Abstract
Functional redundancies, generated by gene duplications, are highly widespread throughout all known genomes. One consequence of these redundancies is a tremendous increase to the robustness of organisms to mutations and other stresses. Yet, this very robustness also renders redundancy evolutionarily unstable, and it is, thus, predicted to have only a transient lifetime. In contrast, numerous reports describe instances of functional overlaps that have been conserved throughout extended evolutionary periods. More interestingly, many such backed-up genes were shown to be transcriptionally responsive to the intactness of their redundant partner and are up-regulated if the latter is mutationally inactivated. By manual inspection of the literature, we have compiled a list of such “responsive backup circuits” in a diverse list of species. Reviewing these responsive backup circuits, we extract recurring principles characterizing their regulation. We then apply modeling approaches to explore further their dynamic properties. Our results demonstrate that responsive backup circuits may function as ideal devices for filtering nongenetic noise from transcriptional pathways and obtaining regulatory precision. We thus challenge the view that such redundancies are simply leftovers of ancient duplications and suggest they are an additional component to the sophisticated machinery of cellular regulation. In this respect, we suggest that compensation for gene loss is merely a side effect of sophisticated design principles using functional redundancy.
Keywords: evolution, gene duplications, modeling, systems biology, noise
Duplicate genes and paralogous gene families long have been perceived as genomic sources of genetics robustness (1–5). The assumption is that a functional overlap of these genes acts to compensate against mutations. Yet, this very fact also renders redundancy evolutionarily instable (5, 6), and functional overlaps, typically, are rapidly lost because of divergence (7).
Nevertheless, numerous examples of paralogs retaining their functional overlap for extended evolutionary periods (for examples, see refs. 6 and 8–12) suggest that, at least for a fraction of gene pairs, redundancies are conserved throughout evolution despite their predicted instability. One such example is the pair of O-acyl-transferases isozymes, redundantly catalyzing the conjugation of sterols to fatty acids, for which functional overlap has been conserved all of the way from yeast (Are1 and Are2) to mammals (ACAT1 and ACAT2) (13). In fact, although retention of redundancy is much less frequent than its loss, its widespread existence is nontrivial and cannot (6) be dismissed as leftovers of recent duplication events. In one study, modeling of evolutionary dynamics suggested that this conservation may be the result of asymmetries in the functional efficiencies or mutation rates between the redundant pair members (6). Alternatively, redundancies were suggested to be selected for their contribution to robustness and evolvability (3, 14). In this work, we wish to adapt the view that, at least in some pathways, redundancies are selected for based on some evolutionary advantage that they confer to the wild-type organism. In particular, we suggest the existence of regulatory designs that exploit redundancy to achieve functionalities such as control of noise in gene expression or extreme flexibility in gene regulation. In this respect, we suggest that compensation for gene loss is merely a side effect of sophisticated design principles using functional redundancy.
Clues for regulatory designs controlling redundancy were obtained first in a recent study (15) that explored the dispensability of gene duplicates with various degrees of coregulation. The underlying assumption of this study was that in order for one duplicate copy to compensate against the loss of its partner, both duplicates must not only perform the same function, but do so at the same place and time. In other words, coregulation of duplicates was perceived as a prerequisite for functional compensation (4, 15). In reality, however, similarly expressed paralogs were found to almost never back each other up, as evident from their high essentiality. In fact, tightly coregulated gene duplicates were found more essential for viability than singleton genes. Functional redundancy and compensation were found to be most prevalent among gene duplicates that are regulated differently from one another (15). Further insight was provided by the observation that some differentially regulated duplicates maintain the ability to become coregulated under certain environmental conditions. Such conditional coregulation or partial coregulation of these genes within the transcriptional network was shown to be very strongly negatively correlated with the severity of the knockout phenotypes of these genes. Thus, the paradigm that has emerged is that genes that are functionally redundant are not often independently controlled but rather they are regulated by a system that both monitors and responds to their intactness. In this study, we will survey examples of such responsive backup circuits (RBCs) and draw a general outline for their function, design, and evolution.
Analysis of Literature
RBCs.
Two lines of evidence could indicate a function’s direct benefit from existing redundancy: first is the evolutionary conservation of the functional overlap, and second is a nontrivial regulatory design that utilizes it. Many well known examples meet both these criteria, one of which is that of the 1,3-β-glucane synthase catalytic subunit in yeast that is encoded by the two alternative, functionally redundant and synthetically lethal genes Fks1 and Fks2 (16). The evolutionarily selectable advantage of this redundancy can be inferred from the fact that both isozymes are found as duplicates in all 12 sequenced yeast species except for the Yarrowia lipolytica (17). Furthermore, in Saccharomyces cerevisiae, these two genes obey a particular regulation whereby Fks2 transcriptionally responds to the intactness of Fks1 and is up-regulated upon Fks1 mutational inactivation (18). Numerous other examples describing such RBCs exist and cover a wide variety of organisms ranging from bacteria to mammals (see Table 1, which is published as supporting information on the PNAS web site, for more examples). In fact, the observed prevalence of this particular regulatory design for control of genetic redundancy raises questions as to the specific selectable functions that it performs. In what follows, we will highlight common denominators of known RBCs and use these to suggest principles that govern the utilization of redundancy and its evolution.
Conditional Coregulation and the Maintenance of Metabolic Fluxes.
A key requirement of the metabolic regulation is the maintenance of metabolite fluxes, despite the sometimes extreme changes in the external conditions and nutrient availability. On evolutionary time scales, adaptation to extreme environmental changes is sometimes achieved by preservation of gene duplicates (19), as illustrated by the numerous observations of adaptive gene amplifications in response to antibiotics, anticancer drug treatments, nutrient limitations, and more (see ref. 19 for examples and references). Also, it was suggested from a genomewide analysis that enzymes corresponding to reactions with higher metabolic fluxes are more likely to have duplicate partners (20). For a single cell, the ability to quickly and efficiently respond to fluctuating environments is crucial and offers an obvious evolutionary advantage. One avenue through which functional redundancy is used to facilitate this ability is by exploiting the differential efficiencies generated by divergence. For example, in yeast, the Hxt gene family encodes a redundant set of membrane hexose transporters with varying affinities toward glucose and, consequently, different transport efficiencies (21). This variation together with glucose-tuned regulation enables the control of glucose flux through the expression of high-affinity transporters when glucose is limited and of low-affinity transporters when glucose is abundant (21), thus allowing the cell to adapt to different external glucose availability. Other examples that fall under this principle include the transport of iron, copper, manganese, zinc, and other metals in yeast (22). Interestingly, some of these transport systems are further regulated by RBCs (23, 24), which, as we will show later, may serve the beneficial role of providing robustness of the flux control to internal noise arising from genetic variation.
Within the metabolic network, fluxes are governed and regulated by the concentration of active enzymes catalyzing the different reactions. Although the detailed contribution of functional redundancy to this regulation is not fully established, such a contribution is highly expected given the large amount of isozymes and other redundancies that exist within these networks (Fig. 4, which is published as supporting information on the PNAS web site). Fig. 4 shows that individual isozymes are less essential and produce less deleterious effects upon deletion than enzymes existing in single copy. An interesting twist to this account comes from the fact that isozymes, although redundant and consequently dispensable, obey different regulatory programs and are transcribed at different times in response to environmental pressures (15, 25). A recent finding supplying the compromise between these two seemingly opposing observations shows that many differentially regulated genes can be induced for coexpression given that particular environmental stress stimuli were applied (15). More so, gene pairs that maintain this capacity for conditional coexpression were shown to be the most likely candidates for compensating against deletion mutations (15). This conditional coregulation [referred to as PCoR (partial coregulation) in ref. 15)] may provide essential clues for the function of these redundancies in the regulation of metabolic fluxes. The model that emerges is that although many isozymes are specialized for different environmental regimes, alarm signals induced by particular stress stimuli may call for their synergistic coexpression. Here, RBCs provide functional specialization together with extreme flexibility in gene control that could be activated when sufficient stress has been applied. For example, in yeast, glucose serves as a regulatory input for alternating between aerobic and anaerobic growth. Its presence is detected by two separate and independent signaling pathways, one probing intracellular glucose concentrations and the other probing extracellular concentrations (23). This differential sensing enables some genes to be separately regulated by either intracellular or extracellular glucose. One consequence of this differential sensing shows effect in the responsive backup circuit composed of Hxt1 and Htx2. Here, feedback is made possible by having Hxt2 controlled by two opposing signals. One is its induction by extracellular glucose and the second is its repression by intracellular glucose (Fig. 1A; ref. 23). The consequence of this distinction is that although high glucose concentrations result in repression of Hxt2 expression, its induction could be triggered either by low environmental sugar, or alternatively, by mutations in genes responsible for glucose influx (23). Other similar examples include the isocitrate dehydrogenases idp2 and Idh, where the glucose repression of idp2 is reversed in the Δihd mutant (26), and for the pair Acs1 and Acs2, where Acs2’s expression is induced in the ΔAcs1 mutant (27). In all these cases, the common denominator is that one of the two duplicates is under repression in wild type and that that repression is relieved upon its partner’s mutation.
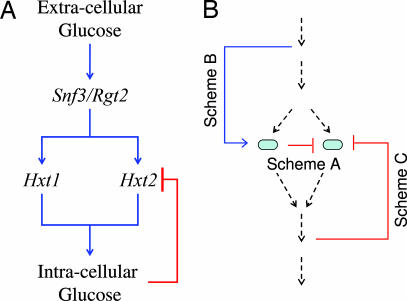
Fig. 1.
Specific (A) and general (B) responsive back up circuitries. (A) The Hxt1/Hxt2 responsive backup circuit. Extracellular glucose concentration is sensed by two membrane receptors on the outer yeast membrane, Rgt2 and Snf3. These receptors, once activated by glucose, initiate a signal cascade that induces the transcription of the Hxt gene family of hexose transporters encoding membrane channels for glucose intake. The flux of incoming glucose generates an increase in intracellular glucose concentration, that, in turn, represses the transcription of Hxt2. (B) Three possibilities for feedback in RBCs. For one duplicate gene to sense and respond to its partners’ intactness, feedback mechanisms must be at play. In this diagram, duplicates are represented as ovals that lie embedded within a reaction pathway illustrated by the consecutive arrows. Lines A, B and C represent the three feedback possibilities, namely, simple negative regulation (A), substrate induction (B), and end-product regulation (C).
Redundancies of Developmental Regulators.
The extent to which genomic functional redundancies have influenced the way we think about biology can be appreciated simply by inspecting the vast number of times the word “redundancy” is specifically referred to in the biomedical literature (Fig. 5, which is published as supporting information on the PNAS web site). Particularly interesting is the abundance with which it is addressed in studies of developmental biology (Fig. 5). In fact, it is here that concepts such as “genetic buffering” and “canalization” (28) first had been suggested. Furthermore, the robustness of the developmental phenotypes such as body morphologies and patterning have been repeatedly demonstrated (14). So the question is, are these redundancies simply leftovers of ancient duplications, or are they an additional component to the sophisticated machinery of cellular regulation?
In criticism, one may argue that many of the reported redundancies do not actually represent functionally equivalent genes but rather reflect only partial functional overlap. In fact, knockout phenotypes have been described for a number of developmental genes that have redundant partners (29–31). For these reasons, it has been suggested to define redundancy as a measure of correlated, rather than degenerate, gene functions (32). Although these facts may suggest that redundancies have not evolved for the sake of buffering mutations, it has, in our opinion, little relevance to the question of whether they serve a functional role. The interesting question is, then, can such a functional role for the duplicated state be inferred from the way the two genes are regulated?
For most cases of developmental redundancies, redundant partners are either temporally or spatially distinct in their expression patterns (Table 1); however, some level of expression overlap is usually observed. Cross-regulation of redundancies has been tested for in only a relatively small number of cases, yet from those, a few persuasive recurring themes do emerge. One of the better known cases of cross-regulated developmental regulators is that of the four master regulators of vertebrate skeletal muscle development: MyoD, Myf-5, myogenin, and MRF4, collectively known as the MRF gene family (33). These four basic helix–loop–helix transcription factors specify and execute the process through which naïve mesoderm cells differentiate to form distinct skeletal muscles (for review, see ref. 34) and are activated sequentially during myogenesis. The myogenic pathway consists of two separate phases. In the first phase, MyoD and Myf-5 specify the myogenic progenitors in the somites into myoblasts, which are cells that are committed to become muscle fibers. In the second phase, myoblasts develop into myofibers, a process initiated by myogenin and MRF4 (35).
Sequence similarity between the myogenic transcription factors suggests that they have evolved through multiple gene duplication events early in the evolution of vertebrates, approximately with the appearance of fish (36–40). Interestingly, despite their long evolutionary separation, these regulators largely have conserved their functional redundancy. In fact, experiments on mice where MyoD was completely inactivated resulted in viable and fertile mice that exhibited phenotypically normal skeletal muscles (41). In strong contrast, mice lacking both MyoD and Myf-5 lack skeletal muscle altogether and die soon after birth (42).
From the perspective of this review, myogenesis is a particularly interesting process because it harbors two responsive backup circuits. The first is manifested by the up-regulation of Myf-5 in response to mutations in MyoD (41). The second is described by the induction of myogenin in response to mutations in MRF4 (35). An additional interesting feature of the MRF RBCs is that the up-regulatory response induced by a heterozygote mutation is approximately half that of the homozygote one, we term this behavior “dosage-dependent linear response.” In particular, for MyoD and Myf-5, mutations in one of the two MyoD alleles results in an 1.8-fold up-regulatory response of Myf-5, whereas disruption of both alleles results in a 3.5-fold response (41). This type of linearity may hold clues as to the both function and regulation of these genetic circuits. One attractive possibility that may be suggested by this linearity is that the process carried out by these redundant regulators benefits from constancy of the sum of their protein concentrations. In other words, although the concentration of the MyoD protein may fluctuate because of noise in gene expression or false induction, the sum of MyoD plus Myf-5 may have evolved to remain constant (see Supporting Text, which is published as supporting information on the PNAS web site).
An additional example illustrating dosage-dependent linear response constitutes the Pax1 and Pax9 regulators of sclerotome development. Here, functional redundancy has been established at the phenotypic level from mutant mouse experiments, showing that Pax1 can fully rescue Pax9 mutants and, conversely, Pax9 can offset the Pax1-null phenotype to a substantial degree (43). In line with what seems to be the general case for numerous examples of developmental redundancies (see Table 1; see also Table 2, which is published as supporting information on the PNAS web site), Pax1 and Pax9 have partially overlapping expression domains during early development, particularly in the sclerotomes (43). However, this overlap decreases in the later stages of development. The responsive circuitry of these regulators was established by using the lacZ/gal system to show an up-regulation and spatial expansion of Pax9 expression in the sclerotomes of the Pax1 mutant (43). Thus, Pax9 expression in the Pax1 mutants was observed in cells that in wild type exhibit only Pax1 expression. Dosage dependency was observed by comparing phenotypes of combinations of wild-type, heterozygous, and homozygous mutants of Pax1 and Pax9 (43). It is worth noting that functional redundancy also was suggested for other members of the Pax gene family, in particular for the two pairs Pax2/Pax5 (11) and Pax3/Pax7 (12). All nine family members of the Pax transcription factors carry roles in the genetic control of mammalian organogenesis (for review, see ref. 44).
Other examples of responsive circuits of redundant developmental regulators include as follows: the closely related homeobox gene pair Gsh1 and Gsh2, for which mutational inactivation of Gsh1 resulted in a pronounced expansion of Gsh2 expression in the cerebral cortex and olfactory bulb of mice with an apparently normal phenotype (45); the vertebrate Distal-less-related regulators dlx3 and dlx7, where morpholino-induced inactivation of dlx3 resulted in a strong induction of dlx7 mRNA expression in zebrafish embryos (46); the two functionally overlapping E3 ligases, Smurf-1 and Smurf-2 (47) for which knockouts of Smurf-1 were shown to result in an up-regulatory response of Smurf-2 (48–50); the midkine and pleiotrophin cytokines for which functional redundancy was observed and a strong up-regulatory response of pleiotrophin was shown to result from a double knockout of the midkine gene (51); and other examples listed in Tables 1 and 2.
The abundance of redundancies occurring in genes related to developmental processes, and their functional role as master regulators (Fig. 5) may be taken to suggest their utilization in either the flexibility or robustness of regulatory control. In fact, redundancies among high-level regulators often have been reported outside the context of developmental pathways (Fig. 5 and Table 1). Although for most examples the regulation that they confer on one another was not assessed, some have been specifically identified as displaying negative cross-regulatory inhibitions (see Table 1). One such example existing in Escherichia coli is that of the pair stpA/HN-S, which regulate genome-scale transcriptional response to DNA damage (52). This pair of regulators displays an additional complexity, where its regulation is induced by pairwise associations to either form homodimers composed of either of the pair members or heterodimers containing both (52). Nevertheless, mutational inactivation of HN-S induced an up-regulatory response of its partner, stpA, with only a marginal effect on phenotype (53). A more recent example indicates that the multidrug resistance phenomenon in S. cerevisiae is also regulated by an RBC encoding for the up-regulation of the transcription factor YRR1 in response to the deletion of its partner, YRM1 (54).
Results
Recurring Regulatory Patterns.
Two architectures of cross-regulated redundancies may exist. According to the first, inactivation of each of the redundant genes from a given pair would result in the induction of the other (Fig. 6A, which is published as supporting information on the PNAS web site), and according to the second, only one of the pair members is responsive (Figs. 2 and 6 B and C). We therefore suggest the terminology bidirectional and unidirectional RBCs. This distinction is important because from the current literature review, all but two of the examples seem to fall into the unidirectional category. We further suggest, for unidirectional RBCs, the distinction between the responsive gene and the controller gene.
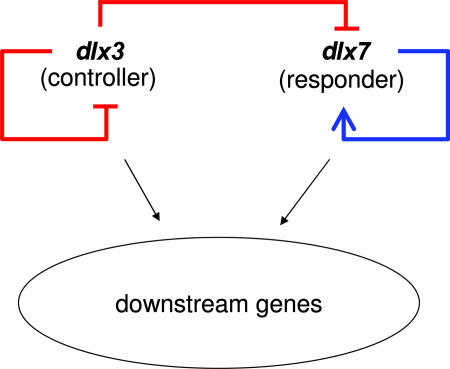
Fig. 2.
The regulatory wiring for the two distalless developmental regulators dlx3 and dlx7 as deduced from morpholino antisense translation inhibitions (46).
The above is but one of several asymmetries and regulatory patterns that systematically recur throughout the literature. An additional example is the classification of redundant pair members into a ubiquitously expressed gene partner and a sporadically expressed one (see Table 1). One of the most profound and insightful of these recurring regulatory themes is that, although both genes are capable of some functional compensation, disruption of the responder produces a significantly less deleterious phenotype than disruption of the controller (Table 1). An insightful example illustrating this theme entails the pair of genes Fks1 and Fks2 redundantly encoding the catalytic subunit of the yeast 1,3-β-glucan synthease (55). This enzyme is responsible for the generation of cross-links within the 1–3-β-glucan matrix comprising the major structural component of the yeast cell wall. Being such, this process requires very tight regulation with cell wall degradation and cell cycle to enable budding and isotropic cell growth. This fact also is suggested from the numerous associations of Fks1 within the genetic interaction network, illustrating its linkage to processes such as cell cycle control, environmental stress responses, and mating (56). The surprising aspect of this story is that despite this high connectivity of Fks1, Fks2 is only sparsely connected (56). More so, although deletion of Fks1 induces an up-regulatory response of Fks2 with mild phenotypic effects, deletion of Fks2 induces no regulatory response of Fks1 but also no detectable effect on the phenotype (55). This result may seem counterintuitive because it is Fks2 that is up-regulated to rescue against deletion of Fks1 and not vice versa, yet Fks2 is the more dispensable gene within this pair. (see Table 1 for more examples). A simple potential interpretation may suggest that although the controller is the key player performing some essential biological role, the responder is merely a less efficient substitute. Yet, accepting the notion that redundancy could not have evolved for the sake of buffering mutations, this interpretation still is severely lacking.
A different, and more biologically reasonable, hypothesis accounting these asymmetries is that one of the functions of the responder is to buffer dosage fluctuations of the controller. This buffering capacity requires a functional overlap that also manifests itself in compensations against the more rare event of gene loss. Other models accounting for this assymetry are discussed further in this work, but our main point of argument is that this complex regulation of functionally redundant, yet evolutionarily conserved genes, strongly indicates utilization of redundancy.
Regulatory Designs.
What regulatory design could account for a gene sensing and responding to its redundant partner’s intactness? From the most general perspective, there are three possible regulatory schemes that could answer this question. Scheme A (Fig. 1B) entails a direct negative regulation of a gene by its functionally redundant partner. Scheme B (Fig. 1B) uses the substrate abundance as a proxy for its partner’s activity. In other words, overaccumulation of substrate, potentially caused by reduced or abolished efficiency of one of the RBC pair members, signals for overproduction of the second member. Scheme C employs end-product inhibition. Assuming that an end-product may inhibit both redundant partners, the lack of function of one of the partners would result in the absence of the product and, hence, relief of repression from the second partner. Conceptually, schemes B and C are symmetric.
One instance of an RBC that relies on a direct regulatory interaction between redundant partners without involving either substrate or end-product regulation constitutes the two vertebrate Distal-less-related regulators, dlx3 and dlx7 (Fig. 2). These paralogous transcriptional regulators are both expressed in embryonic development and are involved in the development of auditory and olfactory placodes (57–59). By injecting anti-dlx3 and anti-dlx7 morpholino oligonucleotides (MO) in zebrafish, it was showed that although the simultaneous inhibition of both genes (dlx3+7-MO) resulted in embryos having severe defects in the auditory and olfactory placodes, dlx7 loss-of-function embryos appeared phenotypically normal, and dlx3 MO embryos exhibited only smaller auditory placodes and inner ear structures than normal (or wild-type) embryos (46). An increase in dlx7 mRNA was observed in the dlx3 MO embryos (46).
The regulatory relationships between the dlx3 and dlx7 were tested by measuring the mRNA content of the different MO-treated embryos (46) and are summarized in a network diagram (Fig. 2) featuring both cross and auto regulation. Thus, the lesson is that, for this case, redundancy is embedded within a more complex interaction network that includes a unidirectional responsive circuit in which the controller (dlx3) also represses its own transcription, whereas the responder (dlx7) is a positive autoregulator.
Another interesting example for which the regulatory pathway leading to induction was well characterized constitutes the unidirectional RBC of Fks1 and Fks2 in yeast (discussed above in a different context). Here, the responder (Fks2), in addition to being activated in the fks1Δ mutant, also is induced by heat shock, cell wall damage, pheromone, and Ca+2 (18). The intricate design of this circuitry is realized by the fact that there are two different, alternative, signaling pathways that operate synergistically to control Fks2 expression. Whereas response to Fks1 deletion is activated through a calcineuerin/Ca2+-dependent pathway, response to heat shock and cell wall damage in induced by both the former and the Roh1-dependent cell integrity pathway (16). Even more interestingly, it was found that these two pathways induce different and complementary dynamics of the Fks2 response (16). Specifically, whereas the calcineuerin-dependent pathway induces a rapid but transient response, the Roh1-dependent pathway induces a delayed response that is sustained for longer time scales.
An end product-activated feedback mechanism is demonstrated by the hexose transporters Hxt1 and Hxt2 in yeast (Fig. 1A), where the expression of both genes is repressed by the level of intracellular glucose. Thus, once the flux of glucose from the environment to the yeast’s cytoplasm decreases, an additional glucose pump is induced for expression.
Functional Consequence of Cross-Regulated Redundancies.
In principle, the up-regulation of the responsive gene in RBCs could be encoded for in a number of different mechanisms. Several such mechanisms are schematically depicted in Table 3, second column, which is published as supporting information on the PNAS web site. However, these different regulations are not equivalent and result in differences in both dynamics and steady states of the response. Similar to refs. 60 and 61, we quantified the different RBCs shown in Table 3 for their steady-state characteristics (see Supporting Text for detailed calculations). Our basic hypothesis was that if there are biological functions that exploit redundancies between RBC pair members, such functions, to a first approximation, would be proportional to either the sum (independent functions) or the product (synergistic functions) of the concentrations of the two redundant proteins (see Fig. 7, which is published as supporting information on the PNAS web site). Examples of the first include reactions that are catalyzed by two independently functioning isozymes. In such cases, the total rate of product production catalyzed by the pair of isozymes would be equal to the production rate contributed by the first isozyme plus that of the second. Examples of the second, i.e., synergistic exploitation of redundancies, could be understood by reactions that exploit cooperativity between RBC pair members. Examples are biological functions that are carried out more efficiently by heterodimers of the partially redundant proteins (see HN-S and StpA for an illustrative example).
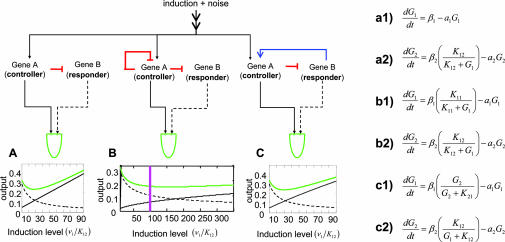
Table 3 shows the steady-state solution for differential equations that describe the dynamics of RBCs with three different circuitries. We examined the capacity of different RBCs (“simple repression,” “dampened controlled,” and “cycled feedback”; see Table 3 for definitions) to sustain constant steady-state concentrations of G1 + G2 or G1 x G2 with respect to variations in the concentration of G1. In other words, we computed the steady-state susceptibility (62), Hss, of both synergistic and additive functions of RBCs to variations in the controller G1 (see Fig. 8, which is published as supporting information on the PNAS web site). We then asked whether a RBC may serve to filter the downstream processes from variation and fluctuations arising from nongenetic noise (63), genetic variability of G1, or genetic variability effecting G1’s regulation. Following our expectations, results illustrated in Fig. 3 and shown in Table 3 and Fig. 8 indicate that fluctuations of the controller are counteracted reciprocally by the responder for both independent and synergistic RBC functions. It is evident further that whereas the efficiency of RBC noise reduction for independent functions is parameter dependent, synergistic functions employ an almost absolute buffering capacity regardless of parameter choice. For independent RBC functions, Fig. 3 and Table 3 show that the efficiency of noise reduction largely depends not only on the model parameters but also on the regulatory circuitries defining the different RBCs. Notably, the most efficient RBC for buffering independent functions is the “dampened inducer” (defined in Table 3), instantiated by dlx3/dlx7. This increased efficiency results from two fundamental advantages of this RBC: First, the strength of the restoration response, but not the inductive response, can be fine tuned by the level of induction of the responsive gene. Second, it has the additional advantage of the negative autoregulation of the controller.
Fig. 3.
Signal robustness provided by RBCs. Three general responsive backup circuitries are examined as follows: simple repression, modeled by equations a1 and a2 (A); dampened controller, modeled by equations b1 and b2 (B); and cycled feedback modeled by equations c1 and c2 (C). β and α represent the rates of protein synthesis and protein degradation, respectively; Kij is a constant quantifying the regulatory control i has over j. The RBCs are examined for their efficiency in filtering variations of the regulatory input, v1, of the controller gene, G1. For each RBC, a diagram is shown describing the regulatory interactions between the responsive and controlling gene. The plots show the dependency of the controller (solid black), the responder (broken black), and their sum, f2 = G1 + G2, (green) on G1’s induction level, v1. Induction level is shown in units of K12, such that an induction level of 1 corresponds to 50% saturation of the K12 promoter element. The purple vertical line in B corresponds to v1/K12 = 100 to help with the comparisons. For a more rigorous treatment, see Supporting Text and Fig. 8.
Discussion
One of the central open questions that still must be addressed is just how common are RBCs in biology and how frequently do they appear in the different genomes. Also, we can ask whether this frequency, be it low or high, can be faithfully estimated from the amount of times RBCs are reported in the literature. In other words, do RBCs represent a “genomewide” phenomenon or a collection of rare incidents? It is unfortunate that at this point no conclusive answer can be provided to that question, mainly because of the very limited number of studies that have specifically probed for cross-regulation among redundant protein pairs. Also, studies aimed at a genomewide establishment of “synthetic lethal” gene pairs (64) are far from sufficient because these interactions only rarely result from overlaps in function (65).
Furthermore, we argue that although the question of prevalence of RBCs is important, one cannot judge their biological significance solely based on that criteria. This fact is emphasized, for example, by the pair of genes utrophin and dystrophin associated with structural components of muscle fiber. This pair of genes constitutes an RBC in that utrophin was found to be up-regulated in the absence of its homolog dystrophin (66). This pair has attracted particular attention because dystrophin mutations were found causative of the Duchenne muscular dystrophy condition in human patients. Yet, in mice, utrophin has shown remarkable ability to compensate for dystrophin knockouts, and it is estimated that partial compensation also occurs in humans (67). Inspired by the compensatory effect demonstrated by this RBC in mice, its artificial induction in humans by means of gene therapy has been suggested (66–68). Although such modalities have not yet been realized, they suggest a fruitful possibility.
Supplementary Material
Acknowledgments
We thank the Y.P. laboratory and A. Bar-Even for helpful discussions; R. Levy for discussions and critical reviews of the manuscript; and E. Tzahor, T. Brody, S. Shen-Orr, and R. Milo for commenting on the manuscript. Y.P. is an incumbent of the Rothstein Career Development Chair in Genetic Diseases. We thank the Ben May Charitable Trust and the Tauber Fund for grant support.
Abbreviations
- MO
morpholino oligonucleotides
- RBC
responsive backup circuit
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Conant G. C., Wagner A. Proc. R. Soc. London B; 2004. pp. 89–96. [Google Scholar]
- 2.Wagner A. Nat. Genet. 2000;24:355–361. doi: 10.1038/74174. [DOI] [PubMed] [Google Scholar]
- 3.Kirschner M., Gerhart J. Proc. Natl. Acad. Sci. USA. 1998;95:8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu Z., Steinmetz L. M., Gu X., Scharfe C., Davis R. W., Li W. H. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 5.Ohno S. Evolution by Gene and Genome Duplication. New York, NY: Springer; 1970. [Google Scholar]
- 6.Nowak M. A., Boerlijst M. C., Cooke J., Smith J. M. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- 7.Lynch M., Conery J. S. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 8.Hughes A. L. Proc. Biol. Sci.; 1994. pp. 119–124. [DOI] [PubMed] [Google Scholar]
- 9.Maconochie M., Nonchev S., Morrison A., Krumlauf R. Annu. Rev. Genet. 1996;30:529–556. doi: 10.1146/annurev.genet.30.1.529. [DOI] [PubMed] [Google Scholar]
- 10.Weiss K., Stock D., Zhao Z., Buchanan A., Ruddle F., Shashikant C. Eur. J. Oral Sci. 1998;106(Suppl. 1):55–63. doi: 10.1111/j.1600-0722.1998.tb02154.x. [DOI] [PubMed] [Google Scholar]
- 11.Schwarz M., Alvarez-Bolado G., Urbánek P., Busslinger M., Gruss P. Proc. Natl. Acad. Sci. USA. 1997;94:14518–14523. doi: 10.1073/pnas.94.26.14518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mansouri A., Gruss P. Mech. Dev. 1998;78:171–178. doi: 10.1016/s0925-4773(98)00168-3. [DOI] [PubMed] [Google Scholar]
- 13.Yang H., Bard M., Bruner D. A., Gleeson A., Deckelbaum R. J., Aljinovic G., Pohl T. M., Rothstein R., Sturley S. L. Science. 1996;272:1353–1356. doi: 10.1126/science.272.5266.1353. [DOI] [PubMed] [Google Scholar]
- 14.Gerhart J., Kirschner M. Cells, Embryos, and Evolution: Toward a Cellular and Developmental Understanding of Phenotypic Variation and Evolutionary Adaptability. Oxford, U.K.: Blackwell Science; 1997. [Google Scholar]
- 15.Kafri R., Bar-Even A., Pilpel Y. Nat. Genet. 2005;37:295–299. doi: 10.1038/ng1523. [DOI] [PubMed] [Google Scholar]
- 16.Zhao C., Jung U. S., Garrett-Engele P., Roe T., Cyert M. S., Levin D. E. Mol. Cell. Biol. 1998;18:1013–1022. doi: 10.1128/mcb.18.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leon M., Sentandreu R., Zueco J. Yeast. 2002;19:1003–1014. doi: 10.1002/yea.893. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Rodriguez L. J., Trilla J. A., Castro C., Valdivieso M. H., Duran A., Roncero C. FEBS Lett. 2000;478:84–88. doi: 10.1016/s0014-5793(00)01835-4. [DOI] [PubMed] [Google Scholar]
- 19.Kondrashov F. A., Rogozin I. B., Wolf Y. I., Koonin E. V. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-research0008. RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papp B., Pal C., Hurst L. D. Nature. 2004;429:661–664. doi: 10.1038/nature02636. [DOI] [PubMed] [Google Scholar]
- 21.Ozcan S., Johnston M. Microbiol. Mol. Biol. Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eide D. J. Annu. Rev. Nutr. 1998;18:441–469. doi: 10.1146/annurev.nutr.18.1.441. [DOI] [PubMed] [Google Scholar]
- 23.Ozcan S. J. Biol. Chem. 2002;277:46993–46997. doi: 10.1074/jbc.M208726200. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson B., Sawamura T., Masaki T., MacLeod C. L. J. Biol. Chem. 1998;273:14663–14666. doi: 10.1074/jbc.273.24.14663. [DOI] [PubMed] [Google Scholar]
- 25.Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCammon M. T., McAlister-Henn L. Arch. Biochem. Biophys. 2003;419:222–233. doi: 10.1016/j.abb.2003.08.022. [DOI] [PubMed] [Google Scholar]
- 27.van den Berg M. A., de Jong-Gubbels P., Kortland C. J., van Dijken J. P., Pronk J. T., Steensma H. Y. J. Biol. Chem. 1996;271:28953–28959. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- 28.Waddington C. H. Nature. 1942;150:563–565. [Google Scholar]
- 29.Qiu M., Bulfone A., Martinez S., Meneses J. J., Shimamura K., Pedersen R. A., Rubenstein J. L. Genes Dev. 1995;9:2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- 30.Qiu M., Bulfone A., Ghattas I., Meneses J. J., Christensen L., Sharpe P. T., Presley R., Pedersen R. A., Rubenstein J. L. Dev. Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- 31.Enns L. C., Kanaoka M. M., Torii K. U., Comai L., Okada K., Cleland R. E. Plant Mol. Biol. 2005;58:333–349. doi: 10.1007/s11103-005-4526-7. [DOI] [PubMed] [Google Scholar]
- 32.Tautz D. BioEssays. 1992;14:263–266. doi: 10.1002/bies.950140410. [DOI] [PubMed] [Google Scholar]
- 33.Sabourin L. A., Rudnicki M. A. Clin. Genet. 2000;57:16–25. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- 34.Molkentin J. D., Olson E. N. Curr. Opin. Genet. Dev. 1996;6:445–453. doi: 10.1016/s0959-437x(96)80066-9. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W., Behringer R. R., Olson E. N. Genes Dev. 1995;9:1388–1399. doi: 10.1101/gad.9.11.1388. [DOI] [PubMed] [Google Scholar]
- 36.Krause M., Fire A., Harrison S. W., Priess J., Weintraub H. Cell. 1990;63:907–919. doi: 10.1016/0092-8674(90)90494-y. [DOI] [PubMed] [Google Scholar]
- 37.Michelson A. M., Abmayr S. M., Bate M., Arias A. M., Maniatis T. Genes Dev. 1990;4:2086–2097. doi: 10.1101/gad.4.12a.2086. [DOI] [PubMed] [Google Scholar]
- 38.Venuti J. M., Goldberg L., Chakraborty T., Olson E. N., Klein W. H. Proc. Natl. Acad. Sci. USA. 1991;88:6219–6223. doi: 10.1073/pnas.88.14.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atchley W. R., Fitch W. M., Bronner-Fraser M. Proc. Natl. Acad. Sci. USA. 1994;91:11522–11526. doi: 10.1073/pnas.91.24.11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holland P. W., Holland L. Z., Williams N. A., Holland N. D. Development (Cambridge, U.K.) 1992;116:653–661. doi: 10.1242/dev.116.3.653. [DOI] [PubMed] [Google Scholar]
- 41.Rudnicki M. A., Braun T., Hinuma S., Jaenisch R. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 42.Rudnicki M. A., Schnegelsberg P. N., Stead R. H., Braun T., Arnold H. H., Jaenisch R. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 43.Peters H., Wilm B., Sakai N., Imai K., Maas R., Balling R. Development (Cambridge, U.K.) 1999;126:5399–5408. doi: 10.1242/dev.126.23.5399. [DOI] [PubMed] [Google Scholar]
- 44.Dahl E., Koseki H., Balling R. BioEssays. 1997;19:755–765. doi: 10.1002/bies.950190905. [DOI] [PubMed] [Google Scholar]
- 45.Toresson H., Campbell K. Development (Cambridge, U.K.) 2001;128:4769–4780. doi: 10.1242/dev.128.23.4769. [DOI] [PubMed] [Google Scholar]
- 46.Solomon K. S., Fritz A. Development (Cambridge, U.K.) 2002;129:3127–3136. doi: 10.1242/dev.129.13.3127. [DOI] [PubMed] [Google Scholar]
- 47.Yamashita M., Ying S. X., Zhang G. M., Li C., Cheng S. Y., Deng C. X., Zhang Y. E. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. Mol. Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 49.Lin X., Liang M., Feng X. H. J. Biol. Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y., Chang C., Gehling D. J., Hemmati-Brivanlou A., Derynck R. Proc. Natl. Acad. Sci. USA. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herradon G., Ezquerra L., Nguyen T., Silos-Santiago I., Deuel T. F. Biochem. Biophys. Res. Commun. 2005;333:714–721. doi: 10.1016/j.bbrc.2005.05.160. [DOI] [PubMed] [Google Scholar]
- 52.Dorman C. J. Nat. Rev. Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 53.Zhang A., Rimsky S., Reaban M. E., Buc H., Belfort M. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- 54.Onda M., Ota K., Chiba T., Sakaki Y., Ito T. Gene. 2004;332:51–59. doi: 10.1016/j.gene.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 55.Inoue S. B., Takewaki N., Takasuka T., Mio T., Adachi M., Fujii Y., Miyamoto C., Arisawa M., Furuichi Y., Watanabe T. Eur. J. Biochem. 1995;231:845–854. doi: 10.1111/j.1432-1033.1995.tb20770.x. [DOI] [PubMed] [Google Scholar]
- 56.Lesage G., Sdicu A. M., Menard P., Shapiro J., Hussein S., Bussey H. Genetics. 2004;167:35–49. doi: 10.1534/genetics.167.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akimenko M. A., Ekker M., Wegner J., Lin W., Westerfield M. J. Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ekker M., Akimenko M. A., Bremiller R., Westerfield M. Neuron. 1992;9:27–35. doi: 10.1016/0896-6273(92)90217-2. [DOI] [PubMed] [Google Scholar]
- 59.Ellies D. L., Stock D. W., Hatch G., Giroux G., Weiss K. M., Ekker M. Genomics. 1997;45:580–590. doi: 10.1006/geno.1997.4978. [DOI] [PubMed] [Google Scholar]
- 60.Tyson J. J., Chen K. C., Novak B. Curr. Opin. Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 61.Shen-Orr S. S., Milo R., Mangan S., Alon U. Nat. Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 62.Paulsson J. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- 63.Elowitz M. B., Levine A. J., Siggia E. D., Swain P. S. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 64.Tong A. H., Boone C. Methods Mol. Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- 65.Lesage G., Shapiro J., Specht C. A., Sdicu A. M., Menard P., Hussein S., Tong A. H., Boone C., Bussey H. BMC Genet. 2005;6:8. doi: 10.1186/1471-2156-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Porter J. D., Rafael J. A., Ragusa R. J., Brueckner J. K., Trickett J. I., Davies K. E. J. Cell Sci. 1998;111:1801–1811. doi: 10.1242/jcs.111.13.1801. [DOI] [PubMed] [Google Scholar]
- 67.Deconinck A. E., Rafael J. A., Skinner J. A., Brown S. C., Potter A. C., Metzinger L., Watt D. J., Dickson J. G., Tinsley J. M., Davies K. E. Cell. 1997;90:717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- 68.Dowling P., Culligan K., Ohlendieck K. Naturwissenschaften. 2002;89:75–78. doi: 10.1007/s00114-001-0289-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.