Abstract
Nuclear transport proceeds through nuclear pore complexes (NPCs) that are embedded in the nuclear envelope of eukaryotic cells. The Saccharomyces cerevisiae NPC is comprised of 30 nucleoporins (Nups), 13 of which contain phenylalanine-glycine repeats (FG Nups) that bind karyopherins and facilitate the transport of karyopherin-cargo complexes. Here, we characterize the structural properties of S. cerevisiae FG Nups by using biophysical methods and predictive amino acid sequence analyses. We find that FG Nups, particularly the large FG repeat regions, exhibit structural characteristics typical of “natively unfolded” proteins (highly flexible proteins that lack ordered secondary structure). Furthermore, we use protease sensitivity assays to demonstrate that most FG Nups are disordered in situ within the NPCs of purified yeast nuclei. The conclusion that FG Nups constitute a family of natively unfolded proteins supports the hypothesis that the FG repeat regions of Nups form a meshwork of random coils at the NPC through which nuclear transport proceeds.
The nuclear pore complex (NPC) spans the nuclear envelope of eukaryotes and serves as the conduit for all nucleocytoplasmic transport (1). In Saccharomyces cerevisiae, the NPC is composed of multiple copies of 30 different nucleoporins (Nups) that form an octagonal pore structure of 60 MDa measuring 95 nm in diameter and 35 nm in height (2). The FG family of Nups contain extensive regions of FG repeats (typically 200–700 aa in length) and facilitate the passage of karyopherin-cargo complexes through the NPC by binding directly to karyopherins (Kaps/importins/exportins/transportins) (3, 4), the soluble receptors that recognize nucleocytoplasmic transport signals within cargo (5).
Although the mechanism of Kap translocation through NPCs is poorly understood, Kap–FG Nup interactions are central to the process. It is therefore likely that the structures of FG Nups play important roles in directing the movement of Kap-cargo complexes as they traverse the NPC; however, little is known about the structure of the individual FG Nups within the NPC. The FG repeat region of the mammalian Nup p62 was initially hypothesized to adopt a β-conformation (6). More recent models of NPC organization have favored disordered conformations for the FG Nups (7, 8), despite a lack of conclusive evidence. In support of these models, Bayliss et al. (9) observed that a fragment of the Nsp1p FG repeat region (a yeast homolog of p62) lacks α-helical structure and may be disordered. Also, our recent biophysical analyses of the full-length S. cerevisiae FG Nup Nup2p characterized it as a natively unfolded protein (10). Natively unfolded proteins, such as Nup2p, lack significant α-helical and β-sheet secondary structure and exhibit high flexibility under physiological conditions. They also exhibit low overall hydrophobicity, high net charge, and low compactness/nonglobularity (11, 12). Here, we perform structural analyses of additional FG Nups to test whether unfolded structures are a conserved feature of all FG Nups.
Materials and Methods
Construction and Purification of Recombinant Proteins.
Recombinant Nups were expressed as GST fusions by using the vector pGEX-2TK (Amersham Biosciences) which incorporates a thrombin cleavage site at the fusion junction. The NUP2, NUP1 (codons 300-1076), YRB2, NUP100 (codons 1–640), NSP1 (codons 1–603), NUP159 (codons 441–881), and NUP85 genes were amplified from yeast genomic DNA (Promega) by PCR. The PCR products were ligated into pGEX-2TK and transformed into BL21 codon plus Escherichia coli (Novagen). Protein expression and purification were performed as described (10, 13). The GST portion of the GST-Nups was cleaved off with thrombin before the final purification of the Nups in gel filtration columns.
Gel Filtration and Calculation of Stokes Radii.
Purified proteins were resolved by FPLC in a Superose-6 gel filtration column (Amersham Biosciences) equilibrated with 20 mM Hepes (pH 6.8), 150 mM KOAc, and 2 mM Mg(OAc)2. The elution volumes (Ve) of proteins were determined by UV absorbance or collection of 0.5-ml fractions followed by SDS/PAGE and staining with Coomassie blue. The Stokes radii (RS) of proteins were calculated by using the method of Porath (14) as described (10).
Sucrose Gradients and Calculation of Sedimentation Coefficients.
Linear sucrose gradients (5–20% sucrose) were poured by using a gradient mixer (Hoefer), and recombinant Nups (5 μg) were layered on top. The gradients were subjected to centrifugation at 259,000 × g for 15 h at 4°C in a TLS-55 rotor (Beckman). Fractions were collected from the top of each gradient, and proteins were resolved by SDS/PAGE and visualized with Coomassie blue. Purified BSA (4.7 S) and aldolase (7.5 S) were used in parallel gradients as standards. Sedimentation coefficients (s20,w) were calculated by the method of McEwen (15) as described (10). Molecular mass estimates based on the s20,w and RS values of each Nup were calculated as described (14) by using the following equation:
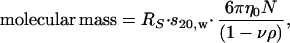 |
where η0 = the viscosity of the solvent (g/cm⋅s); ρ = the density of the solvent (g/cm3); ν = the partial specific volume of the protein (cm3/g); and N = Avogadro's number.
CD and Fourier Transform IR (FTIR) Spectroscopy.
Far-UV CD spectra of purified Nups were obtained on Aviv Associates (Lakewood, NJ) 60 DS and 62A DS spectrophotometers as described (10). Attenuated total reflectance data were collected on a Nicolet 800SX FTIR spectrometer equipped with a mercury cadmium telluride detector as described (10).
Proteinase K Digestions.
Nuclei were purified from a protease-deficient strain of S. cerevisiae as described (16) and resuspended to 4 mg/ml in 20 mM Hepes (pH 6.8), 150 mM KOAc, 250 mM sorbitol, 2 mM MgOAc, and 50% glycerol. Proteinase K (Sigma) was added at 300 ng/ml, and the reaction mixture was incubated at 37°C. Aliquots were removed at intervals, and digestions were quenched with SDS sample buffer plus 2 mM PMSF followed by heating at 95°C for 10 min. The fraction of full-length proteins remaining was detected by SDS/PAGE followed by Western blotting with specific antibodies and 125I-Protein A (Amersham Biosciences). Radioactive blots were exposed to phosphor screens, and the images were quantified by using imagequant software (Molecular Dynamics). Most of the specific polyclonal antibodies were generated in rabbits by using the GST fusions as antigens. Purified proteins (1 mg/ml) were digested with 100 ng/ml proteinase K and processed as above. Samples taken at intervals were resolved by SDS/PAGE and visualized by Coomassie blue.
Results
Amino Acid Composition Analyses of Nups.
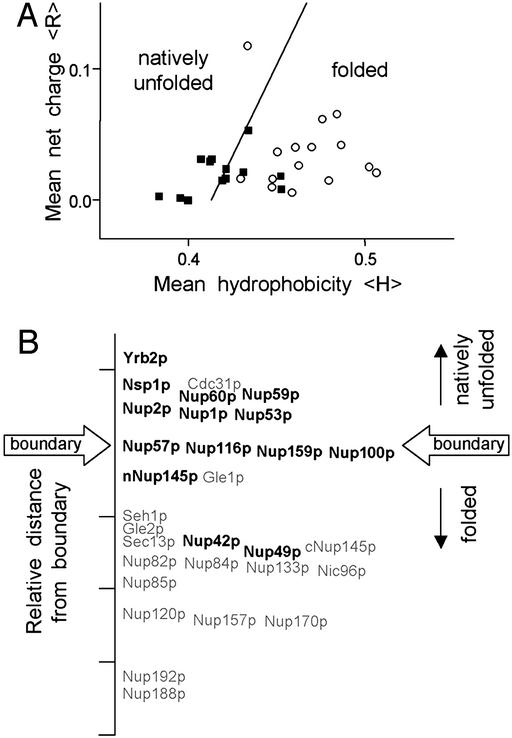
We initially examined the amino acid compositions of all S. cerevisiae Nups for indications of structural disorder by using predictive analyses described by Uversky et al. (11) and Dunker et al. (12). Natively unfolded proteins typically possess a high net charge at neutral pH and low overall hydrophobicity (11, 17). As a result, they generally occupy nonoverlapping regions of charge-hydrophobicity plots in comparison with folded proteins (11); of the 91 natively unfolded polypeptides examined in the original analysis, only three appeared outside of the “natively unfolded” region of the plot (11). A plot of the mean net charges and Kyte–Doolittle hydrophobicities for all S. cerevisiae Nups reveals two distinct populations (Fig. 1A). The relative distance of individual Nups from the folded-unfolded boundary is displayed in Fig. 1B. Most FG Nups cluster in the natively unfolded region of the plot or at the boundary, whereas all non-FG Nups (except Cdc31p) cluster in the folded region (Fig. 1A). Altogether, the charge-hydrophobicity plot indicates that the chemical properties of the FG Nups are similar to those of natively unfolded proteins.
Figure 1.
Computer analysis of the hydrophobicity and net charge of all S. cerevisiae Nups predicts that the FG Nups are unfolded in physiological conditions. (A) Charge versus hydrophobicity plot. The mean net charges and mean hydrophobicities (Kyte-Doolittle, 5-aa window) were calculated and plotted for FG Nups (■) and non-FG Nups (○) as described (11). The line, 〈H〉b = (〈R〉 + 1.151)/2.785, demarcates the observed boundary between folded and natively unfolded proteins in the charge-hydrophobicity plot. The relative distance to the boundary line for individual FG Nups (bold type) and non-FG Nups (gray type) is shown on a linear axis in B.
A database of known protein structures shows that disordered regions are significantly depleted of the amino acids I, L, V, W, F, Y, C, and N and enriched in the amino acids E, K, R, G, Q, S, P, and A (12). Hypothetically, hydrophobic amino acids in the former group contribute order to a protein, whereas charged and polar amino acids in the latter group contribute disorder. Table 1 lists the percent frequency of order- and disorder-conferring amino acids for each S. cerevisiae Nup in comparison with the S. cerevisiae proteome and ranks the Nups according to the difference in frequency of disorder and order amino acids. The non-FG Nups contain slightly more order (2.9%) and fewer disorder (2.4%) residues than the average yeast protein (Table 1). In contrast, FG Nups are significantly enriched in the disorder-conferring amino acids (Table 1), containing on average 9% more disorder amino acids and 8% fewer order amino acids in comparison with the yeast proteome. Analysis of only the FG regions of Nups shows a further bias of 11.5% for the disorder-conferring amino acids compared with the yeast proteome (Table 1). Also, Nups containing mostly the FxFG variant of the FG motif have more disorder amino acids than Nups containing mostly GLFG, PSFG, or xxFG motifs. This finding may be explained by the fact that intervening sequences between FxFG repeats frequently contain charged amino acids, whereas the sequences between GLFG repeats lack acidic residues and are enriched in asparagines (18).
Table 1.
Amino acid composition (% by frequency) of S. cerevisiae Nups
| Protein | FG repeats | % Order amino acids* (N, C, I, L, F, W, Y, V)
|
% Disorder amino acids† (A, R, Q, E, G, K, P, S)
|
||
|---|---|---|---|---|---|
| Full length | FG region | Full length | FG region | ||
| Nup2p | FxFG | 25.6 | 24.8 | 57.8 | 59.5 |
| Nsp1p | FxFG | 26.5 | 22.7 | 58.3 | 63.1 |
| Yrb2p | FxFG | 26.6 | ‡ | 57.9 | ‡ |
| Nup1p | FxFG | 27.0 | 20.8 | 56.6 | 59.9 |
| Nup60p | FxFx | 30.3 | 28.5 | 58.1 | 60.1 |
| Nup42p | PSFG | 29.6 | 28.7 | 56.2 | 56.4 |
| Nup57p | GLFG | 30.4 | 26.1 | 55.3 | 57.1 |
| Nup116p | GLFG | 32.1 | 30.3 | 55.3 | 58.1 |
| Nup159p | PSFG | 30.8 | 30.4 | 53.5 | 55.5 |
| Nup53p | xxFG | 34.0 | ‡ | 53.8 | ‡ |
| Nup49p | GLFG | 33.2 | 30.1 | 52.2 | 58.4 |
| nNup145p | GLFG | 33.6 | 32.9 | 51.6 | 53.8 |
| Gle1p | 35.3 | 52.7 | |||
| Nup100p | GLFG | 35.9 | 34.8 | 50.4 | 53.4 |
| Gle2p | 34.6 | 47.9 | |||
| Seh1p | 34.7 | 48.0 | |||
| Cdc31p | 35.4 | 45.4 | |||
| Sec13p | 37.2 | 46.8 | |||
| Nup59p | xxFG | 38.2 | ‡ | 46.0 | ‡ |
| cNup145 | 40.0 | 46.5 | |||
| Pom34p | 37.8 | 43.2 | |||
| Pom152p | 40.0 | 44.7 | |||
| Nup170p | 41.4 | 44.0 | |||
| Nup157p | 41.9 | 44.3 | |||
| Nup82p | 41.8 | 43.7 | |||
| Nic96p | 42.7 | 43.5 | |||
| Nup85p | 42.2 | 42.4 | |||
| Nup84p | 43.4 | 43.0 | |||
| Ndc1p | 43.0 | 41.0 | |||
| Nup192p | 46.1 | 39.3 | |||
| Nup133p | 46.1 | 39.1 | |||
| Nup188p | 46.7 | 38.9 | |||
| Nup120p | 47.8 | 36.7 | |||
| S. cerevisiae proteome | 38.1 | 46.0 | |||
| FG Nup avg. | 30.9 | 54.5 | |||
| Non-FG Nup avg. | 41.0 | 43.6 | |||
| FG regions only avg. | 28.2 | 57.8 | |||
Amino acid composition is calculated as percent frequency of the primary sequence.
Amino acids typically depleted in disordered regions of proteins (12).
Amino acids typically enriched in disordered regions of proteins (12).
The FG regions of Nup59p, Nup53p, and Yrb2p are too small or dispersed for this analysis.
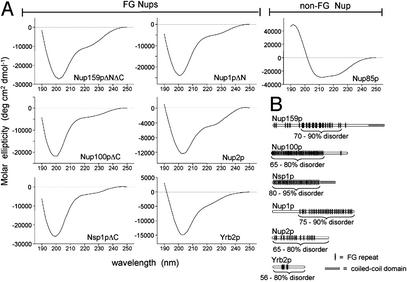
The analyses above predict that all FG Nups are unstructured under physiological conditions, as is the case for Nup2p and a portion of the Nsp1p FG region (9, 10). To test this prediction experimentally, we characterized the biophysical properties of additional recombinant FG regions of yeast Nups (Nup159pΔNΔC, Nup100pΔC, Nsp1pΔC, Nup1pΔN, and Yrb2p) and one non-FG Nup (Nup85p, as a control) by gel filtration, sucrose gradients, CD spectroscopy, and FTIR spectroscopy. Yrb2p is not technically a Nup, but it is included in this study because (i) it is similar to Nup2p (19), (ii) it contains characteristic FG repeats, and (iii) it binds karyopherins (20). The Nup1p, Nup100p, Nup159p, and Nsp1p truncations characterized here (see Fig. 3B) lack their respective N-terminal (ΔN) NPC targeting or C-terminal (ΔC) coiled-coil domains (21, 22), yet all include extensive FG repeat regions. The C-terminal truncations of coiled-coil regions were necessary for expression of the Nups in soluble form in E. coli (23), and the N-terminal truncations were necessary for sufficient yields for purification. In the case of Nup100p, the soluble full-length protein was also purified, and its analysis gave results similar to those shown here for Nup100pΔC (data not shown). For simplicity, we refer collectively to the FG repeat regions of Nups as FG Nups. In all cases, the recombinant FG Nups and Nup fragments used here are active in binding karyopherins (4, 24, 25).
Figure 3.
(A) Far-UV CD measurements of purified Nups or Nup fragments show extensive regions of structural disorder in FG Nups. Shown are the CD spectra of purified recombinant proteins. The portion of each Nup used is noted by brackets in B. All spectra were obtained at 25°C at neutral pH in 1-mm cuvettes as described (10). Protein concentrations ranged from 0.05 to 0.2 mg/ml. (B) Diagram of FG Nups analyzed. The portion of each FG Nup that was purified and analyzed is bracketed; in the case of Yrb2p and Nup2p, the full-length protein was characterized. The content of disordered structure as determined by FTIR analysis (see Table 3) is given as a percentage of the fragment analyzed.
FG Nups Exhibit Large Hydrodynamic Dimensions.
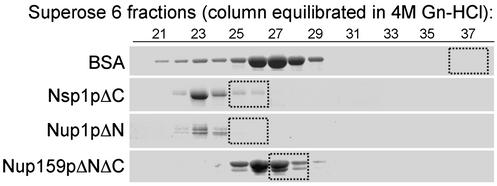
Natively unfolded proteins exhibit large hydrodynamic dimensions and slow sedimentation behavior characteristic of nonglobular structures with low compactness (12, 17, 26). Gel filtration and velocity sedimentation in sucrose gradients were used here to calculate the Stokes radii (RS) and sedimentation coefficients (s20,w) of the purified Nups. We found that all of the FG Nups yield very large RS (54–75 Å) and small s20,w (2.2–3.2 S) values (Table 2). Table 2 also lists the RS and s20,w values of three previously analyzed full-length FG Nups (Nup2p, Nup49p, and Nup57p); these also exhibit large hydrodynamic dimensions (10, 23). For reference, the monomeric globular protein BSA (66 kDa) is roughly equivalent in mass to the FG Nups, yet it sediments faster in sucrose gradients (s20,w = 4.7 S) and has smaller hydrodynamic dimensions in gel filtration columns (RS = 35.5 Å). In contrast with the FG Nups, the non-FG Nup85p possesses RS (51 Å) and s20,w (4.6 S) values indicative of higher compaction and globularity (Table 2). The molecular mass of each Nup was estimated by using the RS and s20,w values from Table 2 as described in Materials and Methods, and the results indicate that the purified recombinant Nups are monomeric (Table 2). This finding implies that the large Stokes radii of recombinant FG Nups in gel filtration columns are not caused by oligomerization or aggregation. To confirm this experimentally, we compared the elution profiles of Nsp1pΔC, Nup1pΔN, and Nup159pΔNΔC in a Superose 6 gel filtration column in physiological buffer and in 4 M Gn-HCl. As expected for unstructured proteins, the elution profiles were similar for FG Nups under native and denaturing conditions (Fig. 2). In contrast, the elution volume of BSA changed significantly, reflecting the conversion from globular to unfolded structure in the chaotroph (Fig. 2).
Table 2.
Stokes radii (RS) and sedimentation efficients (S20,w) of recombinant Nups
| Protein | Predicted molecular mass, kDa* | Stokes radius RS (Å) ± 5%† | Sed. coef. (×10−13 s−1) s20,w ± 7%† | Calculated molecular mass, kDa‡ | Source/ref. |
|---|---|---|---|---|---|
| Nup2p | 78.0 | 79 | 2.9 | 89 | 10 |
| Nup1pΔN | 77.1 | 75 | 2.8 | 81 | This study |
| Yrb2p | 36.1 | 54 | 2.2 | 47 | This study |
| Nup100pΔC | 64.5 | 59 | 3.2 | 67 | This study |
| Nsp1pΔC | 61.9 | 74 | 2.8 | 76 | This study |
| Nup159pΔNΔC | 44.7 | 61 | 2.6 | 59 | This study |
| Nup49p | 49.1 | 41¶ | 1.4 | 36§ | 23 |
| Nup57p | 57.5 | 47¶ | 2.2 | 55§ | 23 |
| Nup85p | 84.9 | 51 | 4.6 | 98 | This study |
RS values were derived from Superose 6 gel filtration data by using the method of Porath (14) as described (10). s20,w values were calculated from linear sucrose gradients (5–20% sucrose) by using the method of McEwen (15) as described (10).
Predicted molecular mass for monomeric proteins.
Estimated error.
Molecular mass calculated from RS and s20,w values as described (14).
Molecular mass determined by equilibrium sedimentation (23).
RS values extrapolated from published gel filtration data (23).
Figure 2.
Elution profiles of FG Nups in a Superose 6 gel filtration column are similar in the presence and absence of 4 M Gn-HCl. Purified FG Nups and BSA (≈100 μg each) were separated by size in a Superose-6 column (24 ml) equilibrated with 4 M Gn-HCl; 0.5-ml fractions were collected, and the eluted proteins were precipitated with trichloroacetic acid, separated by SDS/PAGE, and stained with Coomassie blue. Boxes identify the elution peak of the protein separated in the same Superose 6 column equilibrated with physiological buffer [20 mM Hepes, pH 6.8/150 mM KOAc/2 mM Mg(OAc)2].
Large hydrodynamic dimensions are typical of natively unfolded proteins as well as fibrous proteins that oligomerize to form extended coiled-coil domains. The monomeric state of the purified FG Nups clearly distinguishes them from fibrous proteins, which typically form homodimers (27). Moreover, the Nup fragments analyzed here do not contain predicted coiled-coil domains when analyzed with MACSTRIPE 2.0 software, nor do they contain α-helical structure (see below). Instead, the experimentally determined RS values for the FG Nups compare favorably to theoretical RS values predicted for monomeric proteins in natively unfolded conformations (Table 4, which is published as supporting information on the PNAS web site, www.pnas.org) (17).
Secondary Structure of FG Nups.
Natively unfolded proteins lack ordered secondary structure (α-helix and β-sheet) and yield CD and FTIR spectra distinguishable from folded proteins (17, 26). Far-UV CD measurements of purified FG Nups produce spectra characteristic of unfolded proteins (Fig. 3A). Each FG Nup has an intensive minimum near 200 nm and low ellipticity at 222 nm that reflect extensive contributions of unstructured coil and a lack of α-helical structure. These CD spectra are similar to those reported for Nup2p and a fragment of the FG region of Nsp1p (9, 10). By contrast, the non-FG Nup85p yields minima at 208 and 222 nm that indicate significant contributions of α-helix secondary structures (Fig. 3A).
Because FTIR spectroscopy is more sensitive to β-sheet structure than CD, we also generated FTIR spectra (amide I region) for each FG Nup. These spectra show broad absorbance bands near 1,655 cm−1 corresponding to disordered conformations (Table 3 and Fig. 5, which is published as supporting information on the PNAS web site). Deconvolution (Fourier self-deconvolution and second derivative) and curve fitting of the FTIR spectra permitted quantitative analyses of secondary structure content, confirming that purified FG Nups are composed mostly (≈80%) of disordered structure (Table 3; Fig. 3B). In comparison, Nup85p shows a high propensity for α-helical structure and contains much less disorder (18–35%) (Table 3).
Table 3.
Secondary structure analysis of Nups determined by FTIR spectroscopy
| Structure assignment | Wavenumbers, cm−1 | % Contribution of secondary structure
|
||||||
|---|---|---|---|---|---|---|---|---|
| FG Nups
|
Non-FG Nup
|
|||||||
| Nup2p | Nup1pΔN | Nsp1pΔC | Yrb2p | Nup100pΔC | Nup159pΔNΔC | Nup85p | ||
| Turns | 1682–1689 | 11.8 | 5.1 | 3.8 | 4.9 | 4.4 | 7.4 | 5.3 |
| Loops | 1667–1677 | 22.2 | 18.8 | 33.6 | 18.4 | 20.8 | 23.8 | 22.8 |
| Loops/disordered | 1653–1656 | 42.6 | 58.8 | 47.4 | 37.5 | 42.8 | 47.6 | 49.5† |
| Disordered/β-sheet* | 1634–1640 | 16.2 | 13.7 | 12.5 | 24.4 | 30.8 | 18.2 | 18.1 |
| β-sheet | 1621–1625 | 7.2 | 3.6 | 2.7 | 14.8 | 1.2 | 3.0 | 9.6 |
| Total disorder content | 65–80 | 75–90 | 80–95 | 56–80 | 65–95 | 70–90 | 18–35 | |
Secondary structure content was determined from curve fitting to spectra deconvoluted by using second derivatives and Fourier self-deconvolution to identify component band positions as described (10). Structure assignments are given as percent contribution of total protein secondary structure. The estimated error in the frequencies is ±1.5 cm−1. Source spectra are shown in Table 4.
The peak reflects disordered secondary structure or a combination of extended and disordered structures.
In Nup85p, this peak corresponds to α-helix structures.
Protease Sensitivity of FG Nups Within NPCs.
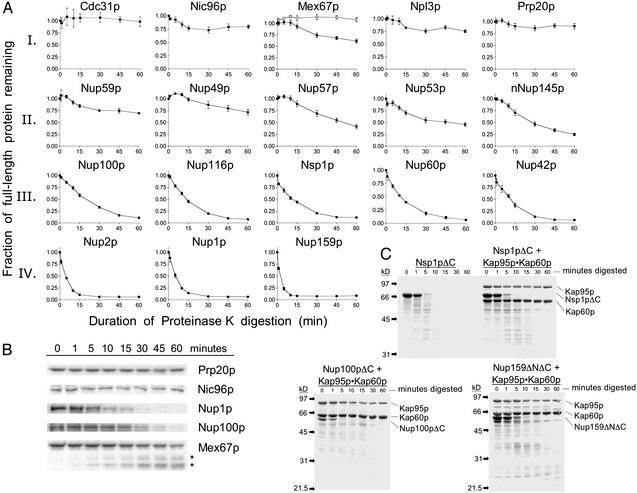
Because of their low compactness and lack of secondary structure, natively unfolded proteins exhibit high flexibility that can impart hypersensitivity to protease digestion (10, 17). To assay the flexibility of yeast Nups within NPCs embedded in nuclear envelopes, we treated purified S. cerevisiae nuclei (16) with low concentrations of proteinase K (300 ng/ml), a protease that is small enough (29 kDa) to diffuse across the yeast NPC. Under these mild conditions, proteinase K is useful in identifying stable, folded domains of proteins. The degradation profiles of many nuclei-associated proteins were quantified by using Western blots with specific antibodies and 125I-Protein A. The protein degradation patterns observed could be grouped into four (I–IV) categories (Fig. 4). We previously showed that the karyopherins Kap95p and Kap60p (importins β and α), and the NPC proteins Nup85p and Pom152p, are insensitive to this proteinase K treatment and are representative of category I proteins, although both Kap60p and Nup85p lost ≈2 kDa from their respective N termini (10). This finding was expected as Kap95p, Kap60p, and Nup85p are folded proteins composed almost entirely of α-helices (Fig. 3) (28, 29). The karyopherins Kap121p and Kap123p are also fully resistant to proteinase K (data not shown). Here, we observed that the non-FG Nups Cdc31p and Nic96p, the mRNA export factor Mex67p, the nucleoplasmic shuttling protein Npl3p, and the chromatin-associated Gsp1p exchange factor Prp20p are similarly insensitive to the protease treatment (Fig. 4A). Minor digestion of Mex67p was observed; however, the disappearance of full-length Mex67p protein (Fig. 4A) correlates with the appearance of two stable Mex67p fragments of slightly faster mobility (Fig. 4B). This result indicates that Mex67p can be accessed by proteinase K and clipped, but is mostly resistant to this protease treatment.
Figure 4.
FG Nups within NPCs in purified nuclei are hypersensitive to protease digestion. (A) Purified nuclei (4 mg/ml) were digested with proteinase K (300 ng/ml) for the indicated times at 37°C. Aliquots were mixed with SDS buffer with 2 mM PMSF to stop the digestion. The degradation of specific proteins was quantified by using Western blots probed with specific antibodies and 125I-Protein A. For each protein, the fraction of full-length protein remaining is shown. Each graph represents the average of three to six independent digestions; error bars represent the SEM. The top line in Mex67p represents the sum of full-length and clipped Mex67p (*); the bottom line represents full-length Mex67p. (B) Representative 125I-Protein A Western blots used to generate graphs shown in A. Note that in the case of Mex67p, the full-length protein is clipped to yield two large, stable fragments (*). (C) Purified FG repeat regions of Nups (1 mg/ml) with and without stoichiometric amounts of Kap95p–Kap60p were incubated with proteinase K (100 ng/ml) for the indicated times at 37°C. Samples were processed as in A, resolved by SDS/PAGE, and visualized by Coomassie blue staining.
In contrast, most FG Nups in nuclei are fully proteolysed within 60 min (category III) or 15 min (category IV) (Fig. 4A). Nups localized to the nuclear basket structure of the NPC (Nup60p, Nup1p, and Nup2p) and to cytoplasmic fibrils (Nup159p and Nup42p) (30–32) are the most sensitive to proteinase K (Fig. 4A). Some FG Nups located at the center of the NPC (Nup57p, Nup53p, and nNup145p) (30) degrade after a lag period of 10 min (category II), suggesting that these Nups also contain highly flexible structures but are not immediately accessible to the protease (Fig. 4A). Finally, Nup49p and Nup59p are the least protease sensitive of the FG Nups (Fig. 4A). Importantly, we did not detect stable fragments of the FG Nups (>15 kDa) in the Western blots (data not shown).
In the above experiments, the protease sensitivity of a protein depends on two factors: the flexibility of its structure and its accessibility to the protease. To control for differences in protease accessibility within the NPC, we correlated the degradation profiles of nuclei-associated proteins with those of purified, recombinant versions of the same proteins. Coomassie-stained gels of protease digestions of purified FG Nups with and without Kap95p-Kap60p confirm that FG Nups are rapidly and completely degraded, whereas Kap95p and Kap60p are protease resistant (Fig. 4C) (10). Of the FG Nups tested, only purified Yrb2p shows partial protease resistance, yielding a stable digestion product of ≈14 kDa (data not shown). This fragment encompasses the Gsp1p binding domain of Yrb2p, which is predicted to form a β-sheet structure (19). We also performed proteinase K digestions of recombinant Nup85p and observed that it is largely resistant to proteinase K, yielding a fragment of ≈83 kDa (data not shown). An identical pattern of degradation was observed with endogenous Nup85p in nuclei digested under similar conditions (10). In addition, purified Sec13p remains intact, and Nup120p and cNup145p yield large, resistant fragments after proteinase K treatment (data not shown).
Proteinase K digestions of SDS-denatured proteins and nuclei were also conducted. In the case of Nsp1p, we observed no significant difference in the rate or pattern of degradation for the purified protein, and only a minor increase in its rate of degradation in nuclei after the SDS treatment (data not shown). In contrast, Nup85p and Kap95p (normally category I proteins) degrade rapidly after the SDS pretreatment, resembling category IV proteins (data not shown). These results confirm that the unstructured characteristics of FG Nups, and not merely their location within the NPC, are responsible for their sensitivity to proteinase K in the experiments with purified nuclei.
Discussion
Purified FG Nups exhibit the biophysical and structural characteristics of natively unfolded proteins as indicated by gel filtration, sucrose gradient, CD and FTIR spectroscopy, and protease digestion experiments (Tables 2 and 3; Figs. 2–4). The large FG regions of Nups are also highly disordered in situ in their native environment within NPCs (Fig. 4), implying that FG Nups exist and function in a disordered state. Although it remains inherently possible that FG Nups possess unconventional structural motifs not detectable by the methods used here, such structures must be flexible and disordered, or in rapid transition between ordered and unstructured states, to account for their protease hypersensitivity (Fig. 4).
Despite their large content of disordered structure, specific domains of FG Nups fold on incorporation into NPCs. Indeed, Nsp1p, Nup57p, and Nup49p form a complex via coiled-coil interactions between their C termini (21, 23). Nup159p and Nup42p also use their C termini to form putative coiled-coil interactions with Nup82p (33). However, these interaction domains are small in comparison to the FG repeat regions (Fig. 3B). Based on the hypersensitivity of nuclei-associated FG Nups to proteinase K (Fig. 4), we speculate that their large FG regions retain considerable disorder and flexibility at the NPC, whereas their non-FG regions are responsible for structural interactions via coiled-coils or other folded domains.
Based on the relative abundance of FG Nups in the NPC (30) and the size of their FG regions (typically 200–700 aa), we estimate that up to 30% of the yeast NPC mass may be composed of extensive regions of unstructured protein. These data support the hypothesis that FG Nups form a flexible and amorphous meshwork of filaments at the NPC that surrounds or engulfs macromolecules of heterogeneous shapes and sizes during transport (7, 8). In principle, the density of random coils in this meshwork could dictate the size exclusion limit of the NPC permeability barrier. Molecules smaller than the average distance between random coils would easily diffuse across the meshwork. For macromolecules larger than the size exclusion limit, the intertwined collection of unstructured FG regions would form an entropic barrier impermeable to proteins that cannot interact with FG Nups (30). In contrast, karyopherin–cargo complexes, which are larger than the size exclusion limit, likely gain access to the NPC by binding to FG Nups. They may then traverse the meshwork via a stochastic mechanism of repeated associations and dissociations with FG repeats (3).
The unordered structure of FG Nups may be intimately related to the mechanism of nuclear transport. Natively unfolded proteins exhibit biochemical features that may be important for FG Nup function at the NPC. These features include (i) multiple domains that allow simultaneous interactions with multiple binding partners (17, 34), (ii) nonrigid binding domains that can accommodate a variety of interacting partners (including different members of a protein family) (12, 26), and (iii) fast molecular association and dissociation rates (12). Indeed, FG Nups display all three characteristics, as they interact simultaneously with multiple binding partners (e.g., Nup2p in the tetrameric Nup60p–Gsp1p–Nup2p–Kap60p complex) (13), bind many different members of the karyopherin family (4), and exhibit fast association and dissociation rates with Kaps (35).
Despite considerable sequence divergence among the S. cerevisiae FG Nups, the overall amino acid composition that results in structural disorder appears to be well conserved (Table 1), highlighting its significance to FG Nup function. Paradoxically, the phenylalanine residue so prevalent in FG Nups is not a disorder-conferring amino acid, but is in fact order-conferring (12). The selective pressure to retain a high frequency of this residue is explained by the observation that phenylalanines in FG repeats are the key binding determinant in Kap–Nup interactions (36).
We suspect that structural disorder is conserved in mammalian FG Nups. In the case of p62, a far-UV CD analysis indicates that 47% of its structure is random coil (6). Also, a p62-containing complex exhibits hydrodynamic dimensions (s20,w = 7.0 S; RS = 81 Å) larger than predicted (11.5 S; 52 Å) for a globular complex of its mass (234 kDa) (37). However, a preliminary charge-hydrophobicity analysis of human FG Nups reveals a higher mean hydrophobicity than yeast FG Nups (data not shown), making it likely that mammalian FG Nups possess more domains with folded structure than their yeast orthologs. Nevertheless, most mammalian FG Nups (e.g., hNup153, p62, hNup214/CAN, hNup358, and hNup98) contain a high frequency of disorder-conferring amino acids similar to their yeast counterparts (data not shown). It is therefore likely that the FG regions of human Nups are natively unfolded as well.
Supplementary Material
Abbreviations
- NPC
nuclear pore complex
- Nup
nucleoporin
- FTIR
Fourier transform IR
References
- 1.Rout M, Aitchison J. J Biol Chem. 2001;276:16593–16596. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- 2.Yang Q, Rout M P, Akey C W. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 3.Rexach M, Blobel G. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 4.Allen N, Huang L, Burlingame A, Rexach M. J Biol Chem. 2001;276:29268–29274. doi: 10.1074/jbc.M102629200. [DOI] [PubMed] [Google Scholar]
- 5.Gorlich D, Kutay U. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 6.Buss F, Kent H, Stewart M, Bailer S M, Hanover J A. J Cell Sci. 1994;107:631–638. doi: 10.1242/jcs.107.2.631. [DOI] [PubMed] [Google Scholar]
- 7.Ribbeck K, Gorlich D. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macara I G. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayliss R, Kent H M, Corbett A H, Stewart M. J Struct Biol. 2000;131:240–247. doi: 10.1006/jsbi.2000.4297. [DOI] [PubMed] [Google Scholar]
- 10.Denning D P, Uversky V, Patel S S, Fink A L, Rexach M. J Biol Chem. 2002;277:33447–33455. doi: 10.1074/jbc.M203499200. [DOI] [PubMed] [Google Scholar]
- 11.Uversky V N, Gillespie J R, Fink A L. Proteins Struct Funct Genet. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 12.Dunker A K, Lawson J D, Brown C J, Williams R M, Romero P, Oh J S, Oldfield C J, Campen A M, Ratliff C M, Hipps K W, et al. J Mol Graphics Modeling. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 13.Denning D, Mykytka B, Allen N, Huang L, Burlingame A, Rexach M. J Cell Biol. 2001;154:937–950. doi: 10.1083/jcb.200101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel L M, Monty K J. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 15.McEwen C R. Anal Biochem. 1967;20:114–149. doi: 10.1016/0003-2697(67)90271-0. [DOI] [PubMed] [Google Scholar]
- 16.Aris J P, Blobel G. Methods Enzymol. 1991;194:735–749. doi: 10.1016/0076-6879(91)94056-i. [DOI] [PubMed] [Google Scholar]
- 17.Uversky V N. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rout M P, Wente S R. Trends Cell Biol. 1994;4:357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 19.Dingwall C, Kandels-Lewis S, Seraphin B. Proc Natl Acad Sci USA. 1995;92:7525–7529. doi: 10.1073/pnas.92.16.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taura T, Krebber H, Silver P A. Proc Natl Acad Sci USA. 1998;95:7427–7432. doi: 10.1073/pnas.95.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailer S M, Balduf C, Hurt E. Mol Cell Biol. 2001;21:7944–7955. doi: 10.1128/MCB.21.23.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Priore V, Heath C, Snay C, MacMillan A, Gorsch L, Dagher S, Cole C. J Cell Sci. 1997;110:2987–2999. doi: 10.1242/jcs.110.23.2987. [DOI] [PubMed] [Google Scholar]
- 23.Schlaich N L, Haner M, Lustig A, Aebi U, Hurt E C. Mol Biol Cell. 1997;8:33–46. doi: 10.1091/mbc.8.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L, Baldwin M A, Maltby D A, Medzihradszky K F, Baker P R, Allen N, Rexach M, Edmondson R D, Campbell J, Juhasz P, et al. Mol Cell Proteomics. 2002;1:434–450. doi: 10.1074/mcp.m200027-mcp200. [DOI] [PubMed] [Google Scholar]
- 25.Allen N P, Patel S S, Lutzmann M, Hurt E, Huang L, Burlingame A, Rexach M. Mol Cell Proteomics. 2002;1:930–946. doi: 10.1074/mcp.t200012-mcp200. [DOI] [PubMed] [Google Scholar]
- 26.Uversky V N. Eur J Biochem. 2002;269:1–10. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 27.Cohen C, Parry D A D. Proteins Struct Funct Genet. 1990;7:1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- 28.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 29.Cingolani G, Petosa C, Weis K, Muller C W. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 30.Rout M P, Aitchison J D, Suprapto A, Hjertaas K, Zhao Y, Chait B T. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hood J K, Casolari J M, Silver P A. J Cell Sci. 2000;113:1471–1480. doi: 10.1242/jcs.113.8.1471. [DOI] [PubMed] [Google Scholar]
- 32.Solsbacher J, Maurer P, Vogel F, Schlenstedt G. Mol Cell Biol. 2000;20:8468–8479. doi: 10.1128/mcb.20.22.8468-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belgareh N, Snay-Hodge C, Pasteau F, Dagher S, Cole C N, Doye V. Mol Biol Cell. 1998;9:3475–3492. doi: 10.1091/mbc.9.12.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dyson H J, Wright P E. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 35.Gilchrist D, Mykytka B, Rexach M. J Biol Chem. 2002;277:18161–18172. doi: 10.1074/jbc.M112306200. [DOI] [PubMed] [Google Scholar]
- 36.Bayliss R, Littlewood T, Stewart M. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 37.Guan T, Muller S, Klier G, Pante N, Blevitt J M, Haner M, Paschal B, Aebi U, Gerace L. Mol Biol Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.