Abstract
Cellular Src and epidermal growth factor receptor (EGFR) collaborate in the progression of certain human malignancies, and their cooverexpression characterizes relatively aggressive animal tumors. Our study addressed the mode of oncogenic cooperation and reports that overexpression of c-Src in model cellular systems results in the accumulation of EGFR at the cell surface. The underlying mechanism involves inhibition of the normal, c-Cbl-regulated process of ligand-induced receptor down-regulation. In response to activation of c-Src, c-Cbl proteins undergo tyrosine phosphorylation that promotes their ubiquitylation and proteasomal destruction. Consequently, ubiquitylation of EGFR by c-Cbl is restrained in Src-transformed cells, and receptor sorting to endocytosis is impaired. In conclusion, by promoting destruction of c-Cbl, c-Src enables EGFR to evade desensitization, which explains Src-EGFR collaboration in oncogenesis.
Keywords: cancer‖oncogene‖signal transduction‖tyrosine kinase‖ubiquitin
The essential role of growth factors in inductive cell fate determination and in tissue remodeling is reflected in neoplastic processes. One example is the ErbB family of growth factor receptors and their ligands, all containing an epidermal growth factor (EGF) motif (reviewed in ref. 1). Several types of human tumors, including glioblastomas and carcinomas, display increased expression of the EGF receptor (EGFR; also called ErbB-1). The mitogenic ability and transforming potential of EGFR are enhanced by heterodimerization with other ErbBs. Another oncogenic partner of EGFR is the cytoplasmic tyrosine kinase c-Src and its family members (reviewed in refs. 2 and 3). Elevated levels of Src family kinases, and mutational activation were observed in colon carcinoma (4), and similar findings were reported for lung, breast, and brain tumors. Evidence derived from experimental models indicates that Src activation potentiates EGF-induced mitogenesis (5, 6). Likewise, cooverexpression of c-Src and EGFR characterizes human breast cancers, and it leads to a synergistic increase in tumor formation in mice (7).
The mechanisms underlying Src-EGFR collaboration are incompletely understood, although it is clear that the two kinases maintain bi-directional interactions: stimulation of EGFR increases the catalytic activity of c-Src (8), and c-Src phosphorylates EGFR at sites that stimulate the intrinsic catalytic activity of EGFR (7). On the other hand, another set of reports attributed to c-Src restraining effects on EGFR. Accordingly, overexpression of c-Src accelerates clathrin-mediated internalization of EGFR (9), probably because clathrin is tyrosine-phosphorylated by c-Src (10). The present study addresses the mechanism of Src-EGFR collaboration and identifies c-Cbl, a regulator of EGFR endocytosis (reviewed in ref. 11), as a target of c-Src: activation of Src leads to phosphorylation and polyubiquitylation of c-Cbl, and subsequent degradation of c-Cbl enables EGFR to escape desensitization.
Materials and Methods
Materials, Buffers, and Antibodies.
mAbs to EGFR were generated in our laboratory. Other antibodies were obtained from Santa Cruz Biotechnology. SYF cells were obtained from the American Type Culture Collection. The composition of buffers has been described (12, 13). EGF was labeled with 125I by using the Iodogen reagent (Pierce).
Plasmid Construction, Transfection, and Preparation of Cell Extracts.
EGFR mutants, hemagglutinin (HA)-tagged Cbl mutants, and a GFP-Cbl construct have been described (12, 13). An HA-Ub expression vector was obtained from Dirk Bohmann (European Molecular Biology Laboratory, Heidelberg). Src expression vectors and the control pUSE vector were purchased from Upstate Biotechnology (Lake Placid, NY). Cells were transfected by using the Lipofectamine 2000 method. Unless otherwise indicated, we used the following amounts of plasmid DNA for transfection of cells in 90-mm plates: EGFR and HA-tagged ubiquitin, 1 μg per plate; c-Src and c-Cbl, 2 μg per plate. The total amount of plasmid DNA in each transfection was normalized with the respective empty plasmid.
Immunoprecipitation and Immunoblotting Analyses.
Essentially, we used described protocols (12). Immunoprecipitates were resolved by gel electrophoresis and transferred to a nitrocellulose membrane. Membranes were blocked in PBS containing 0.5% Tween 20 and 1% milk, blotted for 2 h with a primary antibody (1 μg/ml), followed by a secondary antibody (0.5 μg/ml) linked to horseradish peroxidase. Immunoreactive protein bands were detected with an enhanced chemiluminescence reagent (Amersham Pharmacia).
Receptor Down-Regulation Assay.
Cells were treated without or with EGF for various time intervals at 37°C. The medium was then removed, and the cells were washed once with binding buffer and twice with an acidic buffer (135 mM NaCl/2.5 mM KCl/50 mM acetic acid) to remove surface-bound ligand. The cells then were washed twice in ice-cold binding buffer and incubated with [125I]EGF (0.5 nM) for 4 h at 4°C. Thereafter, cells were washed twice and solubilized, and their radioactivity was determined.
Pulse–Chase Analysis of Protein Turnover.
Cells were pulse-labeled at 37°C by incubation with [35S]methionine [250 μCi/ml (1 Ci = 37 GBq)] in methionine-free medium supplemented with 2% (vol/vol) dialyzed FBS. Thereafter, cells were washed, cultured in chase medium containing unlabeled methionine, and extracted for immunoprecipitation.
Northern Blot Analysis.
Total RNA extracted by using the RNeasy kit from Qiagen (Hilden, Germany) was fractionated on a 1.2% agarose gel containing 1.1 M formaldehyde and blotted onto a membrane. The membrane was prehybridized for 4 h at 65°C in a 1% (vol/vol) SDS solution containing 10% dextran sulfate, 5.8% NaCl, and 100 μg/ml salmon sperm DNA. cDNA probes were labeled with [32P]dCTP.
Immunofluorescence.
Cells were fixed for 30 min with 3% (wt/vol) paraformaldehyde and permeabilized for 10 min at 22°C in saline containing 1% albumin and 0.2% Triton X-100. Cover slips were then incubated for 1 h at room temperature with antibodies directed to HA or to c-Src (each at 10 μg/ml). Antibody detection was performed by using fluorescently labeled secondary antibodies (from Jackson ImmunoResearch). Finally, cover slips were mounted in Elvanol (Hoechst, Frankfurt) and examined by using a Zeiss Axiovert confocal microscope.
Results
Src Up-Regulates EGFR Expression by Inhibiting Cbl-Induced Receptor Ubiquitylation and Endocytosis.
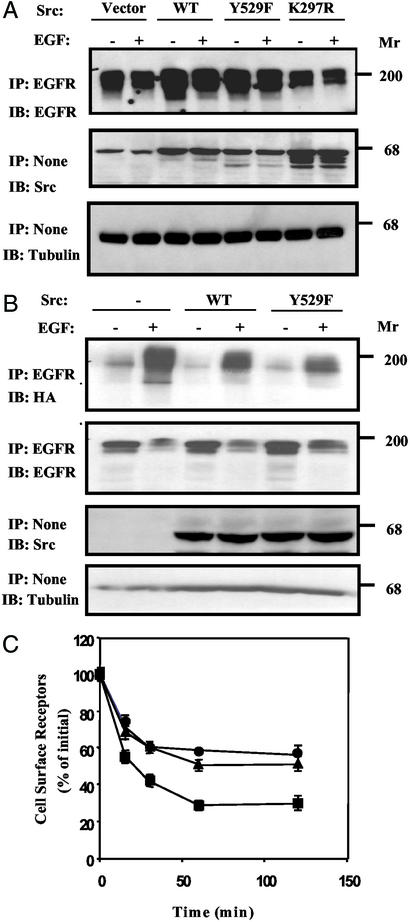
To examine the possibility that c-Src collaborates with EGFR by modulating receptor desensitization, we coexpressed in receptor-negative Chinese hamster ovary cells the human EGFR together with one of the following forms of human c-Src: wild type, an active mutant (Y529F), or a kinase-dead mutant (K297R). Forty-eight hours after transfection, cells were stimulated with EGF for 15 min, and EGFR levels were determined (Fig. 1A). The results showed that EGFR levels were significantly increased (70–110%) in cells overexpressing either wild-type c-Src or an active mutant, but cells expressing a kinase-defective mutant of c-Src displayed reduced expression relative to cells transfected with an empty vector. Quantification of the results revealed that, despite Src-induced up-regulation of EGFR, EGF was still able to reduce, in part, the level of its receptor.
Figure 1.
Src up-regulates EGFR expression by inhibiting c-Cbl-induced receptor ubiquitylation. (A) Chinese hamster ovary cells coexpressing EGFR and the indicated Src proteins were untreated or treated with EGF (100 ng/ml) for 15 min; thereafter, cell extracts were analyzed by immunoprecipitation (IP) and immunoblotting (IB) with the indicated antibodies. (B) SYF cells coexpressing EGFR, HA-tagged ubiquitin, c-Cbl, and the indicated forms of c-Src were untreated or treated with EGF (100 ng/ml) for 15 min; thereafter, cell extracts were analyzed as indicated. (C) Down-regulation of EGFR was tested in SYF cells ectopically expressing EGFR alone (●), in combination with c-Cbl (■), or in combination with both c-Cbl and an active c-Src (Y529F; ▴). Cells were incubated at 37°C with an unlabeled EGF (100 ng/ml) for the indicated time intervals, and the residual level of surface EGFR was determined by binding of a radioactive EGF at 4°C. Averages and the corresponding SDs of triplicates (bars) are shown.
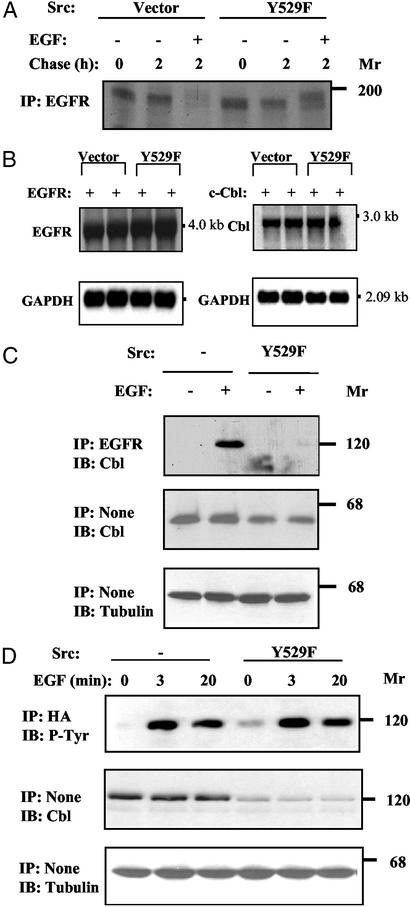
To analyze the interaction between c-Src and EGFR in a null cellular background, we used mouse embryo fibroblasts, denoted SYF cells (14), lacking the three ubiquitous Src-family kinases: Src, Yes, and Fyn. Consistent with the effects observed in Chinese hamster ovary (CHO) cells, overexpression of wild-type c-Src or an active mutant (Y529F) significantly diminished ligand-induced ubiquitylation of EGFR (Fig. 1B), a process mediated by c-Cbl (13, 15, 16). In addition, as we previously reported for CHO cells (12), ectopic expression of c-Cbl accelerated removal of EGFR from the surface of SYF cells (Fig. 1C), but this effect was inhibited by a coexpressed Y529F-Src. Next, we analyzed the effect of c-Src by using pulse–chase metabolic labeling of SYF cells (Fig. 2A). Evidently, an active form of c-Src exerted no significant effect on receptor synthesis or maturation. In this assay system, newly synthesized receptor molecules almost completely disappeared after stimulation with EGF, but c-Src prevented receptor down-regulation while allowing band up-shift due to hyperphosphorylation. Last, to exclude the possibility that Src down-regulates EGFR or c-Cbl by affecting transcription of the respective ectopically introduced gene, we analyzed mRNA isolated from transfected SYF cells. As shown in Fig. 2B, overexpression of Y529F-Src exerted no effect on EGFR or c-Cbl transcripts, implying that c-Src stabilizes the mature, cell surface-expressed receptor. Taken together, the results presented led us to conjecture that one of the multiple substrates of c-Src is involved in EGFR endocytosis.
Figure 2.
c-Src inhibits the interaction between c-Cbl and EGFR by down-regulating c-Cbl. (A) SYF cells expressing EGFR and Y529F-Src were subjected to a 4-h-long metabolic labeling with 35S-labeled amino acids. Thereafter, cells were chased for the indicated time intervals in the absence or presence of EGF (100 ng/ml). (B) SYF cells were transfected (in duplicates) with plasmids encoding EGFR or c-Cbl, in the presence of a control vector (pUSE) or the Y529F-Src expression vector. Forty-eight hours later, cells were harvested and total cellular RNA was extracted. Transcripts corresponding to EGFR and c-Cbl were detected in Northern blots by hybridization to the respective cDNA probes. For normalization, we used a glyceraldehyde 3-phosphate dehydrogenase (GAPDH) probe. (C) SYF cells transfected with vectors encoding EGFR, HA-tagged c-Cbl, and either a Y529F-Src vector or an empty plasmid, were treated for 15 min at 37°C with EGF (100 ng/ml), or they were left untreated. Cell extracts were tested for Cbl-EGFR association by using the indicated antibodies. (D) SYF cells were transfected and treated with EGF as in C. The state of c-Cbl was analyzed by immunoblotting with anti-Cbl and anti-phosphotyrosine (P-Tyr) antibodies. IP, immunoprecipitation; IB, immunoblotting.
c-Src Inhibits Physical Interactions Between c-Cbl and EGFR by Reducing Expression Levels of c-Cbl.
To ubiquitylate EGFR, c-Cbl must associate physically with the receptor and subsequently undergo phosphorylation (15, 17). By using SYF cells, we confirmed EGF-induced physical associations between EGFR and c-Cbl. However, we found that this interaction was abolished upon expression of an active c-Src (Fig. 2C). In addition, we observed both EGF- and Src-induced phosphorylation of c-Cbl in SYF cells (Fig. 2D), which is in line with reports that identified c-Cbl as a major tyrosine phosphorylation substrate of Src family kinases (18). It is important, however, to note that the levels of c-Cbl were down-regulated upon coexpression of Y527F-Src (Fig. 2 C and D). Because no parallel change in tubulin expression was detectable, we inferred specificity to c-Cbl. Indeed, when expression of an active mutant of c-Src was gradually increased, we observed a parallel decrease in c-Cbl (data not shown), in line with negative regulation of c-Cbl by c-Src. Taken together, the results presented in Fig. 2 implied that c-Src up-regulates EGFR by reducing c-Cbl expression and inhibiting formation of a Cbl-EGFR complex.
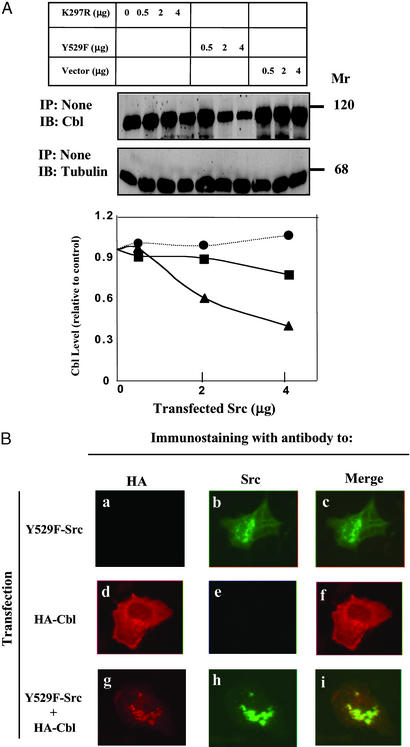
Down-Regulation of c-Cbl Involves the Catalytic Activity of c-Src and Colocalization to Vesicular Structures.
Next, we compared the effect of Y529F-Src with that of a kinase-defective c-Src (K297R; Fig. 3A). Evidently, the kinase activity of c-Src is essential for down-regulation of c-Cbl expression. Previous reports have localized both c-Cbl and c-Src to cytoplasmic vesicular structures (12, 19), but colocalization has not been reported. In the absence of an active c-Src, c-Cbl displayed diffuse cytoplasmic distribution (Fig. 3B). However, c-Cbl translocated to Src-containing vesicular structures when coexpressed with an active mutant of c-Src (note the merge panel i in Fig. 3B). This translocation is reminiscent of the recruitment of c-Cbl to early endosomes containing another active kinase, namely EGFR (12), suggesting that c-Src similarly recruits c-Cbl into vesicular structures before protein destabilization.
Figure 3.
Down-regulation of c-Cbl involves the catalytic activity of c-Src and colocalization to vesicular structures. (A) SYF cells were cotransfected with a c-Cbl expression vector along with increasing amounts of plasmids encoding the indicated mutants of Src or an empty vector. Whole-cell lysates were probed 48 h later with antibodies to c-Cbl or to tubulin. IP, immunoprecipitation; IB, immunoblotting.(B) SYF cells grown on glass slides were transfected with plasmids encoding Y529F-Src (a–c), HA-tagged c-Cbl (d–f), or their combination (g–i). After 24 h, cells were fixed, permeabilized, and incubated with murine antibodies to c-Src and a rat antibody to HA. FITC-conjugated antibody specific to mouse immunoglobulins (green) and Cy3-conjugated antibodies to rat immunoglobulins (red) were used for fluorescent confocal microscopy.
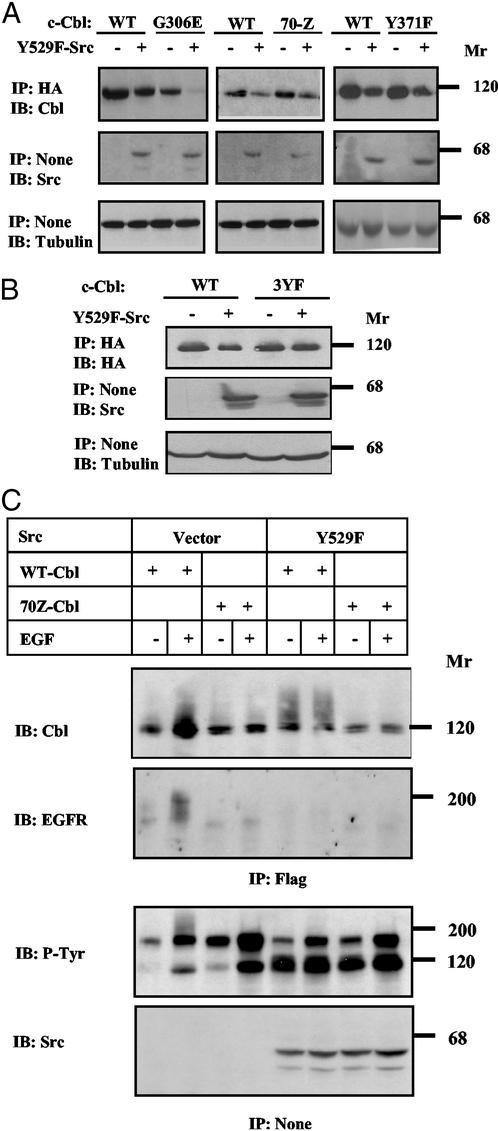
Phosphorylation and Ubiquitylation of c-Cbl Precede the Destructive Effect of c-Src.
To study the structural requirements of c-Src-induced destabilization, we examined a series of mutant c-Cbl proteins. In the G306E-Cbl mutant, a critical glycine residue of the phosphotyrosine-binding (PTB) domain is substituted (20). This mutant retained susceptibility to c-Src (Fig. 4A), indicating that the PTB domain is not involved. 70Z-Cbl is an oncogenic mutant whose RING finger cannot bind E2 enzymes (16). We found that 70Z-Cbl underwent down-regulation by c-Src (Fig. 4A) and completely disappeared from SYF cells expressing very high levels of Y529F-Src (data not shown), suggesting that the intrinsic ubiquitin ligase activity of c-Cbl is unnecessary for the effect of c-Src. This conclusion was further supported by testing Y371F-Cbl, whose critical tyrosine involved in receptor desensitization (15) was mutated (Fig. 4A). Phosphorylation by c-Src affects three major sites of c-Cbl, and the corresponding 3YF mutant (mutated Tyr-700, -731, and -774) displays a reduction in c-Src binding (21). Unlike other mutants of c-Cbl, expression of 3YF-Cbl was not affected by an active c-Src (Fig. 4B). Likewise, v-Cbl, an oncoprotein lacking the whole carboxyl-terminal half of c-Cbl, displayed resistance to c-Src (data not shown). In conclusion, phosphorylation by, or stable binding to, c-Src is essential for accelerated turnover of c-Cbl, but neither phosphotyrosine binding to c-Cbl nor ubiquitin ligase activity are necessary.
Figure 4.
c-Src elevates phosphorylation and self-ubiquitylation of c-Cbl. (A and B) SYF cells were transfected with plasmids encoding the indicated HA-tagged forms of c-Cbl, together with a plasmid encoding Y529F-Src, or the respective empty plasmid. Forty-eight hours after transfection, cell extracts were analyzed as indicated. (C) SYF cells were transfected with plasmids encoding EGFR and a Flag-tagged ubiquitin, along with the indicated forms of Src and Cbl. Forty-eight hours after transfection, cells were untreated or treated for 15 min with EGF (100 ng/ml), and cell extracts were analyzed as indicated. IP, immunoprecipitation; IB, immunoblotting.
Because polyubiquitylation sorts proteins for proteasomal destruction, we predicted that c-Src-induced degradation is preceded by polyubiquitylation of c-Cbl. In line with a previous report (22), we found that EGF enhances polyubiquitylation of both EGFR and c-Cbl (Fig. 4C). Neither modification was detectable when testing 70Z-Cbl, indicating that the RING finger of c-Cbl mediates both self- and transubiquitylation. In analogy, expression of an active c-Src elevated polyubiquitylation of c-Cbl in the absence of EGF, but no increased modification of 70Z-Cbl was detectable (Fig. 4C). These observations imply that c-Src, like EGFR, activates the intrinsic ubiquitylating function of c-Cbl, consistent with a recent in vitro study (17). Taken together, the results presented in Fig. 4 envisage the following scenario: c-Src phosphorylates the carboxyl terminus of c-Cbl and, consequently, c-Cbl undergoes polyubiquitylation and degradation. However, because a ubiquitylation-defective mutant of c-Cbl (namely 70Z-Cbl) retains susceptibility to the destructive action of c-Src, we predict that c-Src recruits not only c-Cbl but also an alternative degradation machinery. This hypothesis and its relevance to oncogene cooperation in human cancer are discussed below.
Discussion
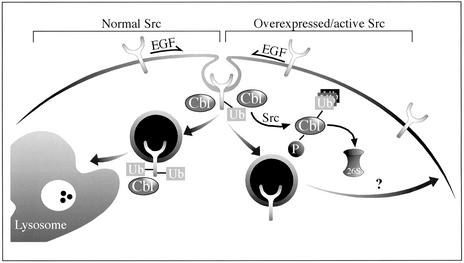
Our results may be summarized as follows: c-Src up-regulates EGFR (Fig. 1A) by inhibiting receptor ubiquitylation (Fig. 1B) and endocytosis (Fig. 1C), and both effects are attributable to accelerated destruction of c-Cbl (Figs. 2 and 3A). One interpretation of these observations is presented in Fig. 5. Normally, activation of EGFR is followed by recruitment of c-Cbl, primarily through a single phosphotyrosine of EGFR, and subsequent sorting for endocytosis and lysosomal degradation (23). This robust desensitization process is inhibited when c-Src is activated or overexpressed: Src binds to and phosphorylates c-Cbl (21, 24, 25). In addition, it enhances self-ubiquitylation of c-Cbl (Fig. 4C and ref. 17) and leads to accelerated destruction by proteasomal proteinases (Fig. 2 and unpublished observations, demonstrating that Src-induced destruction of c-Cbl is sensitive to proteasome inhibitors). Therefore, the efficacy of receptor down-regulation is reduced, and signaling by mitogens like EGF is enhanced in cells whose Src is active (e.g., adherent cells or Src-transformants).
Figure 5.
Proposed mode of interactions between c-Src and c-Cbl and their effect on EGFR trafficking. Normally, EGF promotes receptor phosphorylation, followed by recruitment of c-Cbl, receptor ubiquitylation, and sorting of EGFR to lysosomal degradation. In the presence of an active/overexpressed c-Src, both phosphorylation and ubiquitylation of c-Cbl are enhanced, and the protein is subsequently degraded by the 26S proteasome. Because of Src-induced elimination of c-Cbl, sorting of EGFR to lysosomal degradation is reduced, and the receptor is diverted to the recycling pathway. This regulatory loop may explain the association between Src activation and EGFR over-expression in tumor cells.
Src family members are commonly activated by growth factors like EGF, and this activation involves formation of a physical receptor-Src complex (7). Within the complex, Src phosphorylates the associated receptor at a site located in the kinase domain (26), a modification known to enhance catalytic activity of growth factor receptors. In line with the possibility that these interactions enable synergy between c-Src and various receptors (4, 27), mutational inactivation of the c-Src-specific phosphorylation site on EGFR ablated EGF-induced mitogenicity (5). Thus, by blocking receptor degradation (Fig. 5), Src-receptor complexes gain lasting activity, which may explain why several types of advanced tumors exhibit simultaneous activation of both EGFR and Src family members. Given the role of c-Cbl in enhancing receptor internalization (reviewed in refs. 11 and 28), our results predict an inhibitory effect of c-Src on receptor endocytosis. However, studies performed with 10T1/2 cells detected no effect of c-Src on the half-life of EGFR, but accelerated internalization was observed when the endocytic apparatus was under-saturated (9). Interestingly, this effect disappeared at high receptor occupancy, which is closer to the conditions we used in the present study. Another study concluded that activation of c-Src by EGFR is needed for subsequent phosphorylation of clathrin, which then redistributes to the cell periphery and enhances receptor internalization (10). Notably, those authors reported that the effect of Src is limited to the first two min of ligand internalization, a time window we have not addressed in the present study.
A series of recent reports unveiled complex interactions between c-Src and c-Cbl (reviewed in ref. 29). Two lines of evidence indicate that the interactions involve physical contacts: c-Cbl and c-Src colocalized to vesicular structures (Fig. 3B), and at least three regions of c-Cbl bind to different domains of c-Src, including phosphorylated Tyr-418 (30). This interaction underlies short-term catalytic inactivation of c-Src, but long-term inactivation involves degradation of active Src family kinases by c-Cbl (31). In analogy, our results depict a reciprocal pattern in which c-Src negatively regulates c-Cbl. Nevertheless, the exact mechanism by which c-Src sorts c-Cbl for degradation is still unclear: c-Src increases ubiquitylation of c-Cbl both in vitro (17) and in living cells (Fig. 4C). Moreover, ubiquitylation of c-Cbl is likely mediated by its own RING finger, and it may require prior phosphorylation of a proximal tyrosine residue (Tyr-371) by either c-Src or EGFR (15, 17). However, unlike Src-induced ubiquitylation of c-Cbl, which requires an intact RING finger and a tyrosine at position 371, c-Cbl mutants defective at Tyr-371 or at the RING domain retain sensitivity to active Src proteins (Fig. 4A). It is conceivable, therefore, that c-Src sorts c-Cbl to proteasomal destruction by mobilizing both the ubiquitin ligase function of c-Cbl and a mechanism independent of the RING finger.
Src-transformed cells exhibit a variety of phenotypic characteristics, which may reflect the multiple phosphorylation targets of Src family kinases. These targets are involved in the regulation of cell cycle entry, actin cytoskeleton, and adhesive properties (2). Likewise, the pleiotropic cellular responses to growth factors like EGF resemble many characteristics of the Src-induced phenotype. Our present study explains this similarity by the ability of c-Src to block a major pathway leading to desensitization of growth factor signaling. Evidently, Src executes this function by enhancing destruction of c-Cbl, an evolutionary conserved regulator of receptor endocytosis. Interestingly, c-Src accelerates destruction of another negative regulator, namely protein kinase C-Δ (32), raising the possibility that blocking negative regulatory pathways may be a common feature of Src family kinases.
Acknowledgments
We thank Yaron Mosesson, Sara Lavi, Tona Gilmer, Wallace Langdon, Alexander Tsygankov, and Dirk Bohmann for plasmids. This work was supported by National Cancer Institute Grant CA72981 and U.S. Army Grant DAMD 17-00-1-0499.
Abbreviations
- EGF
epidermal growth factor
- EGFR
EGF receptor
- HA
hemagglutinin
References
- 1.Yarden Y, Sliwkowski M X. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Abram C L, Courtneidge S A. Exp Cell Res. 2000;254:1–13. doi: 10.1006/excr.1999.4732. [DOI] [PubMed] [Google Scholar]
- 3.Biscardi J S, Tice D A, Parsons S J. Adv Cancer Res. 1999;76:61–119. doi: 10.1016/s0065-230x(08)60774-5. [DOI] [PubMed] [Google Scholar]
- 4.Irby R B, Mao W, Coppola D, Kang J, Loubeau J M, Trudeau W, Karl R, Fujita D J, Jove R, Yeatman T J. Nat Genet. 1999;21:187–190. doi: 10.1038/5971. [DOI] [PubMed] [Google Scholar]
- 5.Tice D A, Biscardi J S, Nickles A L, Parsons S J. Proc Natl Acad Sci USA. 1999;96:1415–1420. doi: 10.1073/pnas.96.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson L K, Luttrell D K, Parsons J T, Parsons S J. Mol Cell Biol. 1989;9:1536–1544. doi: 10.1128/mcb.9.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maa M C, Leu T H, McCarley D J, Schatzman R C, Parsons S J. Proc Natl Acad Sci USA. 1995;92:6981–6985. doi: 10.1073/pnas.92.15.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weernink P A, Rijksen G. J Biol Chem. 1995;270:2264–2267. doi: 10.1074/jbc.270.5.2264. [DOI] [PubMed] [Google Scholar]
- 9.Ware M F, Tice D A, Parsons S J, Lauffenburger D A. J Biol Chem. 1997;272:30185–30190. doi: 10.1074/jbc.272.48.30185. [DOI] [PubMed] [Google Scholar]
- 10.Wilde A, Beattie E C, Lem L, Riethof D A, Liu S-H, Mobley W C, Soriano P, Brodsky F M. Cell. 1999;96:677–687. doi: 10.1016/s0092-8674(00)80578-4. [DOI] [PubMed] [Google Scholar]
- 11.Thien C B, Langdon W Y. Nat Rev Mol Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 12.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon W Y, Beguinot L, Geiger B, Yarden Y. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waterman H, Levkowitz G, Alroy I, Yarden Y. J Biol Chem. 1999;274:22151–22154. doi: 10.1074/jbc.274.32.22151. [DOI] [PubMed] [Google Scholar]
- 14.Klinghoffer R A, Sachsenmaier C, Cooper J A, Soriano P. EMBO J. 1999;18:2459–2471. doi: 10.1093/emboj/18.9.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levkowitz G, Waterman H, Ettenberg S A, Katz M, Tsygankov A Y, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, et al. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 16.Yokouchi M, Kondo T, Houghton A, Bartkiewicz M, Horne W C, Zhang H, Yoshimura A, Baron R. J Biol Chem. 1999;274:31707–31712. doi: 10.1074/jbc.274.44.31707. [DOI] [PubMed] [Google Scholar]
- 17.Yokouchi M, Kondo T, Sanjay A, Houghton A, Yoshimura A, Komiya S, Zhang H, Baron R. J Biol Chem. 2001;276:35185–35193. doi: 10.1074/jbc.M102219200. [DOI] [PubMed] [Google Scholar]
- 18.Tsygankov A Y, Mahajan S, Fincke J E, Bolen J B. J Biol Chem. 1996;271:27130–27137. doi: 10.1074/jbc.271.43.27130. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan K B, Swedlow J R, Varmus H E, Morgan D O. J Cell Biol. 1992;118:321–333. doi: 10.1083/jcb.118.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thien C B, Langdon W Y. Oncogene. 1997;14:2239–2249. doi: 10.1038/sj.onc.1201193. [DOI] [PubMed] [Google Scholar]
- 21.Feshchenko E A, Langdon W Y, Tsygankov A Y. J Biol Chem. 1998;273:8323–8331. doi: 10.1074/jbc.273.14.8323. [DOI] [PubMed] [Google Scholar]
- 22.Ettenberg S A, Magnifico A, Cuello M, Nau M M, Rubinstein Y R, Yarden Y, Weissman A M, Lipkowitz S. J Biol Chem. 2001;276:27677–27684. doi: 10.1074/jbc.M102641200. [DOI] [PubMed] [Google Scholar]
- 23.Waterman H, Katz M, Rubin C, Shtiegman K, Lavi S, Elson A, Jovin T, Yarden Y. EMBO J. 2002;21:303–313. doi: 10.1093/emboj/21.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka S, Amling M, Neff L, Peyman A, Uhlmann E, Levy J B, Baron R. Nature. 1996;383:528–531. doi: 10.1038/383528a0. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka S, Neff L, Baron R, Levy J B. J Biol Chem. 1995;270:14347–14351. doi: 10.1074/jbc.270.24.14347. [DOI] [PubMed] [Google Scholar]
- 26.Biscardi J S, Belsches A P, Parsons S J. Mol Carcinog. 1998;21:261–272. doi: 10.1002/(sici)1098-2744(199804)21:4<261::aid-mc5>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 27.Brunton V G, Ozanne B W, Paraskeva C, Frame M C. Oncogene. 1997;14:283–293. doi: 10.1038/sj.onc.1200827. [DOI] [PubMed] [Google Scholar]
- 28.Waterman H, Yarden Y. FEBS Lett. 2001;490:142–152. doi: 10.1016/s0014-5793(01)02117-2. [DOI] [PubMed] [Google Scholar]
- 29. Sanjay, A., Horne, W. C. & Baron, R. (2001) Sci. STKE2001, (110) E40. [DOI] [PubMed]
- 30.Sanjay A, Houghton A, Neff L, DiDomenico E, Bardelay C, Antoine E, Levy J, Gailit J, Bowtell D, Horne W C, Baron R. J Cell Biol. 2001;152:181–195. doi: 10.1083/jcb.152.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andoniou C E, Lill N L, Thien C B, Lupher M L, Jr, Ota S, Bowtell D D, Scaife R M, Langdon W Y, Band H. Mol Cell Biol. 2000;20:851–867. doi: 10.1128/mcb.20.3.851-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blake R A, Garcia-Paramio P, Parker P J, Courtneidge S A. Cell Growth Differ. 1999;10:231–241. [PubMed] [Google Scholar]