Abstract
Mismatch repair (MMR) proteins contribute to genome integrity by correcting replication errors. In higher eukaryotes, MMR proteins also regulate the cellular response to DNA lesions such as oxidized, alkylated, or crosslinked bases. Previous studies have linked MMR proteins to the activation of apoptosis through p53-dependent and p53-independent mechanisms. MMR-deficient cells exhibit variable defects in the induction of p53 and its related p73, which are activators of apoptosis. However, the specific role of each MMR protein in the regulation of apoptosis has not been determined. Here, we describe an interaction between PMS2, an MMR protein, and p73. This interaction causes the stabilization of p73 and the redistribution of PMS2 to the nuclear compartment. Exposure to cisplatin enhances the association between PMS2 and p73. Moreover, stimulation of the p73 proapoptotic function by cisplatin requires PMS2. These results suggest that PMS2 contributes to genome integrity not only through DNA repair but also by enhancing DNA damage-induced apoptosis.
Mismatch repair (MMR) is a DNA repair mechanism that ensures the fidelity of DNA replication. The repair of base:base mismatches and small insertion/deletion mispairs generated during DNA synthesis is mediated by a group of highly conserved MMR proteins (1, 2). In prokaryotes, MMR depends on an interaction between the MutS homodimer and the MutL homodimer that subsequently coordinates the activity of other mismatch repair proteins. In eukaryotic cells, MMR involves interactions between two sets of heterodimers, composed of the MutS-homolog (MSH) and the MutL-homolog (MLH) proteins (3, 4). The MSH heterodimers MSH2/MSH6 and MSH2/MSH3 bind to base:base mismatches and small insertion/deletion mispairs. The MLH heterodimers MLH1/PMS2 and MLH1/MLH3 interact with the MSH heterodimers and recruit other repair enzymes to correct replication errors (5–7). The absence of MMR, therefore, results in higher mutation rates. In humans, germ-line mutations in either the MLH1 or the MSH2 gene cause a cancer predisposition syndrome, hereditary nonpolyposis colorectal cancer (1, 2), and a significant proportion of sporadic cancers are also MMR defective (8–10). The tumor suppression function of MMR is also demonstrated by the cancer-prone phenotype of Mlh1, Msh2, Msh6, and Pms2 knockout mice (11–15).
In addition to the repair of replication errors, MMR proteins also interact with base pairs modified by oxidation, alkylation, and cross-linking (16–18). These modified bases can be repaired by several mechanisms, including nucleotide excision repair, base excision repair, and postreplication repair. Whereas MMR does not play an essential role in correcting alkylated or crosslinked bases, the MMR proteins are involved in the activation of apoptosis in response to such lesions (19, 20). Cancer cells deficient in MMR show reduced apoptosis response to alkylating agents such as N-methyl-N′-nitro-N-nitrosoguanidine and crosslinking agents such as cisplatin (21, 22). Because the MSH heterodimers can bind to N-methyl-N′-nitro-N-nitrosoguanidine- and cisplatin-modified DNA, they may function as damage sensors in a signaling pathway that links these lesions to the activation of apoptosis. This notion has been supported by the interaction of MMR proteins with Ataxia telangiectasia-mutated (ATM), breast cancer associated-1 (BRCA1), and checkpoint kinase 2 (CHK2) (23, 24), which are components of the DNA damage signaling network (25). Moreover, MMR-deficient cells exhibit defects in the activation of p53 and p73 after exposure to alkylating agents or cisplatin (26–28).
The tumor suppressor p53 plays an essential role in DNA damage-induced apoptosis. The resistance of p53-deficient cells to DNA damage-induced apoptosis is well established. Two p53-related transcription factors (i.e., p63 and p73) also regulate the apoptosis response to DNA damage. A role for p73 in DNA damage-induced apoptosis was discovered in cell-based studies (27, 29, 30). Recent studies with knockout mice have confirmed that p73 is required for DNA damage to cause apoptosis (31). Cells derived from p73 knockout mice exhibited reduced apoptosis to DNA damage. Interestingly, apoptosis to DNA damage was completely abrogated with the combined knockout of p73 and p63, despite the expression of a functional p53 (31). These genetic studies suggest that p53 is necessary but not sufficient to activate apoptosis in response to DNA damage; p63 and p73 are also required for DNA lesions to activate cell death.
In a previous study, cisplatin was found to induce the p73 protein, and this response was compromised in MLH1-deficient cells (27). In this study, we have uncovered an interaction between PMS2 and p73. Furthermore, we have found that PMS2 can collaborate with p73 to enhance cisplatin-induced apoptosis. These results support a direct role of MMR proteins in the regulation of p73 in DNA damage response.
Materials and Methods
Cell Lines.
HCT116 (MLH1 mutant), HCT116–3 (6) (MLH1 mutant complemented by chromosome 3 transfer), HEC59 (MSH2 mutant), HEC1A (PMS2, MSH6 mutant), HCT15 (MSH6 mutant), Cos-1, and 293T cells were maintained in DMEM supplemented with 10% FBS (HyClone).
Plasmids.
The MLH1 and PMS2 cDNAs from pCI-ML10 (32) and pCI-PM1 (gift from Chikashi Ishioka, Tohoku University, Sendai, Japan), respectively, were inserted between the BamHI and NotI sites of pcDNA 3.1 (+) (Invitrogen). The PMS2 cDNA was also inserted between the KpnI and NotI sites of pCMV-Myc (CLONTECH). The MLH1, PMS2, MSH2, and TRP73-α cDNA were cloned in pEYFP-C1 (CLONTECH).
Immunoblotting.
Cells were lysed in 200 μl of RIPA buffer (50 mM Tris⋅HCl, pH 7.5/1 mM EDTA/150 mM NaCl/1% Nonidet P-40/0.5% deoxycholate/0.1% SDS) at 4°C. Immunoblotting was performed with standard methods by using anti-p73 (clone 429; Imgenex, San Diego), anti-p53 (Ab-6; Calbiochem), anti-PMS2 (Ab-1; Calbiochem), anti-MLH1 (Ab-1; Calbiochem), anti-MSH2 (Ab-1; Calbiochem), anti-MSH6 (PharMingen), and anti-β-actin (Sigma). To load equal numbers of transfected cells, a β-galactosidase expression plasmid (pCMV-β-Gal) was included in the transfection mix, and the resulting enzyme activity was measured in each transfected cell lysate. Equal amounts of β-galactosidase activity were loaded to normalize for the transfection efficiency.
Protein Stability Assay.
HCT116 cells (1.2 × 105) in a 10-cm dish were transfected with 2 μg of pCMV-HA-p73α (27) or pCMV-PMS2 DNA mixed with 6 μl of FuGENE6 (Roche Molecular Biochemicals), followed by incubation at 37°C for 24 h. The cells were trypsinized and replated into five 6-cm dishes. Each plate of cells was treated with 25 μM cisplatin for 24 h, and then 25 μg/ml cycloheximide (Sigma) was added. After incubation for different times as indicated in individual experiments, the cells in each plate were harvested and analyzed by immunoblotting as described above. The levels of p73 at each time point was determined by desitometry and normalized to that of time 0 (relative intensity).
Coimmunoprecipitation.
293T cells (2 × 105) were transfected with 2.5 μg each of pCMV-HA-p73α and pCMV-Myc-PMS2 by using FuGENE6 and harvested after 48 h. Cells were lysed in 1 ml of TNE buffer (100 mM Tris⋅HCl, pH 7.5/0.5 mM EDTA/150 mM NaCl/1% Nonidet P-40) containing protease inhibitor mixture (Roche Molecular Biochemicals), the lysate was clarified by centrifugation at 10,000 × g at 4°C, and 400 μg of lysate was precleared with 10 μl of protein A/G agarose (Santa Cruz Biotechnology). The supernatant was then mixed with 2 μg of anti-hemagglutinin (HA; 12CA5; Roche Molecular Biochemicals) or anti-c-Myc (9E10; Covance, Berkeley, CA) antibody and 20 μl of protein A/G agarose for 3 h at 4°C. The beads were washed three times and then boiled in SDS sample buffer. Immunoprecipitated proteins were then analyzed by Western blotting.
Subcellar Distribution of Yellow Fluorescent Protein (YFP)-Fusion Proteins.
Cos-1 cells on sterile glass coverslips were cotransfected with 0.5 μg each of indicated plasmids by using FuGENE6. At 48 h posttransfection, cells were fixed in 3.7% paraformaldehyde with 1 μg/ml Hoechst 33258. Propidium iodide was also used to stain the DNA. After fixation, cells were permeabilized with 0.3% Triton X-100 for 5 min and incubated in 1.5 mM propidium iodide with 10 μg/ml RNase at 37°C for 30 min in the dark. The coverslips were rinsed with PBS and mounted on microscope slides. Images were captured by using a DeltaVision deconvolution microscope operated by softworx software (Applied Precision, Issaquah, WA) as described (33). To quantitate the nuclear-only YFP signal, cells were counted in an epifluorescence microscope (Zeiss) at ×400 magnification. Cells in 10 randomly chosen fields were scored per experiment.
Apoptosis Assay.
One day before transfection, HCT116 cells were plated on coverslips and transfected with 1 μg each of the p73 or PMS2 expression plasmids along with 0.2 μg of the GFP-H2B plasmid by using FuGENE6. After 24 h of incubation at 37°C, cisplatin (Sigma) was added to the media at a final concentration of 25 μM. After an additional 24 h, cells were fixed as described above and stained with Hoechst 33258 (Sigma). The percentage of apoptotic cells was determined as the percentage of GFP-positive cells with condensed chromatin or fragmented nuclei. At least 300 GFP-positive cells were counted per experiment.
Results
Differential Induction of p73 by Cisplatin in MMR-Deficient Cells.
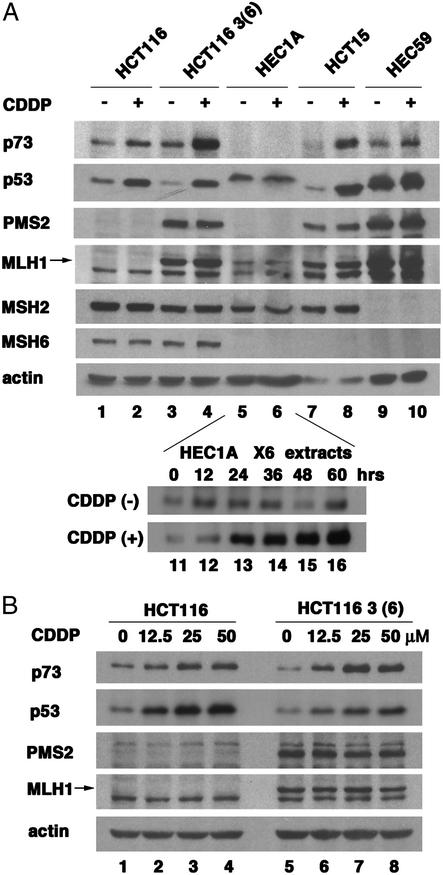
Induction of p73 by cisplatin is compromised in MLH1-deficient cells [e.g., the HCT116 cell line (ref. 27; Fig. 1 A, lanes 1 and 2, and B, lanes 1–4)]. Reconstitution of MLH1 expression in HCT116–3 (6) cells enhanced the p73 induction by cisplatin (Fig. 1 A, lanes 3–4, and B, lanes 5–8). In addition to the loss of MLH1, HCT116 cells also lacked the PMS2 protein (Fig. 1A, lanes 1 and 2), because PMS2 is unstable in the absence of MLH1 (34). Restoration of MLH1 expression, therefore, corrects the defect in MLH1 and allows PMS2 expression (Fig. 1 A, compare lane 3 with 1, and B, compare lanes 5–8 with 1–4). To determine whether PMS2 played a role in p73 induction, we examined HEC1A cells, which express MLH1 and MSH2 but not PMS2 or MSH6 (Fig. 1A, lanes 5 and 6). The HEC1A cells contain a higher basal level of p53 that was not responsive to cisplatin. We could not detect p73 in 30 μg of total protein from HEC1A cells (Fig. 1A, lanes 5 and 6). However, when we loaded 6-fold more HEC1A lysate, p73 became detectable and its induction by cisplatin was also observed (Fig. 1A, lanes 11–16). In HCT15 cells, which lack MSH6, p73 and p53 were both induced by cisplatin (Fig. 1A, lanes 7 and 8), showing MSH6 is dispensable in this response. Previous studies have shown that MSH2 is required for cisplatin to activate p53 (26, 35). We found that p73 induction by cisplatin was reduced, albeit not abolished in MSH2-deficient HEC59 cells (Fig 1A, lanes 9 and 10). Taken together, these results suggest MSH2 and MLH1, but not MSH6, play a role in the induction of p73 by cisplatin. Interestingly, PMS2 is dispensable for the inductive effect of cisplatin but required for the accumulation of p73 protein.
Figure 1.
Induction of p73 by cisplatin in MMR-deficient cells. (A) Deficiency of MLH1, PMS2, and MSH2 affected induction of p73 by cisplatin. (Upper) The indicated cell lines were treated with 25 μM cisplatin for 48 h, and 30 μg of total protein was analyzed by immunoblotting. MMR proteins are displayed to confirm the deficiency. β-Actin is included for the loading control. (Lower) Two hundred micrograms of HEC1A cell lysates collected at the indicated time with or without 25 μM cisplatin was probed for p73. (B) p73 and p53 protein level in HCT116 and HCT116 3 (6) cells that were treated with the indicated doses of cisplatin (12.5, 25, or 50 μM cisplatin) for 48 h.
PMS2 Stabilizes p73.
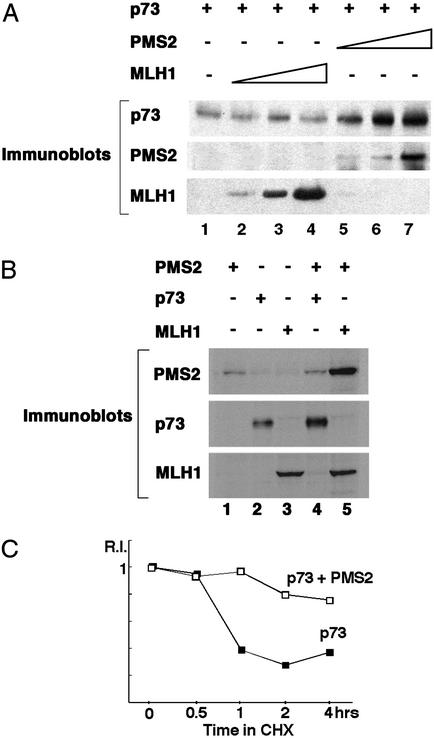
The ability of PMS2 to stabilize the p73 protein was demonstrated in transient coexpression experiments (Fig. 2). The levels of p73 protein increased with increasing amounts of coexpressed PMS2 (Fig. 2A, lanes 5–7). In comparison, MLH1 did not affect the levels of p73 (Fig. 2A, lanes 2–4). Whereas PMS2 caused an increase in p73 (Fig. 2 A and B, lanes 2 and 4), coexpression with p73 did not affect the levels of PMS2 (Fig. 2B, compare lanes 1 and 4). In contrast, coexpression of MLH1 stabilized PMS2 (Fig. 2B, compare lanes 1 and 5), consistent with previous results (34). To determine whether PMS2 prolonged the half-life of p73, transfected cells were treated with cycloheximide to prevent new protein synthesis and the decay of p73 protein was examined (Fig. 2C). The half-life of p73 was about 45 min in the absence of cotransfected PMS2 (Fig. 2C). A significant increase in the p73 half-life was observed with the coexpression of PMS2 (Fig. 2C). It is noteworthy that these transfection experiments were performed with HCT116 cells, which do not express MLH1 (Fig. 1). Therefore, PMS2 could stabilize p73 in the absence of MLH1. Moreover, PMS2 could promote the stabilization of p73 without itself being stabilized.
Figure 2.
PMS2 stabilizes p73 protein. (A) Increase of p73 protein by transient coexpression with PMS2. Different amounts of expression plasmids for PMS2 or MLH1 were introduced with p73 expression plasmid into HCT116 cells along with a pCMV-β-Gal plasmid. Whole-cell extracts were analyzed by immunoblotting after normalized loading by β-galactosidase activity. (B) PMS2 is stabilized by cotransfection with MLH1 but not by p73. The indicated expression plasmids were transfected into HCT116 cells and proteins analyzed as described in A. (C) Half-life of P73 was prolonged by PMS2. Determination of the half-life of p73 is described in Materials and Methods.
p73 Causes the Nuclear Accumulation of PMS2.
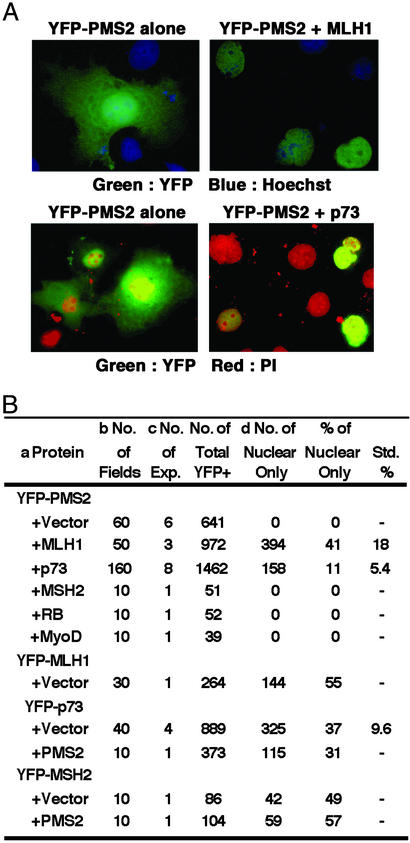
Coexpression with p73 affected the subcellular distribution of the PMS2 protein. PMS2 was fused to YFP to allow the examination of its subcellular localization (Fig. 3). When expressed alone, YFP-PMS2 was distributed throughout the cell (Fig. 3A Left) in over 600 YFP+ cells examined (Fig. 3B). When coexpressed with MLH1 or p73, nuclear accumulation of YFP-PMS2 became evident (Fig. 3A Right). We quantitated the effect of MLH1 or p73 on the redistribution of YFP-PMS2 by counting cells with nuclear-only YFP signal (Fig. 3B). To maintain a consistent scoring criteria, we did not score cells with predominant nuclear and weak cytoplasmic YFP signal. In over 900 YFP+ cells from cotransfections withYFP-PMS2 and MLH1, 30–50% showed an exclusively nuclear localization of YFP-PMS2 (Fig. 3B). In comparison, coexpression with p73 caused 10–20% of the transfected cells to accumulate YFP-PMS2 in the nucleus (Fig. 3B). When YFP-PMS2 was coexpressed with three other nuclear proteins (MSH2, RB, and MyoD), none of the transfected cells showed nuclear-exclusive YFP signal (Fig. 3B). We also fused MLH1, p73, or MSH2 to YFP and examined their distribution (Fig. 3B). The YFP-MLH1, YFP-p73, and YFP-MSH2 proteins were each localized to the nucleus in transfected cells, with 40–60% the cells showing nuclear-only signal (Fig. 3B) and the rest exhibiting predominant nuclear and weak cytoplasmic YFP signal (data not shown). Importantly, coexpression with PMS2 did not alter the subcellular distribution of YFP-p73 or YFP-MSH2 (Fig. 3B). Taken together, these results show that p73 and MLH1, but not MSH2, can alter the subcellular localization of PMS2. The effect of p73 on PMS2 localization was also observed in HCT116 cells (data not shown). Thus, p73 can alter the subcellular distribution of PMS2 without MLH1. A direct interaction between PMS2 and MLH1 is well established (3, 4, 6, 36). The ability of p73 to exert an effect similar to that of MLH on the nuclear accumulation of PMS2 therefore suggests that p73 and PMS2 also interact in vivo.
Figure 3.
MLH1 and p73 can influence the subcellular distribution of PMS2. (A) Merged image of YFP-PMS2 (green) and Hoechst (blue, Upper) or propidium iodide (red, Lower) of Cos-1 cells transfected with the indicated expression plasmids. (B) Quantification of the nuclear localization of YFP-PMS2. a, Each of the indicated proteins was expressed individually (+ vector) or in combination. Transfected cells were stained with Hoechst and visualized with a fluorescent microscope for YFP and nuclei. b, Ten fields were counted per experiment and, on average, each field contained ≈50 cells. c, The number of independent transfection experiments is shown. d, Nuclear-only YFP signal is depicted in A Right.
Cisplatin Augments the Interaction Between PMS2 and p73.
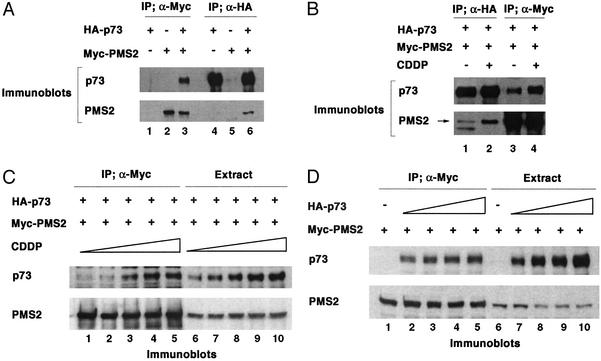
To examine the interaction between PMS2 and p73, we coexpressed HA-tagged p73 and Myc-tagged PMS2 (Fig. 4). Reciprocal coimmunoprecipitation was observed with anti-Myc (Fig. 4A, lanes 1–3) and anti-HA antibodies (Fig. 4A, lanes 4–6) followed by blotting with anti-p73 (Fig. 4A Upper) and anti-PMS2 (Fig. 4A Lower). In the absence of Myc-PMS2, anti-Myc did not precipitate p73 (Fig. 4A, lane 1), and in the absence of HA-p73, anti-HA antibody did not precipitate PMS2 (Fig. 4A, lane 5). The interaction between PMS2 and p73, even under conditions of overexpression, was responsive to cisplatin (Fig. 4B). After transfected cells were treated with 25 μM cisplatin for 24 h, an increased amount of PMS2 was found in the anti-HA immunoprecipitate when compared with untreated cells (Fig. 4B, lanes 1–2). Likewise, anti-Myc immunoprecipitation brought down more p73 from cisplatin-treated than from untreated cells (Fig. 4B, lanes 3–4). In a titration experiment (Fig. 4C), cisplatin caused an increase in the transfected p73 protein (Fig. 4C, lanes 6–10). Cisplatin, however, did not alter the levels of PMS2 (Fig. 4C, lanes 6–10). Despite the constant level of PMS2, we observed an increased association between p73 and PMS2, concomitant with a rise in p73 (Fig. 4C, lanes 1–5). These results suggest that cisplatin, in a dose-dependent manner, causes an increased association between p73 and PMS2, leading to the stabilization of p73. To rule out the alternative interpretation, that the enhanced coimmunoprecipitation was the result of p73 increase, we performed a control experiment in which the levels of p73 protein were raised by increasing the amount of the p73 expression plasmid (Fig. 4D). Under this experimental condition, the amount of p73 found in the anti-Myc immunoprecipitate was constant (Fig. 4D, lanes 1–5), despite increasing p73 levels in the cells (Fig. 4D, lanes 6–10). These observations support the conclusion that p73 and PMS2 interact in vivo and suggest that cisplatin can stimulate this interaction to stabilize p73.
Figure 4.
Cisplatin stimulates the association of PMS2 and p73. (A) Myc-tagged PMS2 and HA-tagged p73 were expressed in 293T cells and immunoprecipitated by anti-Myc and anti-HA antibodies followed by immunoblotting with anti-PMS2 and anti-p73. (B) 293T cells (transfected as described for A) were treated with or without cisplatin, immunoprecipitated by anti-Myc and anti-HA antibodies, and immunoblotted with anti-PMS2 and p73 antibodies. The arrow marks the PMS2 band. (C) 293T cells transfected with 1 μg each of Myc-PMS2 and HA-p73 expression plasmids were treated with cisplatin for 24 h. The cell extracts were precipitated with anti-Myc antibody and then analyzed by anti-p73 and anti-PMS2 immunoblotting. (D) 293T cells were transfected with different amounts of p73 expression plasmids and 1 μg of PMS2 expression plasmids. The cell extracts were precipitated with anti-Myc antibody and then analyzed by anti-p73 and anti-PMS2 immunoblotting.
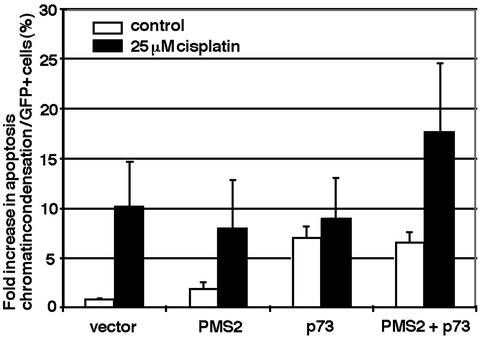
PMS2 Allows Cisplatin to Enhance the Apoptosis Function of p73.
Because the interaction between PMS2 and p73 can be influenced by cisplatin, we examined the effect of PMS2 on the apoptosis-induction function of p73. Transient overproduction of p73 can induce apoptosis in several different cell lines (27, 37). We therefore transfected HCT116 cells with p73 and quantitated apoptosis by counting transfected cells (marked by a cotransfected GFP-H2B, GFP fused to histone H2B) with condensed chromatin (Fig. 5). Transfection with GFP-H2B alone (vector) did not increase apoptosis above the basal level. Transfection with PMS2 did not significantly increase apoptosis either. Transfection with p73 caused apoptosis to rise 7-fold above the basal level, consistent with previous reports (27, 37). Cotransfection with p73 and PMS2 did not increase apoptosis above the level caused by p73 alone (PMS2+p73). Therefore, PMS2 has no effect on p73-dependent apoptosis in the absence of DNA damage.
Figure 5.
Stimulation of p73 apoptosis function by cisplatin requires PMS2. HCT116 cells were transfected with empty vector, PMS2, p73, or both expression plasmids along with GFP-H2B plasmid. GFP-positive cells with condensed chromatin were counted without cisplatin treatment (white bars) or after 24-h cisplatin treatment (black bars). Values represent means and SDs from three independent experiments.
We then measured apoptosis in transfected cells after a 24-h exposure to 25 μM cisplatin (Fig. 5, black bars). Cisplatin caused a 10-fold increase in apoptosis of GFP-H2B-transfected HCT116 cells. Because p53 is induced by cisplatin in HCT116 cells (Fig. 1), it could have accounted for this apoptosis response. Transfection with PMS2 did not increase the apoptosis response to cisplatin. In p73-transfected cells, cisplatin did not stimulate apoptosis caused by the overexpression of p73. This is consistent with the inability of cisplatin to activate p73 in these MLH1-deficient cells. Interestingly, however, cisplatin was able to stimulate apoptosis in cells cotransfected with p73 and PMS2. These results show that overproduction of PMS2 can rescue the activation of p73 by cisplatin.
Discussion
Regulation of p73 in DNA Damage Response.
The stabilization and activation of the p53 family of apoptosis inducers occur in cells exposed to a variety of DNA damaging agents, including cisplatin (DNA cross-linking), doxorubicin (topoisomerase II inhibition), and ionizing radiation (DNA strand breaks; refs. 27, 29, and 30). Interestingly, whereas p53 is frequently mutated in sporadic cancer, p73 does not appear to be inactivated during malignant transformation (38). Given the central role of DNA-damaging agents in cancer therapy, it is important to understand how p73 is regulated by DNA damage, because activation of p73 may be used to kill cancer cells. Previous studies have identified several upstream regulators of p73, including the c-Abl tyrosine kinase, the p300 acetyltransferase, and the p38 stress-activated protein kinase (27, 29, 30, 39, 40). After DNA damage, p73 protein is stabilized and its apoptosis activity is enhanced through the actions of these modifying enzymes. Despite identification of these upstream regulators, the precise mechanism of p73 stabilization and activation has not been elucidated.
In this study, we have identified PMS2 to be another regulator of p73. PMS2 can promote the stabilization of p73. Cisplatin stimulates the interaction between PMS2 and p73, and this interaction is required for the activation of p73. These results establish PMS2 to be an additional component in the p73 regulatory pathway that is activated by DNA damage. Because only p73, but not PMS2, is stabilized through their interaction, the formation of a stable p73/PMS2 complex may not be required to protect p73 from proteolysis. Instead, PMS2 may alter the conformational state of p73, by as-yet-unknown mechanisms, to prolong its half-life. The stabilization of p73 by PMS2 appears to depend on c-Abl, because this effect was not observed in Abl-null cells (data not shown). We could not detect a stable association of PMS2 and c-Abl in transient cotransfection experiments (data not shown). Thus, c-Abl and PMS2 may function in an interdependent manner to stabilize p73, without the formation of a stable trimeric protein complex.
Regulation of Apoptosis by MMR Proteins in DNA-Damage Response.
The involvement of MMR proteins in the regulation of apoptosis is supported by several lines of investigation, with cultured cells and in MMR-defective knockout mice. In this study, we showed that the induction of p73 by cisplatin is compromised in cells lacking MSH2, MLH1, or PMS2. The current evidence suggests that MMR proteins may regulate p73 at multiple steps. The MSH2/MSH6 complex and MSH2 alone have been shown to recognize and bind to cisplatin adducts in DNA (16, 17) and therefore function as the damage sensor. The MSH2/MSH3 complex may also be able to recognize cisplatin adducts because MSH6 is not essential for the induction of p73. The MLH1/PMS2 complex may function as an adaptor, bringing p73 to the site of damaged DNA and allowing p73 to become modified and stabilized, leading to the activation of its apoptosis function.
Our study provides evidence that PMS2 interacts with p73 and stimulates p73-dependent apoptosis. We observed the functional interaction of PMS2 and p73 in cells that do not express MLH1 (HCT116 cells). This raises the interesting possibility that PMS2, at least under conditions of overproduction, can stimulate apoptosis independent of MLH1. Our observation of an MLH1-independent activity of PMS2 is previously unreported but not unprecedented. A recent report (41) describes the colocalization of MLH1 and MLH3 to the meiotic chromosomes. Interestingly, localization of MLH3 to the same meiotic chromosome sites was found in Mlh1-mutant cells, implying an MLH1-independent function for MLH3 (41). It should be noted that the accumulation of p73 protein per se is not sufficient for the activation of its function. A mutant p73 protein lacking three critical lysine residues that are sites of acetylation can accumulate in cells after DNA damage but cannot induce apoptosis (40). Although MLH1 may not be directly responsible for the stabilization of p73 protein, we cannot rule out its role in the regulation of p73 activity.
Implication on the Tumor Suppression Function of PMS2.
Previous studies have shown Pms2−/− mouse cells to exhibit reduced apoptosis response to DNA damage (42). Selection of cisplatin-resistant subclones of osteosarcoma cells has led to the isolation of clones with reduced PMS2 expression (43). However, we showed that PMS2 overexpression did not induce cell death, consistent with a previously published report (44). Instead, PMS2 is required for p73 to be activated by cisplatin. This evidence is the first showing that PMS2 directly regulates the activity of a proapoptotic protein.
The role of PMS2 in hereditary nonpolyposis colorectal cancer has remained controversial, because PMS2 mutation has only been detected in a single hereditary nonpolyposis colorectal cancer family (45). On the other hand, germ-line PMS2 mutations have been detected in several patients with Turcot syndrome, which can also be caused by mutations in the MSH2 gene (46–48). The tumor suppression function of PMS2 has been demonstrated by the increased tumor risk associated with Pms2−/− mice (49). Interestingly, whereas Mlh1−/− mice showed intestinal adenocarcinoma and lymphoma, Pms2−/− mice developed lymphoma, sarcoma, and skin cancer but not intestinal tumors (49). The Pms2−/− cells exhibited a limited spectrum of somatic mutations compared with Mlh1−/− cells. Moreover, Pms2+/− knockout mice developed intestinal adenocarcinoma after methylnitrosourea treatment even though the tumors did not exhibit microsatellite instability and other mutator phenotypes (50). Thus, the tumor suppression function of PMS2 may be associated with its biological function not only in repair but also in the DNA damage response pathway. Our finding that PMS2 interacts with p73 to regulate apoptosis is consistent with the idea that PMS2 stimulates death of damaged cells to suppress tumor formation.
Acknowledgments
We thank Dr. James Feramisco at the University of California at San Diego Cancer Center for assistance in imaging and John Weger and Dana Cassel for performing sequencing. We also thank members of the J.Y.J.W. and R.D.K. laboratories for discussions and technical supports. This work was supported by a center grant from the National Institute on Environmental Health Sciences (to J.Y.J.W. and R.D.K.), the Uehara Memorial Foundation (to H.S.), the Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad (to H.S.), and the Japan Society for the Promotion of Science Research Fellowships for Young Scientists (to A.Y.-Y.).
Abbreviations
- MMR
mismatch repair
- MSH
MutS-homolog
- MLH
MutL-homolog
- HA
hemagglutinin
- YFP
yellow fluorescent protein
References
- 1.Kolodner R D, Marsischky G T. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 2.Harfe B D, Jinks-Robertson S. Annu Rev Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 3.Habraken Y, Sung P, Prakash L, Prakash S. Curr Biol. 1997;7:790–793. doi: 10.1016/s0960-9822(06)00337-x. [DOI] [PubMed] [Google Scholar]
- 4.Habraken Y, Sung P, Prakash L, Prakash S. J Biol Chem. 1998;273:9837–9841. doi: 10.1074/jbc.273.16.9837. [DOI] [PubMed] [Google Scholar]
- 5.Raschle M, Marra G, Nystrom-Lahti M, Schar P, Jiricny J. J Biol Chem. 1999;274:32368–32375. doi: 10.1074/jbc.274.45.32368. [DOI] [PubMed] [Google Scholar]
- 6.Prolla T A, Pang Q, Alani E, Kolodner R D, Liskay R M. Science. 1994;265:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- 7.Lipkin S M, Wang V, Jacoby R, Banerjee-Basu S, Baxevanis A D, Lynch H T, Elliott R M, Collins F S. Nat Genet. 2000;24:27–35. doi: 10.1038/71643. [DOI] [PubMed] [Google Scholar]
- 8.Aaltonen L A, Peltomaki P, Leach F S, Sistonen P, Pylkkanen L, Mecklin J P, Jarvinen H, Powell S M, Jen J, Hamilton S R, et al. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 9.Ionov Y, Peinado M A, Malkhosyan S, Shibata D, Perucho M. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 10.Thibodeau S N, Bren G, Schaid D. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 11.Prolla T A, Baker S M, Harris A C, Tsao J L, Yao X, Bronner C E, Zheng B, Gordon M, Reneker J, Arnheim N, et al. Nat Genet. 1998;18:276–279. doi: 10.1038/ng0398-276. [DOI] [PubMed] [Google Scholar]
- 12.Baker S M, Bronner C E, Zhang L, Plug A W, Robatzek M, Warren G, Elliott E A, Yu J, Ashley T, Arnheim N, et al. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 13.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 14.Reitmair A H, Schmits R, Ewel A, Bapat B, Redston M, Mitri A, Waterhouse P, Mittrucker H W, Wakeham A, Liu B, et al. Nat Genet. 1995;11:64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- 15.Edelmann W, Yang K, Umar A, Heyer J, Lau K, Fan K, Liedtke W, Cohen P E, Kane M F, Lipford J R, et al. Cell. 1997;91:467–477. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- 16.Duckett D R, Drummond J T, Murchie A I, Reardon J T, Sancar A, Lilley D M, Modrich P. Proc Natl Acad Sci USA. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mello J A, Acharya S, Fishel R, Essigmann J M. Chem Biol. 1996;3:579–589. doi: 10.1016/s1074-5521(96)90149-0. [DOI] [PubMed] [Google Scholar]
- 18.Ni T T, Marsischky G T, Kolodner R D. Mol Cell. 1999;4:439–444. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- 19.Aebi S, Kurdi-Haidar B, Gordon R, Cenni B, Zheng H, Fink D, Christen R D, Boland C R, Koi M, Fishel R, et al. Cancer Res. 1996;56:3087–3090. [PubMed] [Google Scholar]
- 20.Drummond J T, Anthoney A, Brown R, Modrich P. J Biol Chem. 1996;271:19645–19648. doi: 10.1074/jbc.271.33.19645. [DOI] [PubMed] [Google Scholar]
- 21.Li G M. Oncol Res. 1999;11:393–400. [PubMed] [Google Scholar]
- 22.Bellacosa A. Cell Death Differ. 2001;8:1076–1092. doi: 10.1038/sj.cdd.4400948. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Cortez D, Yazdi P, Neff N, Elledge S J, Qin J. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 24.Brown K D, Rathi A, Kamath R, Beardsley D I, Zhan Q, Mannino J L, Baskaran R. Nat Genet. 2003;33:80–84. doi: 10.1038/ng1052. [DOI] [PubMed] [Google Scholar]
- 25.Zhou B B, Elledge S J. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 26.Duckett D R, Bronstein S M, Taya Y, Modrich P. Proc Natl Acad Sci USA. 1999;96:12384–12388. doi: 10.1073/pnas.96.22.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong J G, Costanzo A, Yang H Q, Melino G, Kaelin W G, Jr, Levrero M, Wang J Y. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 28.Wu J, Gu L, Wang H, Geacintov N E, Li G M. Mol Cell Biol. 1999;19:8292–8301. doi: 10.1128/mcb.19.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agami R, Blandino G, Oren M, Shaul Y. Nature. 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 30.Yuan Z M, Shioya H, Ishiko T, Sun X, Gu J, Huang Y Y, Lu H, Kharbanda S, Weichselbaum R, Kufe D. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 31.Flores E R, Tsai K Y, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 32.Shimodaira H, Filosi N, Shibata H, Suzuki T, Radice P, Kanamaru R, Friend S H, Kolodner R D, Ishioka C. Nat Genet. 1998;19:384–389. doi: 10.1038/1277. [DOI] [PubMed] [Google Scholar]
- 33.Miller Y I, Chang M K, Funk C D, Feramisco J R, Witztum J L. J Biol Chem. 2001;276:19431–19439. doi: 10.1074/jbc.M011276200. [DOI] [PubMed] [Google Scholar]
- 34.Chang D K, Ricciardiello L, Goel A, Chang C L, Boland C R. J Biol Chem. 2000;275:18424–18431. doi: 10.1074/jbc.M001140200. [DOI] [PubMed] [Google Scholar]
- 35.Toft N J, Winton D J, Kelly J, Howard L A, Dekker M, te Riele H, Arends M J, Wyllie A H, Margison G P, Clarke A R. Proc Natl Acad Sci USA. 1999;96:3911–3915. doi: 10.1073/pnas.96.7.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu L, Hong Y, McCulloch S, Watanabe H, Li G M. Nucleic Acids Res. 1998;26:1173–1178. doi: 10.1093/nar/26.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jost C A, Marin M C, Kaelin W G., Jr Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 38.Melino G, De Laurenzi V, Vousden K H. Nat Rev Cancer. 2002;2:605–615. doi: 10.1038/nrc861. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez-Prieto R, Sanchez-Arevalo V J, Servitja J M, Gutkind J S. Oncogene. 2002;21:974–979. doi: 10.1038/sj.onc.1205134. [DOI] [PubMed] [Google Scholar]
- 40.Costanzo A, Merlo P, Pediconi N, Fulco M, Sartorelli V, Cole P A, Fontemaggi G, Fanciulli M, Schiltz L, Blandino G, et al. Mol Cell. 2002;9:175–186. doi: 10.1016/s1097-2765(02)00431-8. [DOI] [PubMed] [Google Scholar]
- 41.Lipkin S M, Moens P B, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman J W, Green E D, et al. Nat Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- 42.Zeng M, Narayanan L, Xu X S, Prolla T A, Liskay R M, Glazer P M. Cancer Res. 2000;60:4889–4893. [PubMed] [Google Scholar]
- 43.Perego P, Caserini C, Gatti L, Carenini N, Romanelli S, Supino R, Colangelo D, Viano I, Leone R, Spinelli S, et al. Mol Pharmacol. 1999;55:528–534. [PubMed] [Google Scholar]
- 44.Zhang H, Richards B, Wilson T, Lloyd M, Cranston A, Thorburn A, Fishel R, Meuth M. Cancer Res. 1999;59:3021–3027. [PubMed] [Google Scholar]
- 45.Nicolaides N C, Papadopoulos N, Liu B, Wei Y F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, et al. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 46.Hamilton S R, Liu B, Parsons R E, Papadopoulos N, Jen J, Powell S M, Krush A J, Berk T, Cohen Z, Tetu B, et al. N Engl J Med. 1995;332:839–847. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- 47.De Rosa M, Fasano C, Panariello L, Scarano M I, Belli G, Iannelli A, Ciciliano F, Izzo P. Oncogene. 2000;19:1719–1723. doi: 10.1038/sj.onc.1203447. [DOI] [PubMed] [Google Scholar]
- 48.Miyaki M, Nishio J, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Nagato M, Chong J M, Koike M, Terada T, et al. Oncogene. 1997;15:2877–2881. doi: 10.1038/sj.onc.1201668. [DOI] [PubMed] [Google Scholar]
- 49.Prolla T A. Curr Opin Cell Biol. 1998;10:311–316. doi: 10.1016/s0955-0674(98)80005-7. [DOI] [PubMed] [Google Scholar]
- 50.Qin X, Shibata D, Gerson S L. Carcinogenesis. 2000;21:833–838. doi: 10.1093/carcin/21.4.833. [DOI] [PubMed] [Google Scholar]