Abstract
In Escherichia coli, chemoreceptor clustering at a cell pole seems critical for signal amplification and adaptation. However, little is known about the mechanism of localization itself. Here we examined whether the aspartate chemoreceptor (Tar) is inserted directly into the polar membrane by using its fusion to green fluorescent protein (GFP). After induction of Tar–GFP, fluorescent spots first appeared in lateral membrane regions, and later cell poles became predominantly fluorescent. Unexpectedly, Tar–GFP showed a helical arrangement in lateral regions, which was more apparent when a Tar–GFP derivative with two cysteine residues in the periplasmic domain was cross-linked to form higher oligomers. Moreover, similar distribution was observed even when the cytoplasmic domain of the double cysteine Tar–GFP mutant was replaced by that of the kinase EnvZ, which does not localize to a pole. Observation of GFP–SecE and a translocation-defective MalE–GFP mutant, as well as indirect immunofluorescence microscopy on SecG, suggested that the general protein translocation machinery (Sec) itself is arranged into a helical array, with which Tar is transiently associated. The Sec coil appeared distinct from the MreB coil, an actin-like cytoskeleton. These findings will shed new light on the mechanisms underlying spatial organization of membrane proteins in E. coli.
Introduction
The proteins involved in many biological systems are apt to be spatially organized within a cell or even within a smaller compartment rather than to be freely diffusible and encountering each other stochastically. Proper spatial organization of proteins is of vital importance not only in eukaryotes but also in prokaryotes despite their smaller sizes. Bacterial cellular functions that involve localization of proteins include virulence, cell division and chemotaxis. For example, IcsA of Shigella flexneri and ActA of Listeria monocytogenes, which nucleate actin polymerization and are required for the bacterial movement within a host cell and hence virulence, localize to an old pole of the rod-shaped bacterial cell (Goldberg et al., 1993; Smith et al., 1995). CpaE and CpaC, which are required for polar pilus biogenesis in Caulobactor crescentus, localize to one pole and their localization is controlled by PodJ (Viollier et al., 2002). In Escherichia coli, the best characterized bacterial species, many proteins have been shown to localize (for reviews, see Nanninga, 1998; Lybarger and Maddock, 2001; Shapiro et al., 2002; Margolin, 2003). However, little is known about mechanisms underlying protein localization. For example, the cell division machinery of E. coli has to localize to the mid cell position and this localization is regulated by other proteins (the Min system) (Bi and Lutkenhaus, 1991; Akerlund et al., 1992), but it remains to be elucidated whether the localization results directly from interactions with other protein(s), including those between the Min proteins and the division machinery, peptidoglycan (cell wall) or phospholipids. Experimental approaches to examine the mechanisms of their localization are limited, because most of these components are essential for cellular viability and/or morphology.
The chemotactic signalling system of E. coli may be amenable to studying mechanisms underlying spatial organization of proteins because the lack of any chemotactic signalling component protein does not affect cell morphology and viability at least under typical laboratory conditions. Moreover, all of the components (Che proteins and the chemoreceptors) have been identified and well characterized in terms of genetics, biochemistry and structural biology (for reviews, see Parkinson, 1993; Stock and Surette, 1996; Djordjevic and Stock, 1998; Armitage, 1999; Falke and Kim, 2000). The chemoreceptors (also known as transducers or methyl-accepting chemotaxis proteins (MCPs)) form clusters with the histidine kinase CheA and the adaptor CheW at a cell pole and many other Che proteins target to this polar receptor-kinase cluster (Maddock and Shapiro, 1993; Sourjik and Berg, 2000; Shiomi et al., 2002; Cantwell et al., 2003; Banno et al., 2004). Polar localization and clustering have been thought to be critical for signal amplification (Bray et al., 1998; Shimizu et al., 2000; Ames et al., 2002; Gestwicki and Kiessling, 2002; Kim et al., 2002; Homma et al., 2004) and adaptation (Li and Weis, 2000; Barnakov et al., 2001; 2002; Levit and Stock, 2002; Shiomi et al., 2002; Banno et al., 2004).
It has been shown that polar localization of the chemoreceptors depends partially on CheA and CheW (Maddock and Shapiro, 1993; Skidmore et al., 2000; Shiomi et al., 2005). However, it is not known how the chemoreceptor localizes to a cell pole. To target to a pole, a nascent chemoreceptor protein might be inserted into the cytoplasmic membrane (i) at or near a cell pole thereby staying in the vicinity of the insertion point (the direct membrane insertion model) or (ii) at random positions thereafter migrating or diffusing away from the insertion point (the indirect membrane insertion model) (Fig. 1A). Both of the mechanisms have been shown to operate in other systems: (i) IcsA of S. flexneri employs a direct mechanism when targeting directly to an old pole in E. coli, which does not have an IcsA homologue (Charles et al., 2001) and (ii) SpoIVFB of Bacillus subtilis utilizes an indirect mechanism, being randomly inserted into the cytoplasmic membrane and then diffused to and captured in the outer forespore membrane (Rudner et al., 2002).
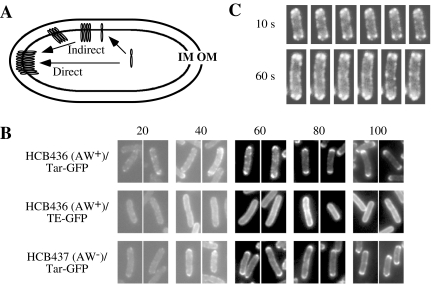
Fig. 1. Polar localization of Tar–GFP.
A. Direct and indirect membrane insertion models for polar localization of the chemoreceptors. To target to a cell pole, a nascent chemoreceptor protein might be inserted into the cytoplasmic membrane (i) at or near a cell pole (a direct model), or (ii) at random positions thereafter migrating or diffusing away from the insertion point (an indirect model). IM, the cytoplasmic (inner) membrane; OM, the outer membrane.
B. Time-course of polar localization of Tar–GFP. HCB436 (CheAW+) or HCB437 (CheAW−) cells carrying a plasmid encoding Tar–GFP or Taz1–GFP were observed at indicated time points after the addition of 1 mM arabinose. Numbers above pictures represent harvested time points (min).
C. Time-lapse observation of polar localization of Tar–GFP. HCB436 cells carrying plasmid encoding Tar–GFP were spotted onto a glass slide covered with 0.5% agarose. Fluorescence was observed with 10 or 60 s intervals.
To elucidate the localization mechanism of the chemoreceptor, it is indispensable to identify the site(s) of membrane insertion. It has been reported that the serine chemoreceptor Tsr of E. coli is inserted into the cytoplasmic membrane via a SecA-dependent process (Gebert et al., 1988). SecA is an ATPase that associates with the integral membrane proteins, SecY, SecE and SecG, to constitute a general protein translocase (for reviews, see Driessen et al., 1998; Mori and Ito, 2001). Recent observation of SecY–GFP (green fluorescent protein) suggested that the Sec machinery is evenly distributed throughout the cytoplasmic membrane in E. coli (Brandon et al., 2003; Espeli et al., 2003). It has recently been reported that the Sec translocases of some Gram-positive bacteria show characteristic localization: to a microdomain in Streptococcus pyogenes (Rosch and Caparon, 2004) and to a helical array in B. subtilis (Campo et al., 2004). Thus without any direct evidence it is hard to judge which model can apply to the chemoreceptor localization in E. coli: (i) a subset of Sec translocases at a pole might be devoted to insert the chemoreceptors (and possibly other polar membrane proteins) into the membrane, or (ii) the chemoreceptors might be inserted into the membrane at virtually any position throughout a cell.
Here we report that the aspartate chemoreceptor Tar employs an indirect mechanism for its polar localization: i.e. after induction, Tar–GFP first appeared in lateral membrane regions, clustered in the presence of CheA and CheW, and then seemed to migrate towards a cell pole. Unexpectedly, Tar–GFP formed a helical array when it is located in lateral membrane regions. Further analyses, including observation of GFP–SecE and the mutant versions of MalE–GFP, as well as indirect immunofluorescent microscopy on SecG, suggested that this Tar coil reflects a helical array of the Sec machinery itself, which was shown to be distinct from that of the actin homologue MreB.
Results
The aspartate chemoreceptor Tar is not directly inserted into polar membranes
To visualize the localization pathway of a chemoreceptor, GFP was fused with the C-terminus of the aspartate chemoreceptor Tar (Homma et al., 2004), and localization of the resulting fusion (named Tar–GFP) was observed sequentially after induction. For tight regulation of expression, the tar–gfp gene was placed downstream of the araBAD promoter (pBAD24-Tar–GFP). Cells expressing Tar–GFP swarmed only slightly slower than those expressing wild-type Tar (Shiomi et al., 2005), indicating that the Tar–GFP retains essential receptor function. A chimeric protein named Taz1–GFP that does not localize to a pole was used as a negative control (pBAD24-Taz1–GFP): Taz1 was constructed by replacing the cytoplasmic domain of Tar, which is required for interaction with CheA and CheW and formation of a predicted trimer of the Tar dimers, with the cytoplasmic kinase and phosphatase domains of the osmosensor histidine kinase EnvZ (details will be described elsewhere). It was previously reported that the kinase and phosphatase activities of Taz1 (Utsumi et al., 1989), which was not fused with GFP, are modulated by the Tar-specific ligand aspartate (Utsumi et al., 1989; Yang et al., 1993). We observed that Taz1–GFP is distributed almost evenly throughout the cytoplasmic membrane (Figs 1B and 2F). HCB436 (CheAW+) cells carrying the plasmid encoding Tar–GFP or Taz1–GFP were grown in TG medium. After 3 h, 1 mM arabinose was added to express the GFP fusion proteins (time 0). Cells were harvested at indicated time points (Fig. 1B). At 20 min after induction, in most cells both Tar–GFP and Taz1–GFP appeared in lateral cytoplasmic membrane regions, where the fusion proteins formed small clusters, whereas in the other cells we hardly detected any fluorescence from Tar–GFP. At 60 min, subpopulations of Tar–GFP formed clusters at apparently random positions in the cytoplasmic membrane and others localized to cell poles. At 80 min, Tar–GFP localized almost completely to cell poles, whereas Taz1–GFP was almost evenly distributed in the cytoplasmic membrane, an expected result as it lacks the highly conserved domain critical for polar localization (D. Shiomi and I. Kawagishi, unpubl. results). It should be noted that the expression levels of Tar–GFP was comparable with chromosome-encoded Tar at 60 min after the addition of 1 mM arabinose. In the absence of CheA and CheW, Tar–GFP also formed clusters at apparently random positions (see Results at 20 and 40 min) and eventually localized to cell poles to a lesser extent (100 min) than in the presence of CheA and CheW. Under our experimental conditions, cell-to-cell variations in the localization patterns of Tar–GFP at each time point except for at 20 min were only limited (data not shown). These results are consistent with an indirect model, in which the proper polar localization of Tar is achieved by random insertions into the cytoplasmic membrane followed by the association with CheA and CheW and/or interaction with each other through the highly conserved domain among MCPs (HCD) and the diffusion or transport of the resulting clusters to cell poles.
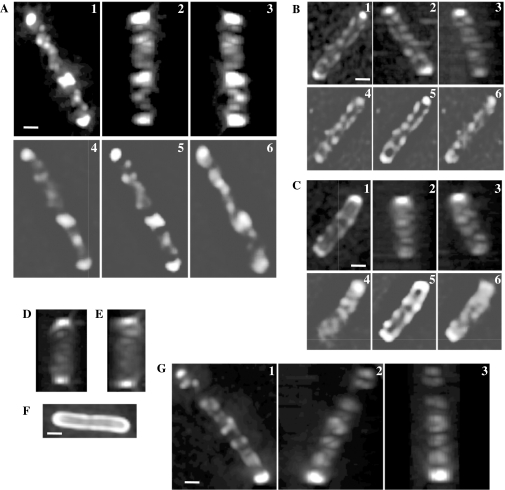
Fig. 2. Helical arrays of the chemotaxis machinery in a cell. Cells were subjected to optical sectioning and processing as described in Experimental procedures. GFP-tagged proteins were induced by 1 mM arabinose for 60 min. Three-dimensionally reconstructed images from different angles (1, 2 and 3) are shown. Each image was rotated around the long axis of the cell to display the helical array. Unprocessed images are also shown (4, 5, 6).
A–C. Tar–GFP in HCB436 (CheAW+) cells.
D and E. Tar–GFP in HCB437 (CheAW–) cells.
F. Taz1–GFP in an HCB436 (CheAW+) cell.
G. GFP–CheA in an HCB437 cell expressing Tar and CheW. Scale bars indicate 1 μm.
To further examine this possibility, we observed the processes of polar localization of Tar–GFP in a single cell. An hour after the addition of 1 mM arabinose, cells were washed and resuspended in TG medium supplemented with 25 μg ml−1 rifampicin to prevent further transcription. Cells were further incubated at 30°C for 10 min and then observed by fluorescence microscopy with 1, 10 or 60 s intervals. Time-lapse images of a representative cell are shown in Fig. 1C. Tar–GFP formed clusters at random positions of the cytoplasmic membrane. At a cell pole, a fluorescent cluster was detected after the addition of rifampicin. Again this observation is consistent with the notion that Tar–GFP clusters at lateral membrane regions and then migrates towards a cell pole. However, we have never seen a cluster travelling from the lateral site to the pole, but this cannot argue against the net migration to the pole: it would be rather hard to imagine that a migrating cluster stays in the same focal plane for a long period. Anyhow, fluorescent spots did not appear to migrate straightforward to the pole. They might move in a random-walk fashion and be trapped when they reach the pole. It should also be noted that this result may not exactly reflect native migration patterns of the chemoreceptor from lateral membrane regions to a pole because cells were attached to a 0.5% agarose layer on a slide glass and therefore nutrients and oxygen must be limited.
Tar–GFP shows a helical arrangement
Careful observation of Tar–GFP clusters at lateral cytoplasmic membrane regions (Fig. 1B and C) led us to suspect that Tar could be organized into a coil-like array in the lateral membrane. The two-dimensional images of lateral clusters of Tar–GFP are reminiscent of those of the bacterial actin homologue MreB that forms a helical structure along the long axis of a cell (Jones et al., 2001; Shih et al., 2003). We therefore wanted to know the three-dimensional arrangement of Tar in lateral cytoplasmic membrane regions. Optical dissection of single cells (more than 30 cells for each condition) was carried out and the resulting series of fluorescent images were processed by using a deconvolution software to remove out-of-focus fluorescence and to reconstitute a three-dimensional fluorescent image. First, we examined Tar–GFP in the presence of CheA and CheW (i.e. in strain HCB436) at 60 min after the addition of the inducer arabinose (Fig. 2A–C). All of the cells checked showed coil- or ring-like fluorescence patterns along their long axes. Similar helical fluorescence patterns were detected even when Tar–GFP was expressed in the absence of CheA and CheW (i.e. in strain HCB437) (Fig. 2D and E). In contrast, Taz1–GFP did not show any characteristic pattern in either host strain (Fig. 2F). We previously observed that GFP–CheA forms clusters at a cell pole and lateral cytoplasmic membrane regions when Tar was overexpressed (D. Shiomi and I. Kawagishi, unpubl. results). Under the same condition, we obtained reconstituted three-dimensional images of GFP–CheA and detected coiled fluorescence patterns (Fig. 2G). These results suggest that Tar and CheA are organized into a helical array in lateral membrane regions. The formation of such an array might involve the HCD-mediated interactions of Tar–GFP with neighbouring Tar–GFP (between dimers) and/or CheA.
We now hypothesized two possibilities about the nature of this helical arrangement of Tar–GFP: (i) Tar might migrate along a helical track within lateral regions of the cytoplasmic membrane; or (ii) the protein translocation machinery that inserts Tar into the cytoplasmic membrane might be arranged into a helical array. Whichever the case, introduction of Cys residues in the periplasmic region of Tar, which is devoid of Cys residue, might result in a better helical image if disulphide cross-linking leads to higher oligomers and occurs immediately after the translocation. Introduction of a Cys residue at an appropriate periplasmic position (e.g. S36C) of the subunit interface within a dimer results in a disulphide bond between subunits. When another Cys residue is introduced at the external surface of the dimer (e.g. D142C), the double Cys mutant would form cross-linked oligomers (Fig. 3A). This has been demonstrated for Tar-S36C&D142C (Homma et al., 2004). Among the double Cys mutant proteins tested, Tar-S36C&A118C was most efficiently cross-linked (H. Irieda, M. Homma, M. Homma and I. Kawagishi, submitted for publication). Immunoblotting analyses of the GFP fusion of the double Cys mutant showed that it is indeed highly cross-linked leaving only a very small amount of the uncross-linked monomer even without the addition of any catalyst for oxidation (Fig. 3B; note that highly cross-linked products could not get into the gel). In HCB436 cells, Tar-S36C&A118C–GFP showed clearer coiled structures (Fig. 3C).
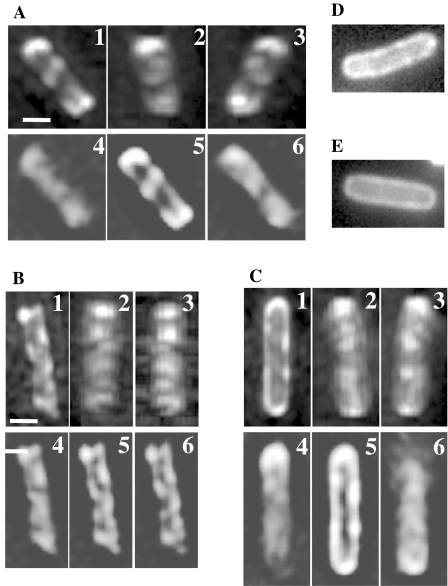
Fig. 3. Helical arrays of disulphide-cross-linked Tar–GFP and Taz1–GFP.
A. Schematic illustration of the inter-dimer cross-linking strategy of Tar–GFP or Taz1–GFP. Two cysteine residues were introduced into the periplasmic domain of the Tar– or Taz1–GFP protein to form higher oligomers with disulphide bonds linking subunits both within and between dimers. IM, the cytoplasmic (inner) membrane; N and C, the N- and C-termini; open and shaded circles, the introduced Cys residues (S36C and A118C).
B. In vivo disulphide cross-linking of the Cys-replaced Tar– and Taz1–GFP proteins. HCB436 cells (ΔMCP ΔCheR ΔCheB) expressing the wild-type (WT) or mutant (S36C&A118C) versions of the fusion proteins (Tar–GFP and Taz1–GFP) were incubated with (+) or without (−) 60 μM Cu(II)(o-phenanthroline)3 for 5 min at 30°C and their whole-cell extracts were subjected to non-reducing (−2ME) and reducing (+2ME) SDS-PAGE followed by immunoblotting with anti-GFP (Molecular Probes).
C and D. Three-dimensionally reconstituted images of HCB436 cells expressing the S36C&A118C mutant versions of Tar–GFP (C) and Taz1–GFP (D). For each cell, reconstituted images form different angles (1–3) and unprocessed images (4–6) are shown. Scale bars indicate 1 μm.
We also constructed and examined Taz1-S36C&A118C–GFP, which lacks HCD, as a negative control. Unexpectedly, however, it also formed a helical array (Fig. 3D), indicating that HCD is dispensable for the helical array. A possible explanation of this phenomenon is that Taz1-S36C&A118C–GFP might be cross-linked immediately after inserted into the cytoplasmic membrane thereby trapped in the vicinity of the sites of insertion. It was previously shown that the serine chemoreceptor Tsr is inserted into the membrane via the general protein translocation apparatus (the Sec machinery) (Gebert et al., 1988). We suspect that cross-linking immediately after the insertion would prevent the mutant protein from diffusing away from the Sec machinery, resulting in better helical images. If cross-linking was extremely efficient, a newly inserted Cys derivative of Tar–GFP or Taz1–GFP might be cross-linked with the neighbouring one, which would clog the Sec machinery. If this is the case, overproduction of these proteins would affect cell growth. As expected, the A118C derivative of Tar–GFP or Taz1–GFP retarded cell growth even when the expression level was adjusted roughly equal to the total amount of chromosome-encoded chemoreceptors, whereas the corresponding wild-type versions did not show any significant effect (data not shown). Therefore, we speculate that the helical arrangements of Tar–GFP and Taz1–GFP within a cell may reflect that of the Sec machinery itself.
Cellular organization of the Sec machinery
The Sec machinery contains, as essential components, three integral membrane proteins, SecY, SecE and SecG (the SecYEG complex) that forms a translocation pore, and a membrane-peripheral ATPase, SecA. To examine subcellular localization of the Sec machinery, we constructed GFP–SecE, which was expressed in strain HCB436 and three-dimensional images were reconstituted (Fig. 4A–C). GFP–SecE was also organized into a helical array. This is inconsistent with recent publications reporting that SecY- and SecE–GFP fusion proteins are evenly distributed throughout the cytoplasmic membrane in E. coli (Brandon et al., 2003; Espeli et al., 2003). This discrepancy could be accounted for by levels of the Sec–GFP fusion proteins: their even distribution might result from the failure of excess Sec–GFP proteins to form a SecYEG complex with their partner Sec proteins as we also observed almost even distribution of GFP–SecE when it was mildly overexpressed (Fig. 4D and E).
Fig. 4. Helical arrangement of the GFP–SecE fusion protein. Images of more than 30 cells were analysed and representative cells are shown.
A–C. Three-dimensionally reconstituted images of HCB436 cells expressing GFP–SecE. Cells were harvested 15 min after the addition of 1 mM arabinose and subjected to optical sectioning and processing as described in Experimental procedures. For each cell, reconstituted images form different angles (1–3) and unprocessed images (4–6) are shown.
D and E. Overexpressed GFP–SecE in HCB436 cells. Cells were harvested 30 min after the addition of 1 mM arabinose.
We also wanted to observe subcellular localization of the Sec machinery without GFP fusion and at the wild-type stoichiometry. First, we employed indirect immunofluorescence microscopy (IFM) to see localization of SecG, which is other component of the Sec machinery. Immunoblotting of wild-type cells (W3110) with anti-SecG antibody detected a major band of approximately 12 kDa, which should correspond to SecG, with low levels of cross-reacting bands (Fig. 5A). Wild-type cells were then fixed, treated with anti-SecG antibody and then with the second antibody labelled with Alexa Fluor 488. In many cells, with significant levels of background, diagonal lines were observed (Fig. 5B). These lines are consistent with the helical arrangement of SecG in a cell.
Fig. 5. Helical arrangement of SecG.
A. Detection of chromosome-encoded SecG by immunoblotting. W3110 (wild-type) cells were subjected to immunoblotting with anti-SecG antibody. Arrow head indicates bands of SecG. Open triangle on the top indicates increasing amounts applied (from left to right, one-, two- and fourfold).
B. Detection of chromosome-encoded SecG by IFM. W3110 (wild-type) cells were subjected to IFM with anti-SecG as the first antibody and Alexa Fluor 488-labelled goat anti-rabbit IgG antibody. Typical cells are shown.
Second, we took advantage of export-defective mutations of maltose-binding protein (MBP or MalE) to observe subcellular localization of the Sec machinery at the wild-type expression level. The mutant MalE proteins with M18R or M19R substitution in the leader peptide sequence are defective in translocation across the membrane, but not in its initiation step, and their precursors are thought to remain trapped in the Sec machinery (Bassford and Beckwith, 1979; Bedouelle et al., 1980). It has been shown that wild-type MalE–GFP is exported to the periplasmic space successfully via the Sec machinery but that the protein is not fluorescent presumably due to misfolding (Feilmeier et al., 2000). We therefore reasoned that fluorescence of the M18R or M19R derivative of MalE–GFP would reflect cytoplasmic localization of the Sec machinery (Fig. 6A). Immunoblotting verified that expression levels of the wild-type and mutant versions of MalE–GFP were comparable with each other and that the mutant versions were not processed (Fig. 6B). Wild-type MalE–GFP was not fluorescent (data not shown) whereas the mutant versions of MalE–GFP formed clusters in the cytoplasmic membrane in strain HCB436 (see Fig. 6C–E). The reconstituted three-dimensional images of MalE-M19R–GFP clearly showed helical arrangements (Fig. 6C–E). In this experiment, no component of the Sec machinery is overproduced and therefore the result argues strongly that the Sec machinery is organized into a helical array. We tried to examine whether MalE-M19R–CFP colocalizes with YFP–SecE, but it was unsuccessful probably because the expression level of the latter fusion was difficult to control (data not shown). Instead, we examined colocalization of the coils of MalE and Tar (Fig. 6A and F). CFP and YFP were fused to MalE-M19R and Tar respectively. MalE-M19R–CFP and Tar–YFP colocalized in the lateral membrane regions (Fig. 6F). We therefore conclude that the Sec machinery is organized into a coil-like structure in a cell.
Fig. 6. Helical arrangement of the translocation-defective mutant MalE–GFP protein.
A. Schematic illustration of double labelling of Tar and MalE. YFP and CFP/GFP were fused with the C-terminus of Tar and MalE to yield Tar–YFP and MalE–CFP/GFP respectively. Exported MalE–CFP was not fluorescent whereas the signal sequence mutant versions of MalE–CFP (M18R and M19R) were fluorescent probably because they remain clogged in the Sec machinery. MalE–CFP and MalE*–CFP represent wild-type and mutant versions of MalE–CFP respectively.
B. Expression of the mutant MalE–GFP proteins. HCB436 cells carrying a plasmid encoding GFP or the wild-type (WT) or mutant (M18R or M19R) version of MalE–GFP were grown in the presence (+) or absence (−) of 1 mM arabinose and their whole-cell lysates were subjected to SDS-PAGE followed by immunoblotting with anti-GFP.
C–E. Three-dimensionally reconstituted images of HCB436 cells expressing MalE-M19R–GFP. For each cell, reconstituted images form different angles (1–3) are shown. Scale bars indicate 1 μm.
F. Tar–YFP and MalE-M19R–CFP in strain HCB436. Individual images of Tar–YFP (images 1, 4 and 7; artificially coloured green) and MalE-M19R–CFP [original (2, 5 and 8) and processed (2′, 5′-and 8′) images; artificially coloured red] were merged (images 3, 6 and 9).
The Sec coil is distinct from the helical filament of MreB
It has recently been shown that E. coli has cytoskeletal proteins that are homologues of actin (FtsA and MreB) and tubulin (FtsZ) (van den Ent et al., 2001). FtsA plays a key role in cytokinesis whereas MreB is critical for the rod shape. More recently, it has been shown that MreB forms a helical filament both in B. subtilis (Jones et al., 2001) and E. coli (Shih et al., 2003). The finding of the helical array of the Sec machinery raised a possibility that it may be associated with the MreB coil. To address this issue, we examined whether Tar–YFP colocalizes with CFP–MreB (Fig. 7). In all of the cells checked, the coils of CFP–MreB and Tar–YFP were not perfectly overlapped with each other. The CFP–MreB coil seemed to have a smaller pitch than the Tar–YFP coil. We therefore conclude that the Sec coil is distinct from the MreB coil.
Fig. 7. Subcellular localization of Tar–YFP with CFP–MreB within the same cells. Tar–YFP and CFP–MreB were expressed in strain HCB436. Images of Tar–YFP (left) and CFP–MreB (middle) were merged.
Discussion
In this study, the pursuit of the mechanisms underlying polar localization of the chemoreceptor Tar leads us to the finding that the general protein translocation machinery (i.e. the SecYEG complex) is organized into a helical array. Sequential observations of cells expressing Tar–GFP supported the indirect model for polar localization (Fig. 1A), in which Tar is inserted into lateral membrane regions and then migrates through a lipid bilayer to a cell pole. Further analyses of the Tar– and Taz1–GFP fusions identified helical arrays of fluorescence, which were deduced to reflect a similar array of the Sec machinery that inserts Tar into the membrane (Gebert et al., 1988). The helical array of the Sec machinery was confirmed by IFM observation of chromosome-encoded SecG and the observation of GFP–SecE and the translocation-defective mutant versions of MalE–GFP. Previous immunoelectron microscopic analyses detected 80% of the cytoplasmic membrane population of Tsr, the serine chemoreceptor, at cell poles (Maddock and Shapiro, 1993). In light of our finding, a minor population of Tsr detected at lateral cytoplasmic membrane regions could represent nascent Tsr inserted via the Sec machinery into the cytoplasmic membrane.
It has recently been reported that the Sec machinery is evenly distributed throughout the cytoplasmic membrane in E. coli (Brandon et al., 2003; Espeli et al., 2003). However, this could be due to overexpression of Sec proteins because GFP–SecE was evenly distributed throughout the cytoplasmic membrane when mildly overexpressed (Fig. 4D and E). To visualize the localization of the Sec machinery without violating wild-type cellular levels and stoichiometry of the components, we took advantage of the mutant versions (M18R and M19R) of MalE–GFP, which are thought to be stuck in the Sec machinery (Bassford and Beckwith, 1979; Bedouelle et al., 1980). Wild-type MalE–GFP is transported through the Sec machinery but is not fluorescent probably because the GFP part cannot be folded properly in the periplasmic space (Feilmeier et al., 2000). In contrast, the mutant versions of MalE–GFP were fluorescent, suggesting that these proteins reside in the cytoplasm probably by associating with the Sec machinery. Fluorescence patterns of the M18R (not shown) and M19R versions of MalE–GFP further supported the notion that the Sec machinery is organized into a helical array. We failed to detect significant effects of the mutant MalE–GFP protein on the processing of other exported proteins and cell growth. Considering its fluorescence images, we gather that the mutant MalE–GFP protein does not occupy all of the translocases. Recently, GFP fusions to SecA, SecY and the precursor of a translocation substrate protein of a Gram-positive bacterium B. subtilis have been reported to localize with similar helical patterns (Campo et al., 2004), raising a possibility that such a cellular organization is a common feature of general protein translocases among rod-shape bacteria.
The size and the subunit stoichiometry of the Sec machinery have not been unambiguously determined. Electron microscopy with negative staining suggested that the Sec machinery may consist of dimeric SecA and four SecYEG complexes (Manting et al., 2000). In contrast, cryo-electron microscopy suggested that the translocation pore consists of dimeric SecYEG complex (Breyton et al., 2002). Based on our fluorescence observation, we roughly estimated the average length of the Sec coil to be ∼10 μm assuming that one coil is continuous from one pole to the other. Given the published value (10.5–12 nm) of the diameter of the SecYEG tetramer (Manting et al., 2000), the maximal number of SecYEG tetramers that can be packed into a helical path would then be 800–1000. Another line of evidence estimates the number of SecY, SecE or SecG monomers per cell to be ∼500 (Matsuyama et al., 1992) and therefore the number of SecYEG tetramers should be ∼125. Considering that negative staining does not reflect the actual size of a protein and that SecYEG tetramers might not be packed tightly within a helical path, our rough estimation (800–1000 tetramers per helix) may be in good agreement with the predicted size of a cellular pool (∼125 tetramers per cell).
How can the Sec machinery, a complex of integral membrane proteins, be organized into a helical array? The Sec machinery (i) might associate with some rigid helical support, such as cytoskeletal protein(s) or peptidoglycan (cell wall), or (ii) might itself assemble into a coil. It has been shown that an integral membrane protein subunit of topoisomerase IV (SetB) interacts with MreB and forms a helical array (Espeli et al., 2003). Although the Sec coil was shown to be distinct from the MreB coil in this study as well as in B. subtilis (Campo et al., 2004), it is still possible that unknown cytoskeletal protein(s) serve to anchor the Sec machinery to form a helical array. It has recently been demonstrated that morphology of cells lacking some penicillin-binding proteins, which are involved in cell wall elongation, is abnormal (de Pedro et al., 2003) and some penicillin-binding proteins localize with a spot-like pattern like MreB (den Blaauwen et al., 2003; Scheffers et al., 2004), raising a possibility that it might form a helical polymer and that E. coli might have cytoskeletal proteins other than the identified ones. Another candidate for a helical support may be peptidoglycan as it is newly synthesized as a helix in B. subtilis (Daniel and Errington, 2003). To our knowledge, there is no experimental evidence that the SecYEG complex forms oligomers higher than a tetramer. However, the native complex within the membrane environment might have a property distinct from detergent-solubilized preparations. Moreover, E. coli shows lateral heterogeneity of phospholipid distribution in the cytoplasmic membrane and such heterogeneity is involved in cell division (Fishov and Woldringh, 1999). Therefore, it is possible that such a certain kind of membrane lipids might help the Sec machinery to form a coil even without a rigid anchoring structure. In B. subtilis, a decrease in phosphatidylglycerol causes delocalization of SecA (Campo et al., 2004). It would be intriguing to see how GFP–SecE and MalE-M19R–GFP localize in round-shaped E. coli mutants (rodA or mreB) or mutants in membrane lipid synthesis.
Physiological significance of the helical array of the Sec machinery is unclear. The helical arrangement of the Sec machinery along the long axis of a cell might have some advantage in distributing translocated proteins. If SecYEG tetramers have to be anchored to some rigid support rather than randomly distributed and freely diffusing, then a helical array would be one of the best solutions to spread proteins efficiently throughout the cytoplasmic membranes as well as the periplasmic space and the outer membrane (Fig. 8). Proteins that localize to certain parts of the cell, such as a pole and a septum, might be actively transported or passively diffused away towards their destinations.
Fig. 8. Model for helically arranged Sec protein translocases within a cell. The Sec machinery is organized into a helical array and translocates proteins from the cytoplasm to the cytoplasmic membrane and the periplasmic space that migrate. Some integral membrane proteins are inserted into the cytoplasmic membrane via the Sec machinery and may be sorted after insertion to migrate towards their destined regions (e.g. polar or random distribution). Outer membrane proteins are omitted for clarity. IM, the cytoplasmic (inner) membrane; OM, the outer membrane.
It is not clear whether a chemoreceptor is actively transported within a lipid bilayer to a cell pole or passively diffused and somehow trapped when it reaches a pole. In either case, the fact that both the Tar- and Taz1–GFP fusions colocalized transiently with the Sec coil indicates that membrane proteins with different destinations must be ‘sorted’ after insertion by the Sec machinery. Such sorting event(s) remain to be characterized but might provide a clue to understand the mechanisms of membrane protein localizations as well as the functions of the Sec machinery.
Experimental procedures
Bacterial strains and plasmids
All strains used in this study are derivative of E. coli K-12. Strain RP437 is wild-type for chemotaxis (Parkinson and Houts, 1982). Strains HCB436 (Wolfe and Berg, 1989) and HCB437 (Wolfe et al., 1987) lack all of the four chemoreceptors. In addition, the former lacks the methylesterase CheB and the methyltransferase CheR, and the latter lacks all of the cytoplasmic Che proteins, including the histidine kinase CheA and the adaptor CheW.
The vector plasmid pBAD24 (Apr) (Guzman et al., 1995) carries the araBAD promoter, the araC gene, which encodes the positive and negative regulator of the araBAD promoter. Plasmids pEGFP, pECFP and pEYFP, which encode the enhanced green, cyan and yellow fluorescent protein, respectively, were purchased from Clontech.
Construction of plasmids encoding fluorescent fusion proteins
For construction of plasmids encoding Tar–YFP under the control of the nahG promoter, the AvaI-BsrGI fragment of pEYFP was subcloned between the AvaI and BsrGI sites of pTrc-Tar–GFP (Homma et al., 2004). A BamHI site was introduced downstream of the tar–gfp/yfp gene by polymerase chain reaction (PCR) and the EcoT22I-BamHI fragment was subcloned into pLC113 (Cmr) (Ames et al., 2002), which is compatible with a pBR322-derivative plasmid, to yield pLC113-Tar–GFP/YFP.
For construction of pBAD24-Tar-S36C&A118C–GFP, the AatII-NdeI fragment of pBAD24-Tar-S36C&A118C (M. Homma and I. Kawagishi, unpublished) was subcloned into pBAD24-Tar–GFP (D. Shiomi and I. Kawagishi, unpublished). The NdeI-HindIII fragment of pBAD24-EnvZ–GFP (D. Shiomi and I. Kawagishi, unpublished) was subcloned into pBAD24-Tar-S36C&A118C–GFP to yield pBAD24-Taz1-S36C&A118C–GFP.
For construction of pBAD24–GFP–SecE or GFP/CFP–MreB, the secE or mreB gene of E. coli was amplified by PCR using chromosomal DNA of strain RP437 as a template. The forward and reverse primers used were designed to introduce unique BsrGI and EcoRI sites at the 5′ and 3′ ends of the secE or mreB coding region respectively. The BsrGI-EcoRI fragments were subcloned into pEGFP or pECFP. The NcoI-HindIII fragments from the plasmids were subcloned into pBAD24 to yield pBAD24–GFP–SecE or pBAD24–GFP/CFP–MreB.
A PstI site was introduced at the 5′ end of the ecfp coding region of pECFP by PCR. The PCR product was digested with PstI and EcoRI and the resulting fragment containing the ecfp gene was subcloned into pTrcHisB. The NcoI-XbaI fragment of the resulting plasmid was subcloned into pBAD24 to yield pBAD24-CFP(NX). For construction of MalE–CFP, the malE gene was amplified from pMalE (S. Banno and I. Kawagishi, unpublished) by PCR to introduce unique EcoRI and XhoI sites at the 5′ and 3′ ends of the malE gene respectively. The PCR product was digested with EcoRI and XhoI and the resulting fragment containing the ecfp gene was subcloned into pBAD24-CFP(NX) to yield pBAD24-MalE–CFP. The M18R or M19R mutation was introduced by two-step PCR to yield pBAD24-MalE-M18R/M19R–CFP.
Immunoblotting
Immunoblotting was carried out as described previously (Shiomi et al., 2002) using anti-GFP antibody (Molecular Probes).
Fluorescence microscopy
Observations of fluorescent proteins were carried out essentially as described previously (Shiomi et al., 2002). For time-course experiments, cells carrying a plasmid encoding Tar–GFP or Taz1–GFP were grown in TG medium (1% tryptone, 0.5% NaCl, 0.5% glycerol) at 30°C and after 3 h, 1 mM arabinose was added to each culture. Cells were harvested with appropriate intervals (see Results), spotted onto a glass slide coated with 0.5% agarose and observed under an inverted fluorescence microscope (Olympus IX71). The fluorescent images were processed by using the software Metamorph 5.0r4 (Molecular Devices) and Photoshop ver.7 (Adobe). For time-lapse experiments, after the addition of 1 mM arabinose, cells were further incubated at 30°C for 1 h, harvested, washed and resuspended in TG medium. Rifampicin (25 μg ml−1) was added to inhibit transcription and cells were further incubated for 10 min, harvested and observed with intervals of 1 s, 10 s or 1 min. For GFP–SecE or CFP–MreB, cells were grown at 30°C and after 3 h, 1 or 10 mM arabinose was added respectively. Cells were further incubated for 15 or 30 min, respectively, harvested and observed. For double labelling with MalE-M19R–CFP and Tar–YFP, cells were grown in TG medium supplemented with 0.5 μM sodium salicylate (to induce Tar–YFP) at 30°C for 3 h. Then, 0.1 mM arabinose was added to induce MalE-M19R–CFP. Cells were further incubated for 15 min, harvested and observed. For double labelling with CFP–MreB and Tar–YFP, cells were grown in TG medium supplemented with 0.5 μM sodium salicylate at 30°C for 3 h. Then, 10 mM arabinose was added to induce CFP–MreB. Cells were further incubated for 30 min, harvested and observed.
For optical sectioning, a fluorescence microscope with piezo drive (Zeiss Axiovert) was used to obtain a series of z-sections with a fixed spacing of 0.2 μm. Each stack of 20–30 sectioned fluorescence images was deconvolved by using CELLscan (Scanalytics). Three-dimensional images were reconstituted by using IPLab (Hamamatsu Photonics). Expressions of fluorescent proteins were carried out under the conditions described above.
Immnofluorescence microscopy
Immnofluorescence microscopy was carried out according to the method of Maddock and Shapiro (1993) with modifications. Cells were harvested and resuspended in MLM and fixed by adding an equal volume of 0.6% folmaldehyde in MLM and by incubating on ice for 2 h. Cells were then washed three times with MLM and resuspended in GTE [50 mM glucose, 20 mM Tris-HCl (pH 8.0), 10 mM EDTA (pH 7.0)] supplemented with 2 mg ml−1 lysozyme. An aliquot was spotted on a cover slip and allowed to dry. PBST (140 mM NaCl, 2 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, 0.05% Tween20, 2% BSA) were then spotted onto the sample. After 15 min of incubation, the sample was treated with the first antibody against SecG (provided by Dr H. Tokuda) for 1 h, washed twice, treated with the second antibody [goat anti-rabbit IgG labelled with Alexa Fluor 488 (Molecular Probes)], washed twice and observed under the fluorescence microscope.
Acknowledgments
We thank Dr Koutarou D. Kimura for technical advice on deconvolution, Dr Toshiharu Yakushi for critically reading the manuscript and invaluable discussion and Dr Hajime Tokuda for invaluable discussion and anti-SecG antibody. This work was supported in part by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (to D.S. and I.K.).
References
- Akerlund T, Bernander R, Nordstrom K. Cell division in Escherichia coli minB mutants. Mol Microbiol. 1992;6:2073–2083. doi: 10.1111/j.1365-2958.1992.tb01380.x. [DOI] [PubMed] [Google Scholar]
- Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage JP. Bacterial tactic responses. Adv Microb Physiol. 1999;41:229–289. doi: 10.1016/s0065-2911(08)60168-x. [DOI] [PubMed] [Google Scholar]
- Banno S, Shiomi D, Homma M, Kawagishi I. Targeting of the chemotaxis methylesterase/deamidase CheB to the polar receptor-kinase cluster in an Escherichia coli cell. Mol Microbiol. 2004;53:1051–1065. doi: 10.1111/j.1365-2958.2004.04176.x. [DOI] [PubMed] [Google Scholar]
- Barnakov AN, Barnakova LA, Hazelbauer GL. Location of the receptor-interaction site on CheB, the methylesterase response regulator of bacterial chemotaxis. J Biol Chem. 2001;276:32984–32989. doi: 10.1074/jbc.M105925200. [DOI] [PubMed] [Google Scholar]
- Barnakov AN, Barnakova LA, Hazelbauer GL. Allosteric enhancement of adaptational demethylation by a carboxyl-terminal sequence on chemoreceptors. J Biol Chem. 2002;277:42151–42156. doi: 10.1074/jbc.M206245200. [DOI] [PubMed] [Google Scholar]
- Bassford P, Beckwith J. Escherichia coli mutants accumulating the precursor of a secreted protein in the cytoplasm. Nature. 1979;277:538–541. doi: 10.1038/277538a0. [DOI] [PubMed] [Google Scholar]
- Bedouelle H, Bassford PJ, Jr, Fowler AV, Zabin I, Beckwith J, Hofnung M. Mutations which alter the function of the signal sequence of the maltose binding protein of Escherichia coli. Nature. 1980;285:78–81. doi: 10.1038/285078a0. [DOI] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Brandon LD, Goehring N, Janakiraman A, Yan AW, Wu T, Beckwith J, Goldberg MB. IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Mol Microbiol. 2003;50:45–60. doi: 10.1046/j.1365-2958.2003.03674.x. [DOI] [PubMed] [Google Scholar]
- Bray D, Levin MD, Morton-Firth CJ. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- Breyton C, Haase W, Rapoport TA, Kuhlbrandt W, Collinson I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature. 2002;418:662–665. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, et al. Subcellular sites for bacterial protein export. Mol Microbiol. 2004;53:1583–1599. doi: 10.1111/j.1365-2958.2004.04278.x. [DOI] [PubMed] [Google Scholar]
- Cantwell BJ, Draheim RR, Weart RB, Nguyen C, Stewart RC, Manson MD. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J Bacteriol. 2003;185:2354–2361. doi: 10.1128/JB.185.7.2354-2361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles M, Perez M, Kobil JH, Goldberg MB. Polar targeting of Shigella virulence factor IcsA in Enterobacteriacae and Vibrio. Proc Natl Acad Sci USA. 2001;98:9871–9876. doi: 10.1073/pnas.171310498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- Den Blaauwen T, Aarsman ME, Vischer NO, Nanninga N. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol Microbiol. 2003;47:539–547. doi: 10.1046/j.1365-2958.2003.03316.x. [DOI] [PubMed] [Google Scholar]
- Djordjevic S, Stock AM. Structural analysis of bacterial chemotaxis proteins: components of a dynamic signaling system. J Struct Biol. 1998;15:189–200. doi: 10.1006/jsbi.1998.4034. [DOI] [PubMed] [Google Scholar]
- Driessen AJ, Fekkes P, van der Wolk JP. The Sec system. Curr Opin Microbiol. 1998;1:216–222. doi: 10.1016/s1369-5274(98)80014-3. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Amos L, Lowe J. Bacterial ancestry of actin and tubulin. Curr Opin Microbiol. 2001;4:634–638. doi: 10.1016/s1369-5274(01)00262-4. [DOI] [PubMed] [Google Scholar]
- Espeli O, Nurse P, Levine C, Lee C, Marians KJ. SetB: an integral membrane protein that affects chromosome segregation in Escherichia coli. Mol Microbiol. 2003;50:495–509. doi: 10.1046/j.1365-2958.2003.03736.x. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Kim SH. Structure of a conserved receptor domain that regulates kinase activity: the cytoplasmic domain of bacterial taxis receptors. Curr Opin Struct Biol. 2000;10:462–469. doi: 10.1016/s0959-440x(00)00115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J Bacteriol. 2000;182:4068–4076. doi: 10.1128/jb.182.14.4068-4076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishov I, Woldringh CL. Visualization of membrane domains in Escherichia coli. Mol Microbiol. 1999;32:1166–1172. doi: 10.1046/j.1365-2958.1999.01425.x. [DOI] [PubMed] [Google Scholar]
- Gebert JF, Overhoff B, Manson MD, Boos W. The Tsr chemosensory transducer of Escherichia coli assembles into the cytoplasmic membrane via a SecA-dependent process. J Biol Chem. 1988;263:16652–16660. [PubMed] [Google Scholar]
- Gestwicki JE, Kiessling LL. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature. 2002;415:81–84. doi: 10.1038/415081a. [DOI] [PubMed] [Google Scholar]
- Goldberg MB, Barzu O, Parsot C, Sansonetti PJ. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J Bacteriol. 1993;175:2189–2196. doi: 10.1128/jb.175.8.2189-2196.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M, Shiomi D, Homma M, Kawagishi I. Attractant binding alters arrangement of chemoreceptor dimers within its cluster at a cell pole. Proc Natl Acad Sci USA. 2004;101:3462–3467. doi: 10.1073/pnas.0306660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Kim SH, Wang W, Kim KK. Dynamic and clustering model of bacterial chemotaxis receptors: structural basis for signaling and high sensitivity. Proc Natl Acad Sci USA. 2002;99:11611–11615. doi: 10.1073/pnas.132376499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levit MN, Stock JB. Receptor methylation controls the magnitude of stimulus-response coupling in bacterial chemotaxis. J Biol Chem. 2002;277:36760–36765. doi: 10.1074/jbc.M204325200. [DOI] [PubMed] [Google Scholar]
- Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- Lybarger SR, Maddock JR. Polarity in action: asymmetric protein localization in bacteria. J Bacteriol. 2001;183:3261–3267. doi: 10.1128/JB.183.11.3261-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Manting EH, van Der Does C, Remigy H, Engel A, Driessen AJ. SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J. 2000;19:852–861. doi: 10.1093/emboj/19.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. Bacterial division: the fellowship of the ring. Curr Biol. 2003;13:16–18. doi: 10.1016/s0960-9822(02)01381-7. [DOI] [PubMed] [Google Scholar]
- Matsuyama S, Fujita Y, Sagara K, Mizushima S. Overproduction, purification and characterization of SecD and SecF, integral membrane components of the protein translocation machinery of Escherichia coli. Biochim Biophys Acta. 1992;1122:77–84. doi: 10.1016/0167-4838(92)90130-6. [DOI] [PubMed] [Google Scholar]
- Mori H, Ito K. The Sec protein-translocation pathway. Trends Microbiol. 2001;10:494–500. doi: 10.1016/s0966-842x(01)02174-6. [DOI] [PubMed] [Google Scholar]
- Nanninga N. Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis function. J Bacteriol. 1982;171:6271–6278. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Young KD, Holtje JV, Schwarz H. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J Bacteriol. 2003;185:1147–1152. doi: 10.1128/JB.185.4.1147-1152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch J, Caparon M. A microdomain for protein secretion in Gram-positive bacteria. Science. 2004;304:1513–1515. doi: 10.1126/science.1097404. [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Pan Q, Losick RM. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci USA. 2002;99:8701–8706. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers DJ, Jones LJ, Errington J. Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol Microbiol. 2004;51:749–764. doi: 10.1046/j.1365-2958.2003.03854.x. [DOI] [PubMed] [Google Scholar]
- Shapiro L, McAdams HM, Losick R. Generating and exploring polarity in bacteria. Science. 2002;298:1942–1946. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- Shih YL, Le T, Rothfield L. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA. 2003;100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu TS, Le Novére N, Levin MD, Beavil AJ, Sutton BJ, Bray D. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- Shiomi D, Zhulin IB, Homma M, Kawagishi I. Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase. J Biol Chem. 2002;277:42325–42333. doi: 10.1074/jbc.M202001200. [DOI] [PubMed] [Google Scholar]
- Shiomi D, Banno S, Homma M, Kawagishi I. Stabilization of polar localization of a chemoreceptor via its covalent modifications and its communication with a different chemoreceptor. J Bacteriol. 2005;187:7647–7654. doi: 10.1128/JB.187.22.7647-7654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skidmore JM, Ellefson DD, McNamara BP, Couto MM, Wolfe AJ, Maddock JR. Polar clustering of the chemoreceptor complex in Escherichia coli occurs in the absence of complete CheA function. J Bacteriol. 2000;182:967–973. doi: 10.1128/jb.182.4.967-973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Portnoy DA, Theriot JA. Asymmetric distribution of the Listeria monocytogenes ActA protein is required and sufficient to direct actin-based motility. Mol Microbiol. 1995;17:945–951. doi: 10.1111/j.1365-2958.1995.mmi_17050945.x. [DOI] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- Stock JB, Surette MG. Chemotaxis. In: Neidhardt FC, Curtiss R III, Ingram JJ, Lin ECC, Low KB, Magasanik B, et al., editors. Escherichia coli and Salmonella Typhimurium: Cellular and Molecular Biology. 2nd edn. Washington, DC: American Society for Microbiology; 1996. pp. 1103–1129. [Google Scholar]
- Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci USA. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Berg HC. Migration of bacteria in semisolid agar. Proc Natl Acad Sci USA. 1989;86:6973–6977. doi: 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Conley MP, Kramer TJ, Berg HC. Reconstitution of signaling in bacterial chemotaxis. J Bacteriol. 1987;169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Park H, Inouye M. Ligand binding induces an asymmetrical transmembrane signal through a receptor dimer. J Mol Biol. 1993;232:493–498. doi: 10.1006/jmbi.1993.1405. [DOI] [PubMed] [Google Scholar]