Abstract
Heparin and heparan sulfate glycosaminoglycans (HSGAGs) mediate a wide variety of complex biological processes by specifically binding proteins and modulating their biological activity. One of the best studied model systems for protein–HSGAG interactions is the fibroblast growth factor (FGF) family of molecules, and recent observations have demonstrated that the specificity of a given FGF ligand binding to its cognate receptor (FGFR) is mediated by distinct tissue-specific HSGAG sequences. Although it has been known that sulfate and carboxylate groups in the HSGAG chain play a key role by interacting with basic residues on the proteins, there is little understanding of how these ionic interactions provide the necessary specificity for protein binding. In this study, using all of the available crystal structures of different FGFs and FGF–HSGAG complexes, we show that in addition to the ionic interactions, optimal van der Waals contact between the HSGAG oligosaccharide and the protein is also very important in influencing the specificity of FGF–HSGAG interactions. Although the overall helical structure is maintained in the FGF-bound HSGAG compared with unbound HSGAG, we observe distinct changes in the backbone torsion angles of the oligosaccharide chain induced upon protein binding. These changes result in local deviations in the helical axis that provide optimal ionic and van der Waals contact with the protein. A specific conformation and topological arrangement of the HSGAG-binding loops of FGF, on the other hand, impose structural constraints that induce the local deviations in the HSGAG structure, thereby enabling maximum contact between HSGAG and the protein.
Recent advances in developmental biology, cancer biology, and other fields have resulted in a dramatic increase in the number of known important roles for the extracellular complex polysaccharides heparin and heparan sulfate glycosaminoglycans (HSGAGs) (1–7). An emerging paradigm in this field is that unique HSGAG sequences specifically bind to a wide range of proteins (2, 5, 6), including morphogens (1, 7), growth factors (8–10), and enzymes (5, 11), and influence the physiological state of cells and tissues. Hence it is important to understand how sequence-specific HSGAG–protein interactions impinge on the biological functions of these important signaling molecules.
HSGAGs are complex acidic polysaccharides that are characterized by a disaccharide repeat unit of α-d-glucosamine (1 → 4) linked to uronic (α-l-iduronic/β-d-glucuronic) acid. Structural heterogeneity within the HSGAG polysaccharide arises from the number of disaccharide repeat units present, as well as four potential sites for chemical modification in the form of acetylation or sulfation. The sites of sulfation include the 2-O position of the uronic acid and the N, 3-O, and 6-O positions of the glucosamine, making HSGAG one of the most acidic biopolymers. Further, the N position of the glucosamine can also be acetylated. Similar to DNA and fibrous proteins like collagen, HSGAGs adopt a helical structure (12–15). The topology and geometry of a HSGAG structure is governed by the backbone torsion angles, which include both the intermonosaccharide glycosidic torsion angles and the pyranose ring torsion angles. Moreover, the pyranose ring of the α-iduronic acid can potentially adopt many conformations, including 4C1, 1C4 chair, and the 2S0 skew boat forms (16–19). Although it has been postulated that conformational flexibility of the iduronate ring enhances the specificity of HSGAG oligosaccharides binding to proteins, it is unclear how this and the ionic charges together translate to the required molecular specificity of HSGAG oligosaccharide–protein interactions.
The fibroblast growth factor receptor (FGF) family presently has 23 members and each retains an identifiable, although varying, HSGAG-binding domain (20). There are numerous levels of evidence implicating the differential ability of HSGAGs in regulating FGF signaling via their cell surface tyrosine kinase receptors (FGFR). For example, it has been observed that the specificity of a given FGF to a FGFR is mediated by what appear to be distinct tissue-specific HSGAG sequences (10, 21–24). In addition to the biochemical evidence for FGF–HSGAG interactions, several cocrystal structures of FGF–HSGAG and FGF–FGFR–HSGAG complexes have been solved (25–28). Although the cocrystal structures illustrate the differences in the interactions between the different FGF–FGFR–HSGAG ternary complexes and diversity in the oligomerization states of FGF-1 and -2 in these complexes, they do not shed much light on the specificity of HSGAG–FGF interactions. Biochemical studies have indicated nanomolar affinity for FGF-2 to heparin and heparin-derived oligosaccharides (29). Such nanomolar affinities can not be explained purely on the basis of nonspecific charge interactions between sulfate groups of HSGAG and the basic amino acids on the protein. Furthermore, the HSGAG-binding sites on the protein have largely been described at the primary sequence level through the Cardin–Weintraub heparin-binding consensus sequences (9). In addition, it is unclear as to how the ionic charges and iduronate conformational flexibility in the HSGAG, taken together with the distribution of basic charges on the protein, provide the necessary protein-binding specificity.
In this study, we performed a detailed conformational analysis of all of the FGF–HSGAG and FGF–FGFR–HSGAG cocrystal structures and identified characteristic conformational properties on the HSGAG oligosaccharide and the HSGAG-binding site on FGF that govern their interactions. The results of our analysis indicate that, in addition to the ionic interactions identified in the crystal structures, optimal van der Waals contact between the oligosaccharide and the protein is also important in influencing the specificity of FGF–HSGAG interactions. Although the overall HSGAG helical structure is maintained when bound to FGF, a kink in the helical axis is induced upon FGF binding. This kink is formed by a change in the backbone torsion angles, which cause local conformational changes in the HSGAG structure. In addition, the ability of the iduronic acid in HSGAGs to adopt multiple ring conformations enhances the formation of the kink and governs the nature and degree of the kink. The kink provides optimal ionic and van der Waals contact with the protein. Furthermore, a specific conformation and topological arrangement of the HSGAG-binding loops imposes structural constraints on the HSGAG backbone and thus induces a kink. Moreover, the distinct spatial distribution of basic amino acids in the HSGAG-binding loops on different FGFs influences the distribution of sulfate groups in the interacting HSGAG chain. Thus, both the loop topology and the spatial distribution of basic residues are key structural features on the protein that provide optimal ionic and surface complementarity for HSGAG binding.
Methods
Crystal Structures from the Protein Data Bank (PDB).
All of the cocrystal structures of FGF–HSGAG complexes including the recently solved FGF-2–FGFR1–HSGAG and FGF-1–FGFR2–HSGAG complexes and the crystal structures of FGF-1, -2, -4, -7 and -9 were obtained from PDB, www.rcsb.org. The PDB identifiers of the cocrystal structures with HSGAG oligosaccharides are as follows: FGF-1, 1AXM and 2AXM; FGF-2, 1BFC and 1BFC; FGF-2–FGFR1, 1FQ9; FGF-1–FGFR2, 1E0O; the PDB identifiers of FGF-4, -7 and -9 are 1IJT, 1QQK, and 1IHK, respectively. The resolution of the crystal structures was between 1.9 and 3.0 Å. All of the FGF and FGF–FGFR complexes were cocrystallized with HSGAG oligosaccharides containing a homogeneous trisulfated disaccharide repeat, [I2SHNS,6S]n (I, α-l-iduronic acid; H, α-d-glucosamine), where most contained a uronic acid with a Δ4,5 unsaturated linkage at the nonreducing end. In the FGF-1–HSGAG and the FGF-1–FGFR2–HSGAG cocrystal structures (27, 28), the stoichiometric ratio FGF-1:HSGAG was 2:1, where a single oligosaccharide bridged two FGF-1 monomers (without any protein–protein contact) related by an approximate twofold symmetry about an axis perpendicular to the helical axis of the oligosaccharide. Obviously, the interactions between each of the FGF-1 monomers with the oligosaccharide were different, and one of the monomers had significantly more favorable contacts with the oligosaccharide than the other. For all of our conformational analysis studies, the FGF-1–HSGAG complex involving the monomer with more favorable contacts was considered. FGF-2 was cocrystallized with a tetra- (PDB ID code 1BFB) and hexasaccharide (1BFC). Because the oligosaccharide chains in the two structures are superimposable, the hexasaccharide cocrystal structure was used in this analysis. In the case of the FGF-2–FGFR1 cocrystal structure, the heparin oligosaccharides were diffused into preformed dimers of FGF-2–FGFR1 complexes, resulting in a 2:2:2 stoichiometric ratio. Consequently, the coordinate data sets for two different HSGAG oligosaccharides (chains E and F), one for each complex, were obtained.
Modeling Unbound Oligosaccharide Structures.
HSGAG disaccharides (I2SHNS,6S;) with the iduronic acid in the 2S0 and 1C4 and the glucosamine in the 4C1 ring conformations were modeled by using the bond lengths, bond angles, and ring geometries obtained from the cocrystal structures. Using the nandh program (30), the glycosidic linkage torsions were explored to generate oligosaccharides (hexa- and octasaccharides) of unbound HSGAG satisfying the 21-helical symmetry with an axial rise of 8.4–8.7 Å per unit [consistent with fiber diffraction and NMR data (12, 14)]. From these oligosaccharide structures, one structure for each iduronate ring conformation was chosen as the reference “unbound” HSGAG structure. Modeling was carried out using the Builder and Viewer Modules of insight ii (Accelrys, San Diego).
Calculation of Local Helical Parameters nL and hL.
For all of the HSGAG oligosaccharides in the cocrystal structures, the successive disaccharide units starting from the nonreducing end (I0-H1, H1-I2, I2-H3, etc.) were repeated in space (Fig. 1A) with the experimentally observed backbone torsion angles. Each disaccharide unit could be repeated in space by using two sets of backbone torsion angles, namely, pij sij pij+1 sij−1 and pij sij pij+1 sij+1 (e.g., H1-I2 could be constructed by using p11 s11 p22 s20 or p11 s11 p22 s22; Fig. 1A). For an infinite helical repeat of every disaccharide unit, the number of units per turn and the axial rise per unit were calculated by using nandh (30). The number of units per turn was designated as local number of turns (nL) and the rise per turn was designated as local axial rise (hL), because these values pertained to only a theoretical repeat of a local disaccharide unit and not the entire HSGAG oligosaccharide.
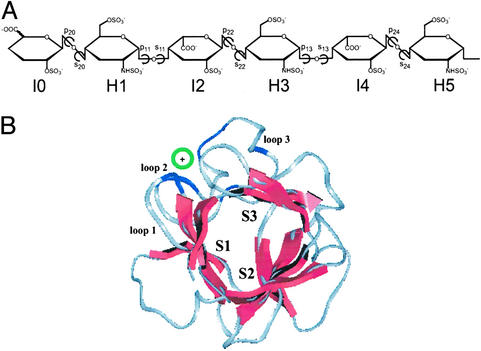
Figure 1.
FGF–HSGAG structural complex. (A) Chemical structure of HSGAG oligosaccharide observed in all of the FGF–HSGAG cocrystal structures. The units are numbered from nonreducing to reducing end. The glycosidic torsion angles are pij (C2j-C1j-Oj-C4j) and sij (C1j-Oj-C4j-C3j), where i = 1 for an H-I linkage and 2 for an I-H linkage, and j is the monosaccharide number. (B) Orientation and chain direction of the HSGAG oligosaccharide relative to sheets S1, S2, and S3 (red) of a β-trefoil scaffold of FGF whose Cα trace is such that the pseudoaxis of threefold symmetry is roughly perpendicular to the plane of the paper. The orientation of the oligosaccharide (indicated by green circle) is at a slight angle to the plane of the paper between S1 and S3. The oligosaccharide interacts with the basic residues (blue) in the loop regions (in gray, numbered 1–3). The observed chain directions are such that either the nonreducing or reducing end is projecting out of the plane of the paper.
Docking Oligosaccharides into HSGAG-Binding Sites of FGF-2 and -7.
The hexasaccharide obtained from FGF-2–HSGAG cocrystal structure (kinked hexasaccharide) and two modeled unbound hexasaccharides (uniform helical structures) with iduronate either in the 1C4 or 2SO conformation were used for the docking studies. The AMBER force field for carbohydrates (31), modified to include sulfate and sulfamate groups (32), was used to assign the potentials for the proteins and oligosaccharides. The starting position and orientation of the modeled hexasaccharides relative to FGF-2 and -7 was fixed by manually docking them onto the HSGAG-binding site of the protein. The manual docking was performed by using the FGF-2–HSGAG cocrystal structure as a guiding framework to ensure that the sulfate groups were positioned to interact with the HSGAG-binding pocket on the proteins. Also, the starting position of the kinked hexasaccharide relative to FGF-7 was obtained by superimposing the β-trefoil scaffold of FGF-7 onto the FGF-2 in the FGF-2–HSGAG cocrystal structure. The interaction energy between the oligosaccharide and protein was minimized by using 200 iterations of steepest-descent minimization, followed by 300 iterations of Newton–Raphson method, which resulted in convergence. During the minimization, a restraining force with a force constant of 5,000 kcal/Å was applied to restrict the ring torsion angles of the oligosaccharides to experimentally observed values, and on the protein side only the loop regions constituting the HSGAG-binding site were allowed to move freely. All of the minimization runs were performed by using the Discover module of insight ii. The minimization parameters for AMBER force field were set according to the insight ii manual. The electrostatic and van der Waals interaction energy between protein and oligosaccharide were computed by using the Docking module of insight ii. Because these calculations are performed for rigid-body docking without any dynamics incorporated, the values of interaction energies reflect only the relative strength of the interaction and not the actual energy of interaction between the molecules in solvent.
Results and Discussion
Despite the varying sequence homologies between the members of the FGF family (20), all of the proteins share a β-trefoil structural fold. The β-trefoil fold comprises of three antiparallel β-sheets, S1, S2, and S3 (each containing four β-strands A, B, C, and D), that are stacked in a triangular fashion to form a pyramid-like arrangement (Fig. 1B). The HSGAG-binding site on FGF contains three surface-exposed loop regions, including one large loop (loop3) between S1 and S3 sheets and two small loops in S1 (loop1) and S3 (loop2) of the β-trefoil scaffold. On the other hand, HSGAGs adopt a 21 symmetry helical structure with two disaccharide units per turn (n = 2) and an axial rise per disaccharide unit of h = 8.3–8.7 Å, as determined by x-ray fiber diffraction of sodium and calcium salts of heparin and heparan sulfate (12, 13) and NMR analysis of heparin (14) and heparin-derived oligosaccharides (33) in solution.
The observed FGF–HSGAG interactions (summarized in Fig. 1B) indicate that the orientation of the oligosaccharide chain relative to the HSGAG-binding site of FGF is approximately the same. However, two different chain directions (nonreducing to reducing and vice versa) relative to FGF have been observed in the cocrystal structures. Importantly, the number of ionic interactions between oligosaccharides and protein are almost identical in both of these chain directions. This emphasizes the need to investigate structural properties beyond ionic interactions to gain a full appreciation of the relative importance of the chain direction in governing FGF–HSGAG interactions. Detailed conformational analysis of the FGF–HSGAG structural complexes showed that in both the chain directions, there are not only optimal ionic, but also van der Waals contacts between the protein and oligosaccharide. These optimal contacts are a result of distinct conformational changes in the backbone of the HSGAG oligosaccharide induced upon protein binding.
Conformational Analysis of Protein-Bound HSGAGs Reveals a “Kink” in the Helical Axis of the Oligosaccharides.
Although the FGF-bound oligosaccharides maintain the overall 21 helical symmetry, a kink was observed closer to the reducing end in the case of the FGF-1-bound oligosaccharide structure and to the nonreducing end in the case of the FGF-2 bound oligosaccharide structure (Fig. 2A). The kink spans three monosaccharide units where an iduronate is flanked by two glucosamines (H-I-H). The end-to-end distance of the trisaccharide-spanning kink is around 11 Å, which is approximately the same dimension as the HSGAG-binding pocket of FGF-1 and -2. The trisaccharide-spanning kink is structurally identical (superposition of the backbone atoms resulted in <1 Å rms deviation) in oligosaccharides bound to FGF-1 and -2. To quantify the kink in the FGF-bound HSGAG oligosaccharides in terms of its effect on the helical symmetry, the local helical propensities of the constituent disaccharide units (nL and hL parameters) were calculated (Table 1). The nL values of all of the constituent disaccharide units are around 2.0 with minor deviations within experimental error indicating that the overall 21 helical symmetry of the HSGAG oligosaccharide is maintained. On the other hand, there is a significant reduction observed in the hL values from 8.4 to 7.7 in the case of the FGF-1-bound and from 8.7 to 7.6 in the case of the FGF-2-bound HSGAG oligosaccharides (Table 1). These lower hL values reflect the higher curvature of the kink compared with that of the uniform helical structure of the unbound HSGAG oligosaccharides (Fig. 2A). The trisaccharide-spanning kink region has been observed to make the most contact with the protein in both the FGF-1 and -2 cocrystal structures.
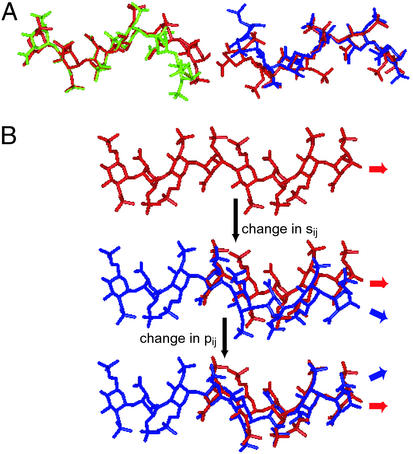
Figure 2.
Kink in FGF-bound oligosaccharides. The direction of the chains shown in A and B is from nonreducing end to reducing end (left to right). (A) Superimposition of FGF-1-bound (green) and modeled unbound oligosaccharide (on the left) and FGF-2-bound (blue) and modeled unbound oligosaccharide (on the right). The ring atoms and the glycosidic oxygens (O1 and O4) of H3 (Fig. 1A) in bound and unbound oligosaccharide structures were superimposed. H3-I4-H5 (Fig. 1A) of FGF-1-bound oligosaccharide and H1-I2-H3 (Fig. 1A) of FGF-2-bound oligosaccharide constitute the trisaccharide-spanning kink. (B) Stepwise formation (top to bottom) of the kink and continuation of helical structure caused by changes in p1j and s1j (according to the boldface values in Table 1) of the oligosaccharide (blue) relative to the unbound oligosaccharide (red).
Table 1.
nL and hL parameters for FGF-1- and FGF-2-bound oligosaccharides
| Oligosaccharides | FGF-1-bound
|
FGF-2-bound
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p | s | p | s | nL | hL | p | s | p | s | nL | hL | |
| H1-I2 | p11 = −171 | s11 = 99 | p22 = 166 | s22 = 136 | −2.1 | 8.4 | p11 = −147 | s11 = 119 | p22 = 172 | s22 = 141 | −2.5 | 7.8 |
| I2-H3 | p22 = 166 | s22 = 136 | p13 = −157 | s11 = 99 | −2.2 | 8.3 | p22 = 172 | s22 = 141 | p13 = −158 | s11 = 119 | −2.3 | 7.6 |
| p22 = 166 | s22 = 136 | p13 = −157 | s13 = 121 | −2.4 | 7.9 | p22 = 172 | s22 = 141 | p13 = −158 | s13 = 80 | 2.1 | 8.3 | |
| H3-I4 | p13 = −157 | s13 = 121 | P24 = 162 | s22 = 136 | −2.2 | 7.7 | p13 = −158 | s13 = 80 | p24 = 166 | s22 = 141 | 2.5 | 8.7 |
The nL and hL values of the constituent disaccharide units of the oligosaccharide bound to FGF-1 and -2. The nomenclature for monosaccharides and the glycosidic torsion angles is shown in Fig. 1A. The two sets of angles (p11, s11 and p13, s13) that result in a kink are shown in boldface.
Of particular interest is the observation that the kink is maintained in the FGF-2–FGFR1–HSGAG cocrystal structure despite the fact that the direction of the chain relative to the FGF-2 is reversed compared with that in the FGF-2–HSGAG cocrystal structure. This observation suggests that irrespective of the chain direction, the geometry of the HSGAG-binding site in FGF-1 and -2 appears to require the formation of a kink in the oligosaccharide to maximize contact with the protein. Taken together these studies indicate that both charge and surface complementarity (quantified by van der Waals contact) of the HSGAG oligosaccharide become important structural features that govern their specific interactions with different proteins.
Kink Is Formed by Local Deviation in Torsion Angles and Degree of Kink Is Enhanced by 1C4 Conformation of Iduronate.
It is important to point out that the kink in FGF-bound oligosaccharides is local, in that the oligosaccharide backbone is eventually restored to the original helical structure. The kink formation is associated with only two torsion angles, p1j and s1j (Table 1). A change in the torsion angle s1j alone caused the oligosaccharide to bend below the helical axis, whereas a change in p1j alone led to a bend above the helical axis (Fig. 2B). Therefore, changes in both of the torsion angles together restore the orientation of the helical axis after forming the kink (Fig. 2B). The degree of the kink in the helical structure is augmented by the 1C4 conformation of the iduronate ring in the H-I-H trisaccharide because this conformation positions the glycosidic bonds axially, thereby causing a sharp bend in the uniform helical structure. Alternative iduronate conformations and glucuronate containing structures provided less specific binding with FGF. For instance, an iduronate in the 2S0 ring conformation positioned the glycosidic bonds more equatorially to the ring, forming a more extended structure with lower curvature and thereby providing lesser contacts with FGF. Similarly, a β-d-glucuronate with 4C1 ring conformation positioned the glycosidic bonds in an equatorial fashion with respect to the ring, thereby forming a more extended helical structure than the 1C4 iduronate ring conformation. Therefore, an analogous trisaccharide containing the glucuronate in the 4C1 conformation (even with the same sulfation state) is expected to have a much lower affinity for FGF-1 and -2 based on the ability to form a kink. The specific structural features determining the shape of the kink observed in the FGF-bound oligosaccharides are attributable to the anomeric configuration, the ring conformation, the linkage between the monosaccharides, and the glycosidic torsion angles. Although dermatan sulfate contains the flexible iduronic acid, it does not form the kink observed in the FGF-bound HSGAG oligosaccharides because of differences in the monosaccharide unit (β-d-galactosamine vs. α-d-glucosamine in HSGAG) and the linkage (1 → 3 vs. 1 → 4 linkage in HSGAG). This provides a plausible explanation for the inability of sulfated chondroitin and dermatan glycosaminoglycan to compete with HSGAG for FGF-2 binding (34).
To understand the role of the iduronic acid in FGF-2 binding, Tabeur et al. (35) performed chemical synthesis to obtain different “structurally constrained” oligosaccharides representing various parts of the FGF-2-binding sequence (X-HNS,6S-Y-HNS,6S-Z; ref. 35). The sequence variation was achieved by substituting X, Y, and Z with iduronic or glucuronic acid. Among all of the oligosaccharides synthesized, it was observed that the one where all three positions X, Y, and Z substituted with iduronic acid had the highest affinity toward FGF-2. Furthermore, NMR analysis of this oligosaccharide indicated that the iduronic acid in the Y and Z position adopted a 1C4 chair conformation. It is important to note that the observed 1C4 conformation of the iduronic acid in position Y that is flanked by the two glucosamine residues is consistent with the trisaccharide-spanning kink defined here.
Implications of Kink: Ionic and van der Waals Contribution to FGF–HSGAG Interactions.
The relative position and sulfation pattern of trisaccharide-spanning kink [HNS,6S-I2S-HNS,6S] is consistent with the high-affinity region on both the FGF-1- and FGF-2-bound HSGAG oligosaccharides (25, 26). Importantly, the N-sulfate of the nonreducing end glucosamine and 2-O-sulfate of the iduronic acid, which are a part of the trisaccharide, contribute to the majority of the interactions with FGF-1 and -2. The ionic interactions are critically dependent on the orientation of these sulfate groups in the HSGAG-binding site of the protein. Projection of the sulfates on a helical wheel (Fig. 3) indicated that their orientation in the protein-bound and unbound oligosaccharides differed significantly. Further, the distance between the N- sulfate of the glucosamine and the 2-O-sulfate of the iduronic acid is lower in the region of the kink in comparison with the uniform helical structure (Fig. 3). Thus, the changes in the orientation of specific sulfate groups to maximize contact with the protein influence the bending of the helical axis of the oligosaccharide structure to form the kink.
Figure 3.
Distribution and orientation of sulfates in FGF-1- and FGF-2-bound oligosaccharides. (A) Designation of sulfates, where the chain direction is nonreducing end (left) to reducing end (right). (B–E) Projection of the sulfates down the helical axis represented on a helical wheel (Upper) and the axial distances between the sulfur atoms mapped on the helix axis (Lower). (B and D) Modeled unbound oligosaccharide structures with ring conformations corresponding to those observed in FGF-1 (25) and -2 (26) cocrystal structures, respectively. (C and E) Bound oligosaccharide structures obtained from the FGF-1 and -2 cocrystal structures, respectively. Note that in C the 2S2 is closer to the 6S2 (Upper) and there is a significant increase in the 6S1–NS2 axial distance and a decrease in the NS2–2S2 axial distance (Lower), compared with the unbound structure shown in B. Note that in E the 6S1 is closer to the NS2 (Upper), there is interchange of the 2S1 and NS1 (Upper), and there is a decrease in the NS2–2S2 axial distance and an increase in the 2S1–6S1 axial distance (Lower), compared with the unbound structure shown in D.
To quantify the optimal surface complementarity provided by the kink, both the kinked oligosaccharide and an oligosaccharide with a uniform helical structure were docked into the binding site of FGF-2. The electrostatic and van der Waals component were extracted from the total interaction energy for each complex. For the complex with kinked oligosaccharide, these values were lower by 11 and 25 kcal/mol, respectively. These results clearly suggest that the kinked oligosaccharide provides more favorable ionic and van der Waals contact with FGF compared with the oligosaccharide with the uniform helical structure.
To understand the biochemical implications of our structural findings, we interpreted the results from a recent study that identified HSGAG sequences with graded affinities (low, moderate, and high) for FGF-1 and -2 (10). The ability of these octasaccharides to form the specific trisaccharide-spanning kink with the appropriate sulfation pattern was evaluated. It is important to point out that the number of sulfate groups is about the same for both the low-affinity and moderately high-affinity binders. However, the affinity of the octasaccharides to FGF-1 and -2 correlated with the propensity of the oligosaccharides to form the specific trisaccharide-spanning kink (see Fig. 6 and Supporting Text, which are published as supporting information on the PNAS web site, www.pnas.org).
Analysis of HSGAG-Binding Sites on FGF.
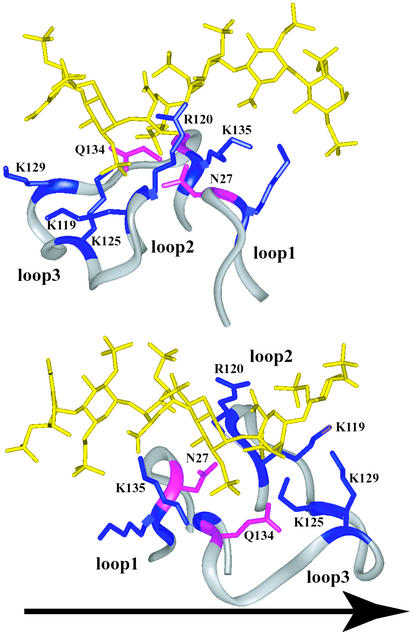
The topology of the HSGAG-binding site in FGF-1 and -2 is that of a narrow pocket containing a lysine at the base of the pocket (toward the N terminus of loop 3) and other basic amino acids (lysines and arginines) on the periphery of the pocket (Fig. 4). Lys-125 in FGF-2 or Lys-118 in FGF-1 has been shown to interact with the critical N-sulfate and 2-O-sulfate groups of the high-affinity region (that forms the kink) of the oligosaccharide chain (25, 26). The distribution of the peripheral basic amino acids becomes important while considering the direction of the HSGAG chain, as discussed above. Analysis of the HSGAG-binding site in FGF-2 revealed that the distribution of the amino acids K135, Q134, N27, R120, K119, and K129 facilitates favorable ionic interactions with the specific sulfate and carboxylate groups of the oligosaccharide for both nonreducing to reducing end and opposite chain directions (Fig. 4). For example, Q134 and N27 are positioned to interact with the critical 2-O-sulfate and N-sulfate groups of the oligosaccharide chain in one direction, as well as with the corresponding N-sulfate and 2-O-sulfate groups of the chain in the opposite direction. Similarly, there is a correspondence between K119 and K135 and between R120 and K129 in terms of their interaction with the sulfate and carboxylate groups of the oligosaccharide chain in either direction. Thus, the overall topology of the HSGAG-binding site (governed by the conformation of loops 1–3) and the distribution of basic residues (such as the location of a lysine at the base of a narrow pocket) could potentially impose specific structural requirements on the oligosaccharide chain, such as the formation of the kink and distribution and orientation of specific sulfate groups.
Figure 4.
HSGAG-binding site in FGF-2 facilitates interaction with both oligosaccharide chain directions. The direction of the oligosaccharide chain (yellow) is fixed from nonreducing to reducing end (see arrow). The topological arrangement of the HSGAG-binding loops in FGF-2 (shown as a ribbon trace in gray) relative to oligosaccharide for both the chain directions, one of which is observed in FGF-2–HSGAG cocrystal structure as shown in A and the other in FGF-2–FGFR1–HSGAG cocrystal structure as shown in B. The critical amino acids are shown numbered according to their sequence in FGF-2 (K and R, colored in blue; Q and N, colored in purple). The kink in the oligosaccharide is induced in both modes of binding (toward the nonreducing end in A and the reducing end in B). The narrow pocket formed by loop2 and loop3, containing the critical K125 residue at the base of the pocket, causes the oligosaccharide chain to kink to provide optimal ionic and van der Waals contacts. K125, Q134, and N27 interact with the critical N-sulfate and 2-O-sulfate groups of the trisaccharide-spanning kink region in both of the chain directions. The other basic residues are distributed on the periphery of the binding site such that they can interact with the oligosaccharide chain in either direction.
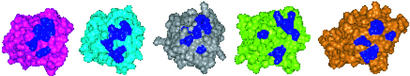
To compare the loop topology and spatial distribution of basic amino acids in other FGFs, we extended our analysis of the HSGAG-binding sites to FGF-4, -7, and -9, whose crystal structures were recently solved (Table 2). Although these FGFs are generally known to bind HSGAGs, there is no direct structural evidence for their interaction with HSGAG oligosaccharides. Most surprisingly, superposition of the Cα trace of the β-trefoil scaffold in all of these FGFs resulted in the HSGAG-binding loops to almost coincide on one another. In fact, the rms deviation for superimposing the Cα trace of loop regions in all of the FGFs was between 0.45 and 0.75 Å. However, the spatial arrangement of basic amino acids and their side chain conformation in the loop regions were significantly different (Fig. 5). These results led us to propose that the specific topology of the HSGAG-binding loops and the spatial arrangement of the basic amino acids in these loops provide a “signature” for the HSGAG-binding surface on different FGFs. The structural signature associated with the HSGAG-binding surface on FGF necessitates the formation of the kink and impinges on the distribution of sulfate groups in the oligosaccharide chain for optimal ionic and van der Waals contact.
Table 2.
Comparison of HSGAG binding loop regions in different FGFs
| FGF | Loop 1 | Loop 2–S3D–Loop 3 |
|---|---|---|
| 1 | CSNG | LKKNGSCKRGPRTHYGQKAIL |
| 2 | CKNG | LKRTGQYKLGSKTGPGQKAIL |
| 4 | CNVG | LGKNGKTKKGNRVSPTMKVTH |
| 7 | CRTQ | LNQKGLPVKGKKTKKEQKTAH |
| 9 | CRTG | LNKDGTPREGTRTKRHQKFTH |
Sequence of the loop regions that were structurally superimposed is shown, where the HSGAG-binding basic residues are indicated in bold. The putative HSGAG-binding residues in FGF-4, -7, and -9 were identified by using the FGF–HSGAG cocrystal structures as a guiding framework.
Figure 5.
Spatial distribution of basic amino acids in FGF. Shows Connolly surface representation of FGF-1 (purple), -2 (cyan), -4 (gray), -7 (green), and -9 (orange) with the basic residues shown in blue. Note that there is a significant difference in the spatial arrangement of the basic residues between the FGFs.
Comparison of FGF-2–HSGAG and FGF-7–HSGAG Interactions.
The putative HSGAG-binding amino acids in FGF-7 (Table 3) were identified by docking the oligosaccharide structure (obtained form FGF-2–HSGAG cocrystal structure) into the proposed HSGAG-binding site (see Methods). As stated earlier, the spatial arrangement of the basic residues in the HSGAG-binding site is remarkably different for FGF-2 and -7. Specifically, the amino acid in FGF-7 corresponding to the critical K125 at the base of the narrow HSGAG-binding pocket in FGF-2 is V143. In addition to the variation in this critical basic amino acid, the peripheral basic amino acids in FGF-7 are more spread out in comparison to the localized distribution of basic amino acids in FGF-2 (Fig. 5). Thus, the differences in the arrangement of the basic amino acids would potentially impose different structural requirements in the HSGAG oligosaccharide chain binding to FGF-2 and -7.
Table 3.
Comparison of HSGAG-binding site on FGF-2 and -7
| FGF-2 | K26 | N27 | N101 | R120 | K125 | Q134 | K135 | A136 | K119 | T121 | K129 |
| FGF-7 | R41 | T42 | N115 | Q138 | V143 | Q152 | K153 | T154 | N137 | K139 | K147 |
Comparison of the structurally homologous amino acids (obtained by superimposing the HSGAG-binding loops) in the HSGAG binding site of FGF-2 and -7. The basic amino acids K and R are bolded.
A quantitative basis for these differential requirements was obtained by comparing the interaction energies of the HSGAG oligosaccharide (obtained from FGF-2–HSGAG cocrystal structure) bound to FGF-2 and -7. These interaction energies were computed by docking the oligosaccharide into the HSGAG-binding site of the FGFs (see Methods). Although the van der Waals component of the interaction energy was roughly the same for both of the FGF–oligosaccharide complexes, the electrostatic component of the FGF-2 complex was lower by 32 kcal/mol than that of the FGF-7 complex. This suggests that although the topology of the HSGAG-binding site on FGF-7 could potentially impose a kink similar to that of the FGF-2 bound oligosaccharide, the distribution of sulfate groups in this oligosaccharide does not provide the optimal charge complementarity with the spatially distributed basic amino acids in FGF-7. Thus, an oligosaccharide comprising a kink forming motif (H-I-H) and a wider spatial distribution of sulfate groups would provide optimal ionic and van der Waals contact with the binding site of FGF-7. A wider spatial distribution of sulfate groups can be achieved by the presence of a glucuronic acid in the sequence. Glucuronic acid residues present in a HSGAG chain provide a longer helical repeat length, because of the equatorial positioning of the backbone oxygen atoms (O1 and O4) and the 4C1 chair conformation of the ring. Although it has been shown that distinct HSGAGs differentially bind to FGF-1/FGF-2 and FGF-7 (36), much less is known about the structural basis for these differential affinities. Our study provides a structural framework to design experiments that compare specific binding affinities of defined HSGAG oligosaccharides to FGF-2 and -7.
Summary.
In this study we demonstrate that FGF–HSGAG interactions are driven by factors that are more profound than simple ionic contacts. These factors are influenced by the structure and conformation of the HSGAG oligosaccharide chain, the topology, and the spatial arrangement of positively charged amino acids in the HSGAG-binding sites of different FGFs. Specifically, we showed that binding to protein causes local conformational changes in the HSGAG backbone, resulting in the formation of a “kink” in an otherwise uniform helical structure. The formation of the kink in the HSGAG structure is associated with changes in the glycosidic torsion angles at a trisaccharide level (H-I-H) and the changes in the iduronate ring conformation. The kink thus represents a “signature” associated with the “fit” between a unique HSGAG sequence and the protein to which it specifically binds. On the protein side, we demonstrated that the topology of the HSGAG-binding site governed by conformation of three specific loop regions in the β-trefoil scaffold is identical in all of the FGF crystal structures. However, the spatial distribution of the basic amino acids in the binding site is different. Thus, the specific topology of the HSGAG-binding loops and the spatial arrangement of the basic amino acids in these loops impose distinctive structural requirements on the oligosaccharide chain for the different FGF-family members.
We also analyzed cocrystal structures of HSGAGs with other proteins, including antithrombin (AT-III), foot and mouth disease virus protein, annexin V, and the NK1 domain of hepatocyte growth factor, to determine whether a kink was induced in these protein-bound oligosaccharides (see Fig. 7 and Supporting Text, which are published as supporting information on the PNAS web site). Our analysis of all these protein–HSGAG cocrystal structures showed a deviation in the protein-bound HSGAG backbone, resulting in the formation of a kink, thus pointing to a broader implication for our findings.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants HL59966 and CA 090940 (to R.S.), the Burroughs Wellcome Foundation (R.S.), and a Merck/Massachusetts Institute of Technology fellowship (to R.R.).
Abbreviations
- HSGAG
heparin and heparan sulfate glycosaminoglycan
- FGF
fibroblast growth factor
- FGFR
FGF receptor
- I
α-l-iduronic acid
- H
α-d-glucosamine
- 2S
6S, NS, and 3S, sulfation at the 2-O position of the uronic acid and the 6-O, N, and 3-O positions of the glucosamine, respectively
References
- 1.Bernfield M, Gotte M, Park P W, Reizes O, Fitzgerald M L, Lincecum J, Zako M. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 2.Casu B, Lindahl U. Adv Carbohydr Chem Biochem. 2001;57:159–206. doi: 10.1016/s0065-2318(01)57017-1. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Thorp S C. Med Res Rev. 2002;22:1–25. doi: 10.1002/med.1026. [DOI] [PubMed] [Google Scholar]
- 4.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Nat Rev Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 5.Conrad H E. Heparin-Binding Proteins. San Diego: Academic; 1998. [Google Scholar]
- 6.Tumova S, Woods A, Couchman J R. Int J Biochem Cell Biol. 2000;32:269–288. doi: 10.1016/s1357-2725(99)00116-8. [DOI] [PubMed] [Google Scholar]
- 7.Dhoot G K, Gustafsson M K, Ai X, Sun W, Standiford D M, Emerson C P., Jr Science. 2001;293:1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- 8.Casu B, Guerrini M, Naggi A, Perez M, Torri G, Ribatti D, Carminati P, Giannini G, Penco S, Pisano C, et al. Biochemistry. 2002;41:10519–10528. doi: 10.1021/bi020118n. [DOI] [PubMed] [Google Scholar]
- 9.Faham S, Linhardt R J, Rees D C. Curr Opin Struct Biol. 1998;8:578–586. doi: 10.1016/s0959-440x(98)80147-4. [DOI] [PubMed] [Google Scholar]
- 10.Kreuger J, Salmivirta M, Sturiale L, Gimenez-Gallego G, Lindahl U. J Biol Chem. 2001;276:30744–30752. doi: 10.1074/jbc.M102628200. [DOI] [PubMed] [Google Scholar]
- 11.Humphries D E, Wong G W, Friend D S, Gurish M F, Qiu W T, Huang C, Sharpe A H, Stevens R L. Nature. 1999;400:769–772. doi: 10.1038/23481. [DOI] [PubMed] [Google Scholar]
- 12.Atkins E D, Nieduszynski I A. Adv Exp Med Biol. 1975;52:19–37. doi: 10.1007/978-1-4684-0946-8_2. [DOI] [PubMed] [Google Scholar]
- 13.Nieduszynski I A, Atkins E D. Biochem J. 1973;135:729–733. doi: 10.1042/bj1350729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulloy B, Forster M J, Jones C, Davies D B. Biochem J. 1993;293:849–858. doi: 10.1042/bj2930849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulloy B, Forster M J. Glycobiology. 2000;10:1147–1156. doi: 10.1093/glycob/10.11.1147. [DOI] [PubMed] [Google Scholar]
- 16.Venkataraman G, Sasisekharan V, Cooney C L, Langer R, Sasisekharan R. Proc Natl Acad Sci USA. 1994;91:6171–6175. doi: 10.1073/pnas.91.13.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ernst S, Venkataraman G, Sasisekharan V, Langer R, Cooney C L, Sasisekharan R. J Am Chem Soc. 1998;120:2099–2107. [Google Scholar]
- 18.Casu B, Petitou M, Provasoli M, Sinay P. Trends Biochem Sci. 1988;13:221–225. doi: 10.1016/0968-0004(88)90088-6. [DOI] [PubMed] [Google Scholar]
- 19.Casu B, Choay J, Ferro D R, Gatti G, Jacquinet J C, Petitou M, Provasoli A, Ragazzi M, Sinay P, Torri G. Nature. 1986;322:215–216. doi: 10.1038/322215b0. [DOI] [PubMed] [Google Scholar]
- 20.Ornitz D M, Itoh N. Genome Biol. 2001;2:reviews3005. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostrovsky O, Berman B, Gallagher J, Mulloy B, Fernig D G, Delehedde M, Ron D. J Biol Chem. 2002;277:2444–2453. doi: 10.1074/jbc.M108540200. [DOI] [PubMed] [Google Scholar]
- 22.Chang Z, Meyer K, Rapraeger A C, Friedl A. FASEB J. 2000;14:137–144. doi: 10.1096/fasebj.14.1.137. [DOI] [PubMed] [Google Scholar]
- 23.Powell A K, Fernig D G, Turnbull J E. J Biol Chem. 2002;277:28554–28563. doi: 10.1074/jbc.M111754200. [DOI] [PubMed] [Google Scholar]
- 24.Pye D A, Vives R R, Hyde P, Gallagher J T. Glycobiology. 2000;10:1183–1192. doi: 10.1093/glycob/10.11.1183. [DOI] [PubMed] [Google Scholar]
- 25.DiGabriele A D, Lax I, Chen D I, Svahn C M, Jaye M, Schlessinger J, Hendrickson W A. Nature. 1998;393:812–817. doi: 10.1038/31741. [DOI] [PubMed] [Google Scholar]
- 26.Faham S, Hileman R E, Fromm J R, Linhardt R J, Rees D C. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 27.Pellegrini L, Burke D F, von Delft F, Mulloy B, Blundell T L. Nature. 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 28.Schlessinger J, Plotnikov A N, Ibrahimi O A, Eliseenkova A V, Yeh B K, Yayon A, Linhardt R J, Mohammadi M. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 29.Thompson L D, Pantoliano M W, Springer B A. Biochemistry. 1994;33:3831–3840. doi: 10.1021/bi00179a006. [DOI] [PubMed] [Google Scholar]
- 30.Pattabiraman N. Ph.D. thesis. Bangalore, India: Indian Institute of Science; 1979. [Google Scholar]
- 31.Homans S W. Biochemistry. 1990;29:9110–9118. doi: 10.1021/bi00491a003. [DOI] [PubMed] [Google Scholar]
- 32.Huige C J M, Altona C. J Comput Chem. 1995;16:56–79. [Google Scholar]
- 33.Guerrini M, Agulles T, Bisio A, Hricovini M, Lay L, Naggi A, Poletti L, Sturiale L, Torri G, Casu B. Biochem Biophys Res Commun. 2002;292:222–230. doi: 10.1006/bbrc.2002.6634. [DOI] [PubMed] [Google Scholar]
- 34.Foxall C, Holme K R, Liang W, Wei Z. Anal Biochem. 1995;231:366–373. doi: 10.1006/abio.1995.0065. [DOI] [PubMed] [Google Scholar]
- 35.Tabeur C, Mallet J M, Bono F, Herbert J M, Petitou M, Sinay P. Bioorg Med Chem. 1999;7:2003–2012. doi: 10.1016/s0968-0896(99)00113-3. [DOI] [PubMed] [Google Scholar]
- 36.Ye S, Luo Y, Lu W, Jones R B, Linhardt R J, Caplia I, Toida T, Kan M, Pelletier H, McKeehan W. Biochemistry. 2001;40:14429–14439. doi: 10.1021/bi011000u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.