (10 μM protein/50 mM Tris, pH 7.5/250 mM KCl/0.2 mM EDTA).
Fluorescence and Circular Dichroism (CD) Spectrometry.
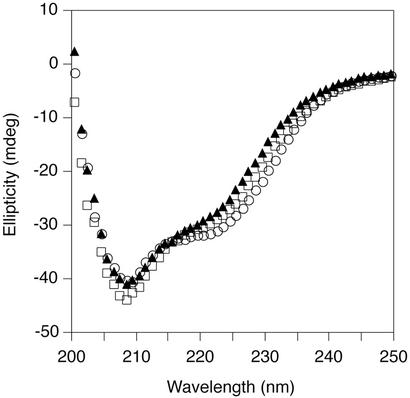
Fluorescence spectra were collected on a Perkin–Elmer LS 50B spectrometer by scanning fluorescence emission from 300 to 420 nm with an excitation wavelength of 280 nm. CD spectra were collected on an Aviv 62A DS CD spectrometer. Far-UV wavelength scans were performed from 200 to 250 nm at 20°C (50 μM protein), and near-UV wavelength scans were performed from 260 to 300 nm at 20°C (100 μM protein). To avoid problems due to buffer absorption at low wavelength, far-UV wavelength scans were performed in 100 mM sodium phosphate (pH 7.0). Wavelength scans performed in 50 mM Tris, pH 7.5/250 mM KCl/0.2 mM EDTA were qualitatively similar but were noisier due to high chloride-ion concentrations. Thermal denaturation was monitored at 222 nm (10 μM protein) and, for A33W/F58D, also at 280 nm (100 μM protein). All denaturation reactions were carried out from 20–80°C and were >95% reversible.
Results and Discussion
An Alanine Residue in the λ Cro Dimerization Interface Was a Tryptophan in a Cro Ancestor.
A likely physical source of the strong dimerization and weak monomer stability of λ Cro is the extensive overlap between the hydrophobic core and the dimerization interface. Phe-58 from one monomer inserts into a hydrophobic pocket in the other monomer, forming a “ball-and-socket” interaction (Fig. 2B; ref. 15). A thermodynamically stable monomeric Cro has been engineered previously by inserting a turn that allows this interaction to be formed in intramolecular fashion (23). In the present study we are interested in the evolutionary origin of this interface. To investigate the possibility that mutations in this region might have led to the unusual oligomerization properties of λ Cro, we examined conservation patterns among side chains participating in the ball and socket. Unfortunately, the known bacteriophage Cro proteins were too diverse in sequence to construct a satisfactory sequence alignment. Reasoning that a better alignment could be obtained with more sequence data, we used sequence similarity searches with known Cro sequences to extract additional Cro sequences from databases, primarily from prophage DNA present in recently sequenced bacterial genomes. We then aligned these sequences to generate the 22-protein alignment in Fig. 2A (see Materials and Methods). The alignment includes one Cro that is known from structural studies to be an all-α monomer (P22 Cro) and one that is known to be an α+β dimer (λ Cro). Although the focus of this article is on oligomerization behavior, the evolutionary secondary structure switching represented by P22 Cro and λ Cro is remarkable and will be discussed in a forthcoming publication (M.H.J.C., unpublished data).
One possible mechanism for evolving a hydrophobic socket involves mutation of a large hydrophobic core side chain to a small one. Examination of conservation patterns among small side chains that line the λ Cro socket revealed that one, Ala-33 (Fig. 2A), is conserved in only 4 of the 22 sequences in our alignment, specifically those most closely related to λ Cro (we will refer to this group as the “λ clade”). In every other sequence in our alignment, the amino acid residue at this position is a tryptophan, the side chain of which occupies 146 Å3 greater volume than alanine (29). Tryptophan is also present at the comparable position in 15 of 22 of the corresponding cI proteins (data not shown), which as paralogues form an outgroup to the Cro proteins. The principle of maximum parsimony argues that the ancestral Cro/cI residue at this position was a tryptophan, which then was mutated to an alanine somewhere in the lineage leading to λ Cro. Given the proximity of position 33 to a gap in the alignment (Fig. 2A), it is ambiguous whether this sequence change was the result of substitutions or small insertion/deletion events. If the result of substitutions, the change from a tryptophan to alanine codon requires at least two point mutations. One plausible pathway would be Trp (TGG) → Gly (GGG) → Ala (GCG), because such a pathway avoids placing a polar residue at a hydrophobic core position.
The Retroevolutionary Mutation A33W Weakens λ Cro Dimerization and Increases Monomer Stability.
What would be the anticipated effect of a “retroevolutionary” mutation changing the alanine at 33 to tryptophan? Modeling of λ Cro-A33W [using SWISS-MODEL calculations based on the refined crystal structure (14)] suggested that the tryptophan side chain would be partly buried within the hydrophobic socket in both the monomeric (data not shown) and dimeric forms (Fig. 2C). The resulting increased burial of hydrophobic surface area within the monomer core might be expected to thermodynamically stabilize the monomeric form. The dimer model also shows a steric clash between Trp-33 and the Phe-58 ball from the other monomer. This interaction might be expected to reduce the strength of dimerization. Thus, we anticipated that the A33W mutation might retroevolve the structural properties of λ Cro toward a stable monomer with a low propensity to dimerize.
Analytical ultracentrifugation of purified λ Cro-A33W confirms the expected weakened dimerization, yielding a Kd of 27.4 ± 6.6 μM compared with 2.4 ± 0.8 μM for the wild-type protein (Fig. 3A). Further, A33W is much more thermally stable (ΔTm ≈ +10°C) than wild-type λ Cro at a protein concentration of 10 μM (Fig. 3B), conditions under which A33W is largely monomeric and wild type is largely dimeric. Because the thermal stability of the wild-type dimer represents a maximal stability for the wild-type folded monomer, it is clear that the A33W mutation leads to substantial stabilization of the monomer. The apparent Tm of ≈52°C indicates that the stability of the A33W monomer is close to the range observed in other monomeric Cro/cI proteins (vide infra) and also close to the value of 58°C reported for the engineered Cro monomer of Mossing and Sauer (23). Clearly, the substitution of an ancestral tryptophan at position 33 with alanine is capable of explaining, at least in part, the mutational origin of the unique oligomerization properties of λ Cro.
Fluorescence spectra of A33W (data not shown) suggest that, consistent with our initial models, packing of Trp-33 into the hydrophobic socket in both the monomeric and dimeric forms is the source of the observed effects. The fluorescence maximum of A33W at 10 μM is 341 nm but undergoes a blue shift of ≈1 nm after increasing the concentration to 100 μM. These values reflect a partial burial of the Trp-33 aromatic ring in both the monomer and dimer and are qualitatively consistent with the extent of burial predicted from models: Trp-33 is predicted to be 15% solvent-accessible in the monomer and 7% accessible in the dimer.
Mutation of the Phe-58 Ball Further Weakens Dimerization.
We next considered whether additional residues in the ball and socket might show conservation patterns suggesting a role in the evolution of this hydrophobic interface. Similar to the socket side chain Ala-33, the Phe-58 ball is conserved only within the λ clade (Fig. 2A). Among other Cro sequences this position is highly variable and is often polar rather than hydrophobic. The high variability at this position prevents a confident reconstruction of its evolutionary history at this time without a more detailed phylogenetic and ancestral state reconstruction analysis. The most commonly observed amino acid type at this position, however, is aspartate, which is incompatible with the structural role played by Phe-58. Replacement of Phe-58 with Asp (Fig. 2D) would both reduce the size of the ball and place a polar side chain at a hydrophobic interface, which might be expected to weaken dimerization. However, because Phe-58 makes no obvious interactions within each monomer of the dimer, such a mutation would not be expected to affect monomer stability significantly, other things being equal.
We introduced the F58D mutation into the λ Cro-A33W mutant background. Analytical ultracentrifugation of purified λ Cro-A33W/F58D shows that it is completely monomeric at 30 (Fig. 3A) and 100 μM and contains only minute traces of higher oligomers at 283 μM. Fit of the 283 μM data to a monomer–dimer equilibrium yields a Kd of 2.3 mM. The error in this value is high because of the low level of oligomers present at 283 μM. However, these data show that if A33W/F58D dimerizes at all, it has a dimerization constant at least on the order of 100-fold higher than A33W. Hence, A33W/F58D appears similar in its weak dimerization propensity to the all-α members of the Cro/cI superfamily. Similar results (not shown) were obtained for A33W/F58A, showing that severe reduction of oligomerization is brought about by the mere absence of a sizable hydrophobic side chain as opposed to requiring the presence of a polar/charged side chain. Thus, introduction of most of the side chains present at position 58 in the Fig. 2A alignment would be likely to impact dimerization in λ Cro severely. By contrast, monomer stability appears unaffected by mutation of Phe-58. Thermal denaturation studies at 10 μM protein show that the monomer stability of A33W/F58D is essentially identical to that of A33W (Fig. 3B). Thus, in accord with our expectations, the identity of the residue at position 58 is clearly a strong determinant of dimerization strength but does not appear to affect the structural integrity of the folded monomer.
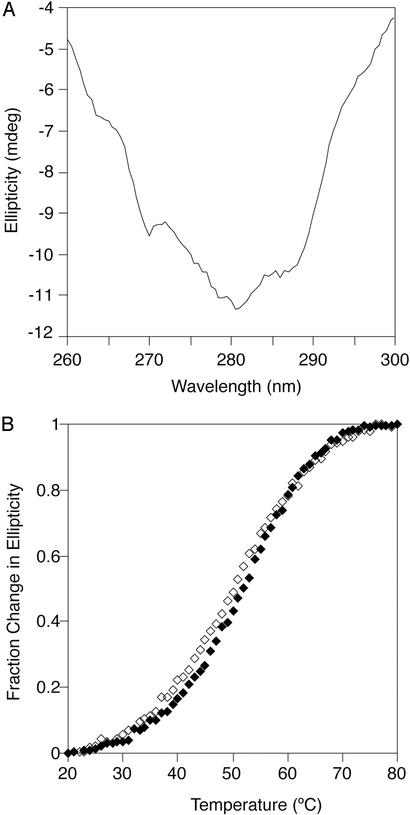
Structural Characteristics of Monomerized Cros.
The far-UV CD spectrum of the A33W/F58D monomer is similar to that of the wild-type dimer, as is the spectrum of a mixture of A33W monomers and dimers (Fig. 4). This suggests that the secondary structure content of the monomerized Cros is similar to that of the monomers within the wild-type dimer. This α+β monomer also appears to have a well defined tertiary structure as judged by the presence of a significant CD signal in the near-UV region for A33W/F58D at 100 μM (Fig. 5A). In addition, thermal melts of A33W/F58D monitored by near- and far-UV CD superimpose well (Fig. 5B), indicating that secondary and tertiary structure unfold coincidentally. This observation is consistent with a cooperative two-state unfolding transition for the monomer, which in turn suggests a “native-like” structure.
Figure 4.
Similar secondary structure content of wild-type and monomerized λ Cro variants. Far-UV CD spectra of wild-type λ Cro (○), A33W (▴), and A33W/F58D (□) at 20°C (50 μM protein/100 mM sodium phosphate, pH 7.0).
Figure 5.
Tertiary structure of monomerized λ Cro variants. (A) Near-UV CD spectrum of A33W/F58D Cro at 20°C (100 μM protein/50 mM Tris, pH 7.5/250 mM KCl/0.2 mM EDTA). (B) Thermal denaturation of A33W/F58D as monitored by CD at 222 nm (♦, 10 μM protein/50 mM Tris, pH 7.5/250 mM KCl/0.2 mM EDTA) and 280 nm (⋄, 100 μM protein/50 mM Tris, pH 7.5/250 mM KCl/0.2 mM EDTA).
Conclusions.
The primary conclusion drawn from the work presented here is that a socket-creating mutation in which a large ancestral hydrophobic core side chain, Trp-33, is substituted with a small one, Ala, accounts for the evolutionary origin of the weak monomer stability and, in part, the strong dimerization exhibited by Cro. Also, the poor conservation of Phe-58 outside the close relatives of λ Cro, and the severe reductions in dimerization observed in λ Cro after substitution by aspartate or alanine, suggest that mutations at this position also played a key role in the development of strong dimerization. The putatively retroevolved monomers appear to retain the α+β fold of the wild-type protein, suggesting that the sequence factors (as yet unknown) that govern the conversion of ancestral α-helix secondary structure to the β-sheet of λ Cro are separable from those that governed the evolution of dimerization/monomer stability. Finally, the monomerized Cros are well folded proteins. Monomeric α+β folds are therefore structurally plausible as intermediates in the evolutionary processes that led to the development of the unique λ Cro fold.
The conservation of Ala-33 and Phe-58 only within the λ clade suggests that members of this subfamily of Cros may all be strong dimerizers and unstable monomers. The strong conservation of Trp-33 and variability at position 58 among other Cros suggest that these family members may be stable monomers with relatively low dimerization propensities in solution, although it is quite possible that some Cro proteins have developed strong dimerization through alternate structural mechanisms. It is known that one of the non-λ clade proteins in Fig. 2A, P22 Cro, has an all-α monomer fold (M.H.J.C., unpublished data). Whether any extant Cros have an intermediate, α+β monomer structure similar to that observed here remains to be determined. If such a structure were found, it could provide support for a model in which the evolution of the α+β dimer fold of λ Cro from an all-α monomer occurred in a two-step process.
Finally, it is worth noting that the evolution of strong dimerization and weak monomer stability in λ Cro carries with it potential functional consequences. Dimerization of λ Cro is critical to function, conferring both enhanced DNA-binding affinity and sequence specificity relative to monomers (23, 30, 31). Although all protein–DNA cocrystal structures in this superfamily show dimer contacts, differences in the strength and structural nature of these interactions could correlate with evolutionary changes in binding affinity and in the sequences of the cognate operators. For instance, it has been noted that in vitro operator binding of P22 Cro is significantly weaker than that of λ Cro (32). In addition, the partial coupling of dimerization with folding stability in λ Cro may have functional consequences. Mossing and coworkers (22) have noted the likelihood that in vivo concentrations of free λ Cro are in the nanomolar range or lower, such that the predominant free species of λ Cro will be a metastable monomer. A weakly stable monomer may exhibit higher susceptibility to degradation by cellular proteases, possibly lowering in vivo Cro concentrations and effectively decreasing operator occupancy. In principle, such an effect could balance increases in binding affinity and/or provide additional levels of regulation. On the whole, it remains in the realm of speculation whether evolutionary changes in the oligomerization behavior of Cro proteins represent adaptation or are more accurately viewed as an alternate structural solution to the same functional problem.
Acknowledgments
We thank Robert Sauer and Megan McEvoy for helpful comments on the manuscript and the Baldwin and Cusanovich laboratories for use of their equipment. We also thank A. J. Clark for supplying us with the sequence of HK620 Cro.
Footnotes
The dimerization constant of λ cI N-terminal domain depends on where the domain boundary is defined. A minimal stable N-terminal domain, consisting of residues 6–85, is monomeric at millimolar concentrations, whereas the dimerization constant of residues 1–102 is ≈0.3 mM (ref. 20), still much weaker than that of λ Cro.
References
- 1.Grishin N V. J Struct Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- 2.Murzin A G. Curr Opin Struct Biol. 1998;8:380–387. doi: 10.1016/s0959-440x(98)80073-0. [DOI] [PubMed] [Google Scholar]
- 3.Cordes M H J, Walsh N P, McKnight C J, Sauer R T. Science. 1999;284:325–328. doi: 10.1126/science.284.5412.325. [DOI] [PubMed] [Google Scholar]
- 4.Cordes M H J, Burton R E, Walsh N P, McKnight C J, Sauer R T. Nat Struct Biol. 2000;7:1129–1132. doi: 10.1038/81985. [DOI] [PubMed] [Google Scholar]
- 5.Green S M, Gittis A G, Meeker A K, Lattman E E. Nat Struct Biol. 1995;2:746–751. doi: 10.1038/nsb0995-746. [DOI] [PubMed] [Google Scholar]
- 6.Huang G S, Oas T G. Biochemistry. 1995;34:3884–3892. doi: 10.1021/bi00012a003. [DOI] [PubMed] [Google Scholar]
- 7.Mondragon A, Subbiah S, Almo S C, Drottar M, Harrison S C. J Mol Biol. 1989;205:189–200. doi: 10.1016/0022-2836(89)90375-6. [DOI] [PubMed] [Google Scholar]
- 8.Mondragon A, Wolberger C, Harrison S C. J Mol Biol. 1989;205:179–188. doi: 10.1016/0022-2836(89)90374-4. [DOI] [PubMed] [Google Scholar]
- 9.Neri D, Billeter M, Wuthrich K. J Mol Biol. 1992;223:743–767. doi: 10.1016/0022-2836(92)90987-u. [DOI] [PubMed] [Google Scholar]
- 10.Pabo C O, Lewis M. Nature. 1982;298:443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- 11.Padmanabhan S, Jimenez M A, Gonzalez C, Sanz J M, Gimenez-Gallego G, Rico M. Biochemistry. 1997;36:6424–6436. doi: 10.1021/bi970085p. [DOI] [PubMed] [Google Scholar]
- 12.Sevilla-Sierra P, Otting G, Wuthrich K. J Mol Biol. 1994;235:1003–1020. doi: 10.1006/jmbi.1994.1053. [DOI] [PubMed] [Google Scholar]
- 13.Matsuo H, Shirakawa M, Kyogoku Y. J Mol Biol. 1994;254:668–680. doi: 10.1006/jmbi.1995.0646. [DOI] [PubMed] [Google Scholar]
- 14.Ohlendorf D H, Tronrud D E, Matthews B W. J Mol Biol. 1998;280:129–136. doi: 10.1006/jmbi.1998.1849. [DOI] [PubMed] [Google Scholar]
- 15.Albright R A, Matthews B W. J Mol Biol. 1998;280:137–151. doi: 10.1006/jmbi.1998.1848. [DOI] [PubMed] [Google Scholar]
- 16.Beamer L J, Pabo C O. J Mol Biol. 1992;227:177–196. doi: 10.1016/0022-2836(92)90690-l. [DOI] [PubMed] [Google Scholar]
- 17.Anderson J E, Ptashne M, Harrison S C. Nature. 1987;326:846–852. doi: 10.1038/326846a0. [DOI] [PubMed] [Google Scholar]
- 18.Mondragon A, Harrison S C. J Mol Biol. 1991;219:321–334. doi: 10.1016/0022-2836(91)90568-q. [DOI] [PubMed] [Google Scholar]
- 19.Wolberger C, Dong Y C, Ptashne M, Harrison S C. Nature. 1988;335:789–795. doi: 10.1038/335789a0. [DOI] [PubMed] [Google Scholar]
- 20.Weiss M A, Pabo C O, Karplus M, Sauer R T. Biochemistry. 1987;26:897–904. doi: 10.1021/bi00377a034. [DOI] [PubMed] [Google Scholar]
- 21.Darling P J, Holt J M, Ackers G K. Biochemistry. 2000;39:11500–11507. doi: 10.1021/bi000935s. [DOI] [PubMed] [Google Scholar]
- 22.Jana R, Hazbun T R, Mollah A K, Mossing M C. J Mol Biol. 1997;273:402–416. doi: 10.1006/jmbi.1997.1256. [DOI] [PubMed] [Google Scholar]
- 23.Mossing M C, Sauer R T. Science. 1990;250:1712–1715. doi: 10.1126/science.2148648. [DOI] [PubMed] [Google Scholar]
- 24.Huang G S, Oas T G. Biochemistry. 1996;35:6173–6180. doi: 10.1021/bi960250l. [DOI] [PubMed] [Google Scholar]
- 25.Padmanabhan S, Laurents D V, Fernandez A M, Elias-Arnanz M, Ruiz-Sanz J, Mateo P L, Rico M, Filimonov V V. Biochemistry. 1999;38:15536–15547. doi: 10.1021/bi991757+. [DOI] [PubMed] [Google Scholar]
- 26.Ruiz-Sanz J, Simoncsits A, Toro I, Pongor S, Mateo P L, Filimonov V V. Eur J Biochem. 1999;263:246–253. doi: 10.1046/j.1432-1327.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- 27.Pakula A A, Sauer R T. Proteins. 1989;5:202–210. doi: 10.1002/prot.340050303. [DOI] [PubMed] [Google Scholar]
- 28.Milla M E, Brown B M, Sauer R T. Protein Sci. 1993;2:2198–2205. doi: 10.1002/pro.5560021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards F M. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 30.Baleja J D, Anderson W F, Sykes B D. J Biol Chem. 1991;266:22115–22124. [PubMed] [Google Scholar]
- 31.Albright R A, Mossing M C, Matthews B W. Protein Sci. 1998;7:1485–1494. doi: 10.1002/pro.5560070701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poteete A R, Hehir K, Sauer R T. Biochemistry. 1986;25:251–256. doi: 10.1021/bi00349a035. [DOI] [PubMed] [Google Scholar]
- 33.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]