Abstract
Cell cycle checkpoints can delay mitotic exit in budding yeast. The master controller is the small GTPase Tem1, with inputs from a proposed guanine nucleotide exchange factor (GEF), Lte1, and a GTPase-activating protein (GAP), Bub2/Bfa1. In this issue, Fraschini et al. (p. 335) show that GAP activity of Bub2/Bfa1 appears to be dispensable for inactivation of Tem1 in cells. Their results call into question the GTP/GDP switch model for Tem1 activity, as have other results in the past. The paper also focuses attention on the two spindle pole bodies as potential sites for regulation of Tem1.
During the cell cycle, budding yeast are able to monitor DNA replication and repair, as well as mitotic spindle assembly and position. If one of these processes has not finished, the cell senses this and delays exit from mitosis, providing extra time to remedy the situation (Fig. 1). The master controller for the decision to exit appears to be a small G-protein, Tem1 (Shirayama et al., 1994). Tem1 appears to collect inputs from various sensors that monitor these processes, integrate that information, and then notify the mitotic exit network (MEN), a cascade of signaling proteins, when it is safe to go ahead and finish mitosis. For a recent review of this area, see Seshan and Amon (2004).
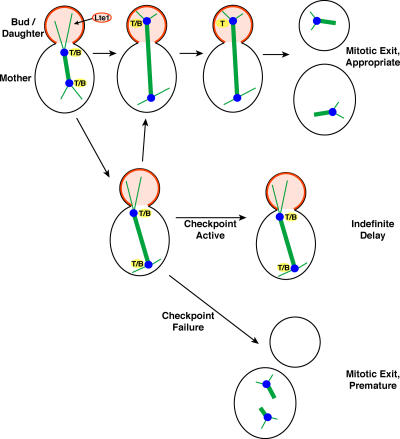
Figure 1.
Stages in progression through mitosis. T/B is Tem1 complexed with Bub2/Bfa1, and the drawing illustrates the timing of their location on SPBs, with respect to spindle position and mitotic exit. In this case, failure of the spindle to move into the neck is what activates the checkpoint. The diffuse cytoplasmic pools of the components are not indicated, but they are likely to be important, as discussed in the text. Over time during anaphase, Tem1 accumulates on the D-SPB, relative to the M-SPB, while Bub2/Bfa1 does the opposite (Molk et al., 2004).
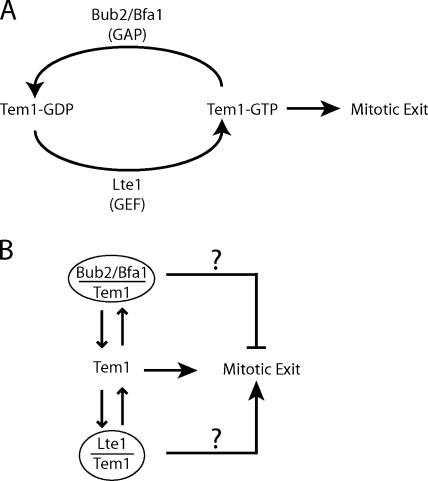
A switch model for Tem1 has been proposed, with the GTP-bound state as ON and promoting mitotic exit and the GDP-bound state as OFF and having no effect on mitotic exit (Fig. 2 A). The ON to OFF transition is proposed to be accelerated by a GTPase-activating protein (GAP), the heterodimer of Bub2 with Bfa1 (Geymonat et al., 2002). In cells, Bub2/Bfa1 clearly inhibits mitotic exit, via Tem1 (Bardin et al., 2000; Bloecher et al., 2000; Pereira et al., 2000), and, in vitro, Bub2/Bfa1 increases GTP hydrolysis by Tem1 (Geymonat et al., 2002). A new study by Fraschini and colleagues in this issue (p. 335) challenges the idea that Bub2/Bfa1 acts as a GAP on Tem1 in cells, based on the finding that the GAP activity of Bub2/Bfa1 appears to be dispensable for inhibiting mitotic exit.
Figure 2.
Schematics of possible models for how Tem1 controls mitotic exit. (A) GTP/GDP switch model. (B) Effector model.
This new study also focuses attention on the mother-bound spindle pole body (M-SPB) as a potential location for interactions that control Tem1 activity, whatever they may be. Several previous observations implicate the daughter-bound SPB (D-SPB) as potentially important. Passage of the D-SPB through the neck appears to be a critical event that sets the cellular clock ticking on the course for mitotic exit (Molk et al., 2004). During the course of a normal cell cycle, Tem1 accumulates on the D-SPB, along with active MEN components (Molk et al., 2004). Bub2/Bfa1 first accumulates and then is lost from the D-SPB, as one might expect an inhibitor to behave.
In this new work, Fraschini et al. (2006) found that a Myc-tagged version of Bub2 was anomalously localized to both SPBs throughout the cell cycle, in contrast to the normal behavior of untagged Bub2 or HA-tagged Bub2, which disappear from the M-SPB over time. Expression of this Myc-tagged Bub2 produced inhibition of mitotic exit, in an appropriate sensitized background. Genetically, the effect was dominant and worked through Tem1. The investigators logically presumed that Bub2-Myc/Bfa1 was exerting excessive GAP activity on Tem1 at the M-SPB, but, surprisingly, they found that the GAP activity of Bub2-Myc/Bfa1 was undetectable in vitro, even with Tem1 as the substrate. Thus, Bub2-Myc displayed loss of function in terms of GAP activity coupled with gain of function in terms of Tem1 inhibition. In addition, Bub2-Myc can inhibit mitotic exit when the spindle assembly checkpoint is activated, shown in previous work by the same group (Fraschini et al., 1999). At face value, the results argue that Bub2/Bfa1 inhibits Tem1 in cells by a biochemical mechanism other than acceleration of GTP hydrolysis.
The authors then deliberately targeted Bub2's GAP activity by mutating a conserved Arg residue in the proposed catalytic site. This mutant, Bub2-R85A, also had undetectable GAP activity in vitro, as expected. However, in cells, Bub2-R85A was not able to inhibit Tem1 and thereby delay mitotic exit when called upon by a checkpoint, in contrast to Bub2-Myc. At this point, one might defend the GAP hypothesis by simply proposing that Bub2-R85A has much less GAP activity than Bub2-Myc, and that the low activity levels of both proteins happen to be undetectable in this particular biochemical assay. On the other hand, in cells, Bub2-R85A showed an important difference compared with Bub-Myc. Bub2-R85A did not recruit Bfa1 to the SPB, while Bub2-Myc did. Bub2 and Bfa1 are both necessary to inhibit Tem1, so the R85 residue of Bub2 may simply be necessary for Bub2 to interact with Bfa1 at the SPB, and thus the heterodimer cannot function. Bub2-R85A did localize to and persist at the M-SPB, as did Bub2-Myc but not wt Bub2. Based on these results, the authors suggest that Bub2's GAP activity might be required for Bub2/Bfa1 to leave the M-SPB, in support of the hypothesis that Bub2/Bfa1 persistence at the M-SPB can inhibit Tem1 and delay mitotic exit.
Other previous results, some of which are also admittedly negative, question whether the GTP/GDP switch model explains the cellular action of Tem1. First, a Tem1 point mutation analogous to Ras Q61L, which should be locked in the GTP state and therefore constitutively active, had no obvious effect on cell growth (Shirayama et al., 1994). However, this mutation remains to be tested in a setting where mitotic exit is delayed. Second, an apparent guanine nucleotide exchange factor (GEF) domain can be found in the sequence of Lte1, a protein that promotes mitotic exit in cells by activating Tem1 (Bardin et al., 2000). However, removing Lte1's GEF domain has little or no effect on Lte1's ability to promote mitotic exit (Jensen et al., 2002; Yoshida et al., 2003). Third, Tem1's intrinsic rates of GTP hydrolysis and release are high (∼0.1–0.2/min; Geymonat et al., 2002), so that increasing them may not be useful.
What alternative mechanisms might one consider? Strong evidence, especially genetic analysis, shows that Bub2/Bfa1 and Lte1 antagonize each other, that they work through Tem1, and that each one is important for cells to thrive, at least under conditions in the wild. For example, at low temperatures, Lte1 is essential for growth and was named as such—low temperature essential (Wickner et al., 1987). What, then, is the active state of Tem1, the state that drives mitotic exit? A mutant lacking Bub2 and Lte1 is viable, under optimal lab conditions, so free Tem1 should be sufficient for mitotic exit (Hofken and Schiebel, 2002; Stegmeier et al., 2002). Perhaps Bub2/Bfa1 sequesters Tem1 in an inactive state, while free Tem1 and Lte1-bound Tem1 bind effectors to activate the MEN (Fig. 2 B). The possibility that the Tem1/Bub2/Bfa1 complex has an independent inhibitory effect on the MEN has not been excluded. Finally, the recent discovery of phosphorylation of Tem1 provides a new factor to consider (Wang and Ng, 2006).
Localization of components has helped to formulate and test models (Fig. 1). As noted above, several results argue that the D-SPB may be a crucial site for regulation. In addition, the activator Lte1 is confined to the bud, positioning it to activate the Tem1 of the D-SPB as the spindle enters the neck (Bardin et al., 2000). The D-SPB often strikes the cortex, where Lte1 is heavily concentrated, but this event does not correlate with the timing of mitotic exit and thus may be incidental (Molk et al., 2004). The Lte1 in the bud cytoplasm may be the form that interacts with Tem1 (Castillon et al., 2003).
The results of Fraschini et al. (2006) suggest that the mother-bound SPB (M-SPB) may also be important, in that the presence of Bub2-Myc/Bfa1 at the M-SPB inhibited mitotic exit, via Tem1. In support of this idea, when mitotic exit is delayed by a checkpoint, Bub2/Bfa1 persists at the M-SPB (Pereira et al., 2001). In addition, a novel kinase, Kin4, which is confined to the mother cortex, has been found to inhibit mitotic exit via Bub2/Bfa1 (D'Aquino et al., 2005; Pereira and Schiebel, 2005). One may need to consider every cellular location of Tem1 as a potentially important place where regulation of Tem1 activity can occur, because Tem1 molecules exchange rapidly between SPB and cytoplasm (Molk et al., 2004). The sum of the inhibiting and activating effects on Tem1 in all its cellular pools may be what tips the balance. On the other hand, scaffolding mechanisms may activate Tem1 and promote its interactions with downstream effectors more effectively in certain locations. A computational analysis of the system should be helpful at this point, perhaps necessary, given the complexity of the signaling pathways (Bosl and Li, 2005).
In sum, the mechanism for controlling the timing of mitotic exit may not be the obvious one suggested by the protein sequences and biochemical activities in vitro. The addition of protein biochemistry to the toolbox of the yeast cell biologist is helping the field to test molecular mechanisms for mitotic exit in new ways, with some unexpected results. Our understanding of what happens in the cell has many gaps to be filled, including the role of the GAP at hand.
Acknowledgments
We are grateful to Ken Blumer, Mark Longtine, Prakash Chudalayandi, Kerry Bloom, and Kelly Tatchell for comments on the manuscript.
Research in this area in our lab is supported by National Institutes of Health grant GM 69895.
Abbreviations used in this paper: D-SPB, daughter-bound spindle pole body; GAP, GTPase-activating protein; MEN, mitotic exit network; M-SPB, mother-bound spindle pole body.
References
- Bardin, A.J., R. Visintin, and A. Amon. 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 102:21–31. [DOI] [PubMed] [Google Scholar]
- Bloecher, A., G.M. Venturi, and K. Tatchell. 2000. Anaphase spindle position is monitored by the BUB2 checkpoint. Nat. Cell Biol. 2:556–558. [DOI] [PubMed] [Google Scholar]
- Bosl, W.J., and R. Li. 2005. Mitotic-exit control as an evolved complex system. Cell. 121:325–333. [DOI] [PubMed] [Google Scholar]
- Castillon, G.A., N.R. Adames, C.H. Rosello, H.S. Seidel, M.S. Longtine, J.A. Cooper, and R.A. Heil-Chapdelaine. 2003. Septins have a dual role in controlling mitotic exit in budding yeast. Curr. Biol. 13:654–658. [DOI] [PubMed] [Google Scholar]
- D'Aquino, K.E., F. Monje-Casas, J. Paulson, V. Reiser, G.M. Charles, L. Lai, K.M. Shokat, and A. Amon. 2005. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol. Cell. 19:223–234. [DOI] [PubMed] [Google Scholar]
- Fraschini, R., E. Formenti, G. Lucchini, and S. Piatti. 1999. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J. Cell Biol. 145:979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini, R., C. D'Ambrosio, M. Venturetti, G. Lucchini, and S. Piatti. 2006. Disappearance of the budding yeast Bub2/Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J. Cell Biol. 172:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat, M., A. Spanos, S.J. Smith, E. Wheatley, K. Rittinger, L.H. Johnston, and S.G. Sedgwick. 2002. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 277:28439–28445. [DOI] [PubMed] [Google Scholar]
- Hofken, T., and E. Schiebel. 2002. A role for cell polarity proteins in mitotic exit. EMBO J. 21:4851–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S., M. Geymonat, A.L. Johnson, M. Segal, and L.H. Johnston. 2002. Spatial regulation of the guanine nucleotide exchange factor Lte1 in Saccharomyces cerevisiae. J. Cell Sci. 115:4977–4991. [DOI] [PubMed] [Google Scholar]
- Molk, J.N., S.C. Schuyler, J.Y. Liu, J.G. Evans, E.D. Salmon, D. Pellman, and K. Bloom. 2004. The differential roles of budding yeast Tem1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol. Biol. Cell. 15:1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., and E. Schiebel. 2005. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol. Cell. 19:209–221. [DOI] [PubMed] [Google Scholar]
- Pereira, G., T. Hofken, J. Grindlay, C. Manson, and E. Schiebel. 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell. 6:1–10. [PubMed] [Google Scholar]
- Pereira, G., T.U. Tanaka, K. Nasmyth, and E. Schiebel. 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 20:6359–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshan, A., and A. Amon. 2004. Linked for life: temporal and spatial coordination of late mitotic events. Curr. Opin. Cell Biol. 16:41–48. [DOI] [PubMed] [Google Scholar]
- Shirayama, M., Y. Matsui, and E.A. Toh. 1994. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol. Cell. Biol. 14:7476–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier, F., R. Visintin, and A. Amon. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 108:207–220. [DOI] [PubMed] [Google Scholar]
- Wang, Y., and T.-Y. Ng. 2006. Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae. Mol. Biol. Cell. 17:80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, R.B., T.J. Koh, J.C. Crowley, J. O'Neil, and D.B. Kaback. 1987. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation of the MAK16 gene and analysis of an adjacent gene essential for growth at low temperatures. Yeast. 3:51–57. [DOI] [PubMed] [Google Scholar]
- Yoshida, S., R. Ichihashi, and A. Toh-e. 2003. Ras recruits mitotic exit regulator Lte1 to the bud cortex in budding yeast. J. Cell Biol. 161:889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]