SUMMARY
The C. elegans somatic gonadal precursor cell (SGP) divides asymmetrically to establish gonad-specific coordinates in both sexes. In addition, the SGP division is sexually dimorphic and initiates sex-specific programs of gonadogenesis. Wnt/MAPK signaling determines the gonadal axes, and the FKH-6 transcription factor specifies the male mode of SGP division. In this paper, we demonstrate that C. elegans cyclin D controls POP-1/TCF asymmetry in the SGP daughters as well as fkh-6 and rnr expression in the SGPs. Although cyclin D mutants have delayed SGP divisions, the cyclin D defects are not mimicked by other methods of retarding the SGP division. We find that EFL-1/E2F has an antagonistic effect on fkh-6 expression and gonadogenesis, which is relieved by cyclin D activity. We propose that cyclin D and other canonical regulators of the G1/S transition coordinate key regulators of axis formation and sex determination with cell cycle progression to achieve the sexually dimorphic SGP asymmetric division.
Keywords: Sex determination, gonadogenesis, cell cycle, cyclin D, C. elegans
INTRODUCTION
Asymmetric cell divisions are a widespread mechanism for generating diverse cell types during animal development (Betschinger and Knoblich, 2004). Model asymmetric divisions include those of the C. elegans zygote, the C. elegans EMS blastomere and the Drosophila neuroblast and sensory organ precursor (Ahringer, 2003; Bei et al., 2002; Cowan and Hyman, 2004; Doe and Bowerman, 2001; Roegiers and Jan, 2004). We have embarked on an in depth analysis of a different asymmetric division – that of the somatic gonadal precursor cell (SGP) in C. elegans. This division establishes the proximal-distal axis of the gonad of both sexes, and it is sexually dimorphic (Kimble and Hirsh, 1979). By teasing apart the molecular regulators of the SGP division, we will learn how precursor cells establish an organ coordinate system and how asymmetric divisions can be modulated during development to generate distinct organs.
The C. elegans embryo generates a four-celled gonadal primordium that appears the same in XX hermaphrodites and XO males (Kimble and Hirsh, 1979; Mathies et al., 2004; Sulston et al., 1983). Within the primordium, one SGP resides at each of two opposite poles, and two germline precursors lie between (Figure 1A). During the first larval stage (L1), the SGP divides asymmetrically in both sexes to generate proximal and distal daughters that establish gonadal axes. The SGP division is also sexually dimorphic with respect to both size and fate of its daughters. The hermaphrodite SGP makes distal and proximal daughters of roughly equal size, but the male SGP produces a smaller distal daughter and a larger proximal daughter (Figure 1A). In addition, the SGP daughters exhibit sex-specific behaviors (e.g. migration) (Figure 1A) and generate sex-specific regulatory cells that control gonad elongation and germline proliferation (Figure 1B). Therefore, the SGP asymmetric division initiates sex-specific programs of gonadogenesis that generate a double-armed ovotestis in hermaphrodites and a single-armed testis in males (Figure 1B).
Figure 1. The q626 mutation affects hermaphrodite and male gonadogenesis.
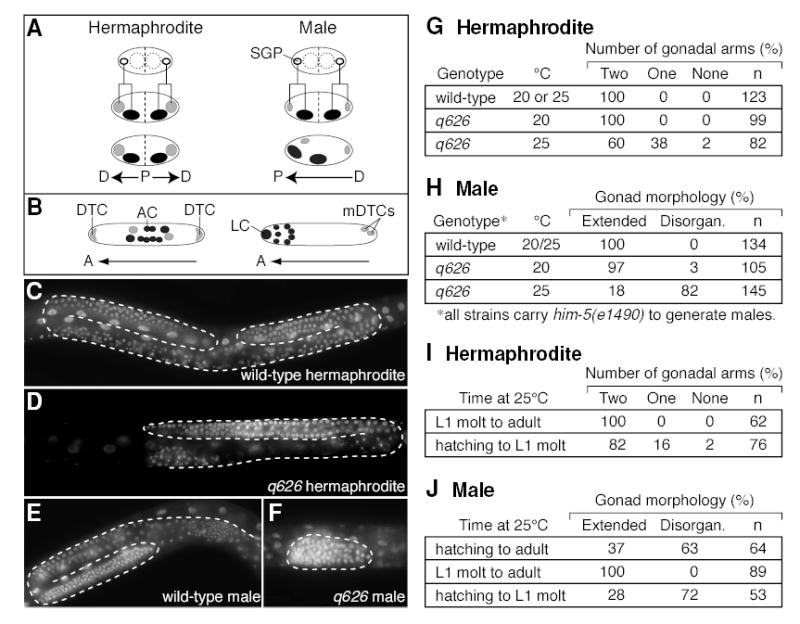
(A) Early events in hermaphrodite and male gonadogenesis. Top diagrams, four-celled gonadal primordium. SGPs (open circles); germline precursors (dashed circles). Middle diagrams, the SGPs divide asymmetrically to produce proximal (dark) and distal (light) daughters. Hermaphrodite proximal and distal daughters are of roughly equal size and do not migrate, creating two proximal-distal (P-D) axes. Male proximal daughters are larger than their distal sisters, and they migrate anteriorly to create a single P-D axis. (B) Gonadal regulatory cells in the L2/L3 developing somatic gonad. Left, hermaphrodite; right, male. Cells derived from distal SGP daughter, light grey; cells from proximal SGP daughter, dark grey. A, Anterior. In hermaphrodites, the distal tip cells (DTCs) control both arm elongation and germline proliferation, and the anchor cell (AC) induces vulval development. In males, the linker cell (LC) controls elongation of the single gonadal arm, and male distal tip cells (mDTCs) control germline proliferation. (C–F) DAPI-stained L4s grown at 25°C. Dashed line, boundaries of gonadal tissue. (C) Wild-type hermaphrodite gonad with two U-shaped gonad arms. (D) q626 hermaphrodite gonad with one gonadal arm. (E) Wild-type male with one J-shaped gonad arm. (F) q626 male with non-extended, disorganized gonad. (G–J) Tables quantifying the penetrance of gonadal defects. Disorgan., disorganized. (I–J) Temperature shift experiments with q626 mutants.
Two major pathways of regulation converge to regulate the SGP division. The first is gender neutral: the Wnt/MAPK pathway specifies the distal SGP daughter fate in both sexes (Kidd et al., 2005; Siegfried and Kimble, 2002; Siegfried et al., 2004; Sternberg and Horvitz, 1988). The terminal regulators of this pathway are POP-1/TCF and SYS-1/β-catenin, a DNA-binding protein and its transcriptional co-activator respectively (Kidd et al., 2005; Lin et al., 1995). In the early embryo, activated Wnt/MAPK signaling promotes nuclear export of POP-1 so that the daughter cell receiving the Wnt signal has less nuclear POP-1 than its sister, a phenomenon called POP-1 asymmetry (Lo et al., 2004; Maduro et al., 2002). A similar situation is observed after the SGP division: the distal daughter is specified by Wnt/MAPK activation and contains less nuclear POP-1 than its proximal sister (Siegfried et al., 2004). In mutants lacking POP-1, SYS-1 or upstream components of the Wnt/MAPK pathway, distal-specific cells are not made and extra proximal-specific cells are sometimes seen (Miskowski et al., 2001; Siegfried and Kimble, 2002; Siegfried et al., 2004; Sternberg and Horvitz, 1988); by contrast, gonads with excess SYS-1 produce extra distal cells and lack proximal cells (Kidd et al., 2005). Therefore, the Wnt/MAPK pathway establishes the proximal-distal axes of both hermaphrodite and male gonads.
The second pathway controls the sexual dimorphism of the SGP division. Most important for this work are two transcription factors. FKH-6 is a forkhead transcription factor that specifies the male-specific SGP division during the first larval stage of development (Chang et al., 2004). FKH-6 controls sex determination specifically in the SGPs and does not affect sex determination in other tissues or at other times of gonadal development; however, it does have a second and more poorly-defined late larval role in hermaphrodite gonadogenesis (Chang et al., 2004). TRA-1 is the C. elegans GLI transcription factor that acts in virtually all tissues to specify the female fate (Hodgkin and Brenner, 1977; Mathies et al., 2004; Zarkower and Hodgkin, 1992). The XO gonad is feminized and disorganized in fkh-6 null mutants, and the XX gonad is masculinized in tra-1 null mutants (Hodgkin and Brenner, 1977; Mathies et al., 2004). The SGP division in tra-1; fkh-6 double mutants is hermaphrodite-like, indicating that TRA-1 acts upstream of FKH-6 (Chang et al., 2004). Therefore, FKH-6 is the terminal regulator of the SGP male fate.
In this paper, we identify the single C. elegans cyclin D gene, cyd-1, as a key regulator of axis formation and sex determination in the gonad. Cyclin D functions during L1, and it appears to be specific to the SGP and its immediate daughters. The cyd-1(q626) mutation is a missense allele, and its effects can be mimicked by cyd-1 RNAi. Cyclin D has its effect on gonadogenesis via the canonical cell cycle machinery, including the cyclin-dependent kinase 4 (cdk-4), Rb (lin-35), E2F (efl-1), DP (dpl-1), the CDK inhibitors (cki-1 and cki-2), and cyclin E (cye-1). Cyclin D affects axis determination in both sexes by promoting POP-1 asymmetry and controls the sex-specific cell size asymmetry of the male SGP division, apparently by relieving E2F repression of the fkh-6 gonad-specific sex determination gene. Cyclin D also affects rnr::GFP expression specifically in SGPs and slows the SGP cell cycle. However, other methods of slowing the SGP cell cycle did not mimic cyd-1 defects. We propose that cyclin D and regulators of G1/S transition coordinate both specification of distal-proximal organ axes and specification of sexual fate with the cell cycle.
RESULTS
Early gonadogenesis defects in q626 mutants
We have isolated a temperature-sensitive mutation, called q626 (see Methods). At the permissive temperature of 20°C, q626 homozygotes of either sex were virtually normal, but at the restrictive temperature of 25°C, they had gonadal defects in both sexes. Normally, hermaphrodites have a gonad with two extended U-shaped “arms”, one anterior and one posterior (Figure 1C, G), but when raised at 25°C, some q626 hermaphrodites were missing either one or both arms (Figure 1D, G). Importantly, q626 hermaphrodites with one gonadal arm were self-fertile, and no other gonadal defects were observed. In contrast to this relatively mild effect on hermaphrodites, q626 had a severe effect on male gonadogenesis. Wild-type adult males and most adult q626 males raised at 20°C have an extended J-shaped gonad (Figure 1E, H). By contrast, most adult q626 males had disorganized gonads that were not extended when raised at 25°C (Figure 1F, H). Furthermore, some male q626 mutants developed a hermaphrodite vulva (3%, n=145; data not shown). These q626 effects were gonad-specific in both sexes: growth was similar to that of wild-type animals, movement seemed unaffected, non-gonadal tissues developed normally, and embryonic survival was normal.
To learn when q626 affects gonadogenesis, we imposed selected temperature regimens on this temperature-sensitive mutant. In both hermaphrodites and males, the gonad developed normally when raised at permissive temperature during the L1 stage and then shifted to restrictive temperature from the L1 molt through adulthood; by contrast, gonad development was defective when L1 larvae were exposed to restrictive temperature and then returned to permissive temperature from the L1 molt through adulthood (Figure 1I, J). We conclude that q626 affects gonadogenesis during the L1 stage in both sexes.
q626 affects generation of regulatory cells in both sexes
Normally, the SGP division generates distal and proximal daughter cells that generate regulatory cells critical for gonad shape and germline proliferation in both sexes (Kimble, 1981; Kimble and White, 1981). Gonadal arm elongation is controlled by “leader cells”, which are distal tip cells (DTCs) in hermaphrodites and the linker cell (LC) in males (Kimble and White, 1981). Leader cells express lag-2::GFP brightly (Blelloch et al., 1999). At restrictive temperature, about one-third of q626 hermaphrodites were missing one lag-2::GFP–expressing DTC (32%, n=91, data not shown), and about one-half of q626 males were missing the lag-2::GFP–expressing LC (58%, n=78, data not shown). Therefore, the lack of arm extension in q626 gonads is due to missing leader cells.
The size of q626 male gonads was variable, apparently due to a varying extent of germline proliferation. In males, germline proliferation is controlled by “male DTCs” (mDTCs), two small cells residing near the distal tip of the wild-type male gonad (Kimble and White, 1981). These mDTCs also express lag-2::GFP, but that expression is much weaker than in leader cells (Blelloch et al., 1999). Whereas all wild-type male gonads had two lag-2::GFP–expressing mDTCs (100%, n=20), only a small percentage of q626 males had detectable lag-2::GFP–expressing mDTCs (23%, n=78). Therefore, mDTCs are not generated normally in q626 XO gonads. We conclude that q626 affects the production of distal SGP daughters in both sexes.
q626 feminizes the XO gonad
The q626 male gonadal defects were reminiscent of fkh-6 XO gonads, which are disorganized, lack linker cells, and sometimes induce vulvae (Chang et al., 2004). Because male fkh-6 gonads are also feminized (or perhaps more correctly hermaphroditized) (Chang et al., 2004), we examined q626 gonads with markers for hermaphrodite and male gonadal differentiation. The hermaphrodite-specific marker cdh-3::GFP labels the hermaphrodite anchor cell, but is not expressed in wild-type male gonads (Figure 2A, B) (Pettitt et al., 1996); however, cdh-3::GFP expression occurred in most q626 males (67%, n=15; Figure 2C). By contrast, the male-specific marker K09C8.2::GFP labels the male seminal vesicle and vas deferens, but is not expressed in wild-type hermaphrodite gonads (Figure 2D, E) (Chang et al., 2004); K09C8.2::GFP was not detected in some q626 males (45%, n=51; Figure 2F). Finally, three other hermaphrodite-specific markers of gonadal differentiation were expressed in most q626 male gonads: lim-7::GFP (sheath cells), fkh-6::GFP (sheath and spermatheca), and ZK813.3::GFP (spermatheca) (data not shown). We conclude that the disorganized gonads in q626 males are feminized.
Figure 2. q626 feminizes the gonad and appears to function like fkh-6.
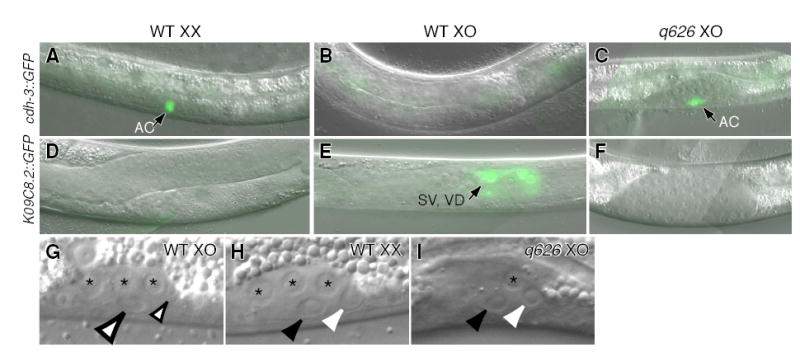
(A–C) L3 animals carrying anchor cell (AC) marker, cdh-3::GFP. (A) Wild-type hermaphrodite. (B) Wild-type male. (C) q626 male raised at restrictive temperature expresses AC marker. (D–F) L4 animals carrying marker for vas deferens (VD) and seminal vesicle (SV), K09C8.2::GFP. (D) Wild-type hermaphrodite. (E) Wild-type male. (F) q626 male raised at restrictive temperature does not express SV/VD marker. (G–I) L1 gonads viewed by Nomarski microscopy. Asterisks, germ cells. (G) Male SGP daughters differ in size. Small open arrow, smaller distal daughter; large open arrow, larger proximal daughter. (H) Hermaphrodite SGP daughters are of roughly equal size. White arrowhead, distal daughter; black arrowhead, proximal daughter. (I) q626 SGP daughters at restrictive temperature are of roughly equal size. Same conventions as (H).
We next asked if the SGP asymmetric division and the behavior of SGP daughter cells were feminized in q626 males. Hermaphrodite SGPs produce daughters of approximately equal size (Figures 1A, 2H), and their proximal daughters do not migrate, but male SGPs produce daughters of unequal size (Figures 1A, 2G), and their proximal daughters migrate towards the anterior. In q626 males, the SGP daughters were of roughly equal size (Figure 2I) and most proximal daughters did not migrate (92%, n=13). We conclude that q626 feminizes both the SGP asymmetric division and later gonadal differentiation.
q626 is a cyd-1 missense mutation
We cloned the gene corresponding to q626 by standard methods (see Experimental Procedures). Three lines of evidence showed that q626 is an allele of cyd-1, which encodes the single C. elegans homolog of cyclin D1 (Boxem and van den Heuvel, 2001; Park and Krause, 1999). First, the cyd-1(q626) genomic sequence revealed a missense mutation just C-terminal to the cyclin box in a region conserved between closely related nematodes, but not in vertebrates (Figure 3A, B). Second, a DNA fragment spanning the cyd-1 locus was able to rescue q626 gonadal defects (see Methods). Finally, q626 failed to complement either of two cyd-1 null mutations (he112 and he116).
Figure 3. q626, a missense mutation in cyd-1 that delays SGP division.
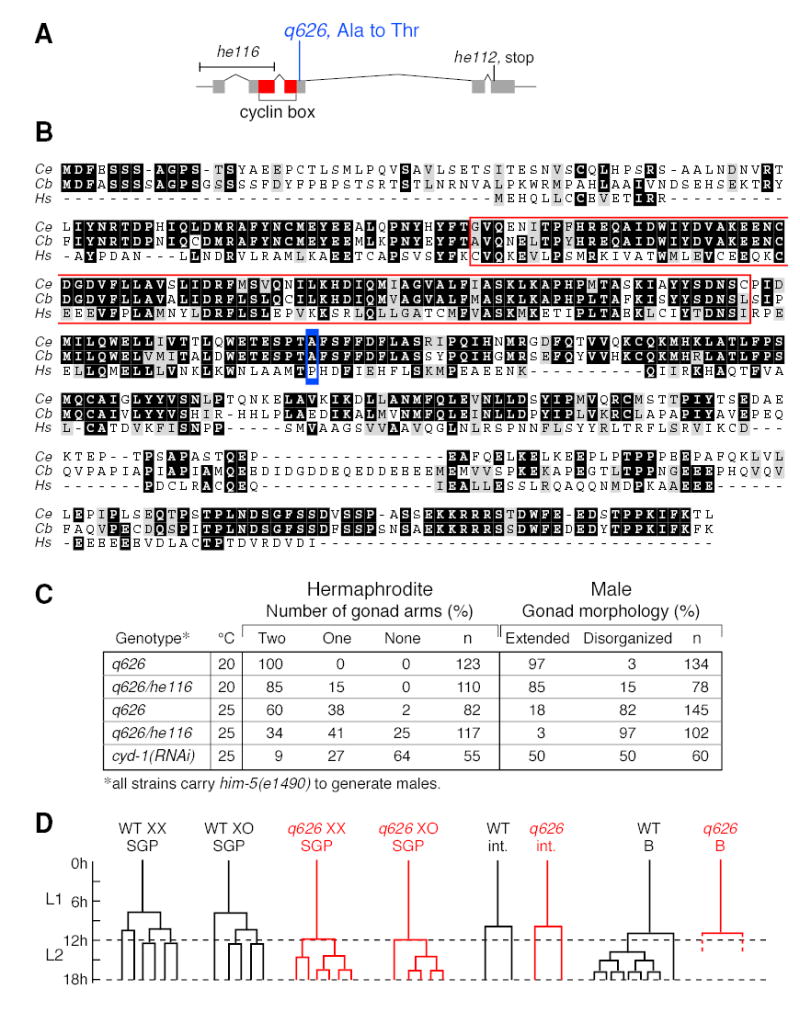
(A) The cyd-1 gene. Boxes, exons; lines, introns; red, cyclin box. q626 is a G to A base change that generates a missense mutation predicted to change an alanine to threonine. he116 and he112 are both predicted cyd-1 null mutations (Boxem and van den Heuvel, 2001). (B) Alignment of C. elegans (top), C. briggsae (middle), and human cyclin D1 (bottom) amino acid sequences (black boxes, identical residues; gray boxes, similar residues). The q626 mutation (blue box) is in a conserved region that lies to the C-terminal side of the cyclin box (red outline). (C) Gonad defects typical of cyd-1(q626) are more severe when placed over cyd-1(0) and are observed after depletion of cyd-1 by RNAi. (D) Comparison of cell divisions in wild-type (WT, black) and cyd-1(q626) mutants raised at restrictive temperature (red). L1, first larval stage; L2, second larval stage. Time scale in hours at 25°C. SGP divisions are delayed in cyd-1(q626) mutants relative to wild-type, but other blast cell divisions are normal.
We next asked if cyd-1(q626) alters a gonad-specific activity or reduces activity more generally. The cyd-1(he116) allele, henceforth called cyd-1(0), deletes the promoter region and first two exons and is thought to be a null allele (Boxem and van den Heuvel, 2001). Most post-embryonic divisions are blocked in cyd-1(0) mutants, but the SGP division is an exception (Koreth and van den Heuvel, 2005). We confirmed that cyd-1(0) SGPs divide once or at most twice, and that gonadal development then arrests in both sexes. Indeed, although later gonadal development did not occur in cyd-1 null mutants, the male SGPs generated daughters of roughly equal size and proximal SGP daughters did not migrate, both diagnostic of a feminized SGP division (n=6). To assay later gonadal development, we turned to cyd-1(q626)/cyd-1(0) transheterozygotes and cyd-1 RNAi. Gonadal defects were more penetrant in cyd-1(q626)/cyd-1(0) transheterozygotes than in cyd-1(q626) homozygotes, at both permissive and restrictive temperatures and in both sexes (Figure 3C). Furthermore, cyd-1 RNAi gonads were affected much like cyd-1(q626) (Figure 3C). The simplest explanation is that the cyd-1(q626) lesion reduces activity of the locus.
SGP divisions are delayed in cyd-1 males and hermaphrodites
We next asked if the timing of cell divisions was impaired in cyd-1(q626) mutants relative to wild type. At restrictive temperature, wild-type SGPs divided between 7 and 9 hours after hatching in both sexes (males, n=9; hermaphrodites, n=13), and two subsequent divisions in the SGP lineage occurred at approximately 10 and 13 hours after hatching, respectively (Figure 3D). By contrast, cyd-1(q626) SGPs divided 12 or more hours after hatching, just prior to or during the L1 molt, in both sexes (males, n=7; hermaphrodites, n=11) (Figure 3D). Subsequent rounds of division occurred during L2, and their timing was normal relative to the SGP division (Figure 3D). Intestinal divisions were not delayed in cyd-1(q626) mutants, and the adult number of intestinal nuclei was normal (n=20; Figure 3D). Similarly, the B blast cell divided normally in cyd-1(q626) males (n=20; Figure 3D), and both cyd-1(q626) hermaphrodites and males had the normal number of coelomocytes (six in hermaphrodites, five in males), using the unc-122::GFP coelomocyte marker (data not shown). Therefore, cyd-1(q626) SGP divisions are delayed specifically, but that delay is not sexually dimorphic.
Control of gonadogenesis by other cell cycle regulators
We next investigated other components of the cell cycle machinery for effects on gonadogenesis (Figure 4A). To learn whether CYD-1 works with its CDK-4 partner to control gonadogenesis, we depleted cdk-4 during larval development using RNAi, but cdk-4(RNAi) effects were minimal (Figure 4B). Nonetheless, cdk-4(RNAi) enhanced cyd-1(q626) gonadogenesis defects: at permissive temperature (20°C), cyd-1(q626); cdk-4(RNAi) hermaphrodite gonads had missing arms and cyd-1(q626); cdk-4(RNAi) male gonads were misshapen (Figure 4B). Most importantly, the cyd-1(q626); cdk-4(RNAi) male gonads expressed hermaphrodite-specific gonadal markers for anchor cell, sheath, and spermatheca (60%, n=33). Therefore, cyd-1 and cdk-4 are likely to work together to control gonadal sex determination.
Figure 4. Regulators of G1 to S transition enhance or suppress cyd-1 gonad defects.
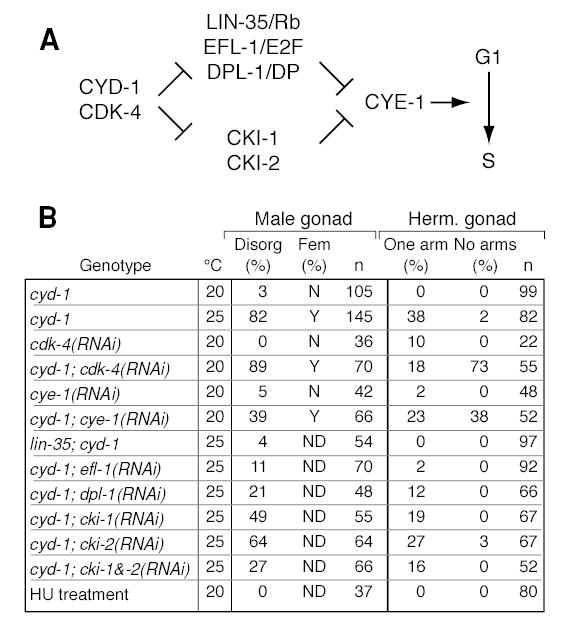
(A) Diagram of G1 to S regulators relevant to this work, and their functional relationships. CYD-1, cyclin D; CDK-4, cyclin dependent kinase; LIN-35/Rb, C. elegans homolog of human retinoblastoma gene; EFL-1, C. elegans homolog of E2F; DPL-1, C. elegans homolog of DP; CKI, inhibitors of CDK; see Koreth and van den Heuvel (2005) for review. (B) Effects of cell cycle regulators on gonadogenesis. RNAi, RNA-mediated interference. HU, hydroxyurea; Disorg, disorganized; Fem, feminized as assessed using three hermaphrodite-specific gonad markers.
CYD-1/CDK-4 promotes S-phase initiation by inhibiting two parallel sets of regulators (Koreth and van den Heuvel, 2005) (Figure 4A). The first set includes LIN-35/Rb, EFL-1,2/E2F, and DPL-1/DP, and the second includes two inhibitors, CKI-1 and CKI-2 (Koreth and van den Heuvel, 2005). We therefore asked if cyd-1 defects could be suppressed by removal or depletion of these downstream genes. Our results are summarized in Figure 4B. Briefly, cyd-1 defects were suppressed by lin-35(0), efl-1(RNAi), dpl-1(RNAi), and cki-1/cki-2(RNAi): XO gonads were fully male, and XX gonads had two arms and were self-fertile. Therefore, cyd-1 appears to be acting through lin-35, efl-1, dpl-1, and the cki genes to control the SGP division.
C. elegans cyclin E is encoded by a single gene, cye-1, which acts downstream of cyd-1 to control entry in S-phase (Koreth and van den Heuvel, 2005) (Figure 4A). Although cye-1(RNAi) male and hermaphrodite gonads were largely normal, cye-1(RNAi) enhanced the cyd-1(q626) defects at the permissive temperature: cyd-1(q626); cye-1(RNAi) animals of both sexes had defective gonads (Figure 4B), and male gonads expressed hermaphrodite-specific gonadal markers (56%, n=25). Therefore, cye-1 appears to affect the SGP division in the same manner as cyd-1. The simplest interpretation is that cyd-1 affects gonadogenesis through the canonical cell machinery that controls the G1 to S phase transition.
Slowing the SGP cell cycle does not mimic cyd-1(q626) defects
Previous studies have shown that a delayed cell cycle can non-specifically alter cell fate decisions or asymmetric divisions (Ambros, 1999; Encalada et al., 2000; Tio et al., 2001). To ask if SGP defects typical of cyd-1(q626) were induced by a lengthened S-phase, we treated L1s with hydroxyurea (HU) prior to the SGP division to slow S-phase. HU treatment did indeed delay the SGP division by four or more hours, but no gonad defects were observed in either sex (Figure 4B). To ask if these SGP defects might be induced by a delay in SGP divisions relative to other cells in the animal, we employed gon-4 RNAi. Previous work showed that SGP divisions are retarded specifically in gon-4 null mutants; other cell divisions were not delayed (Friedman et al., 2000). Using gon-4 RNAi, we found the same effect: most SGP divisions were delayed by ~3 hours, but others (e.g. B, I) were not. We focused on the delayed SGP divisions: hermaphrodite SGPs generated roughly equal-sized daughters that did not migrate (n=14), and male SGPs generated dramatically larger proximal daughters that moved anteriorly (n=8). Therefore, it seems unlikely that an SGP-specific cell cycle delay is responsible for the aberrant SGP divisions in cyd-1 mutants.
cyd-1 promotes POP-1/TCF asymmetry in SGP daughters
The POP-1 and SYS-1 transcription factors control generation of SGP daughters with distal fates in both sexes (see Introduction). Because distal cells are often missing in cyd-1(q626) mutants, we reasoned that cyd-1(q626) might affect the Wnt/MAPK pathway. Loss-of-function sys-1 mutations are haplo-insufficient and dominantly enhance mutations in other Wnt/MAPK components, while ectopic sys-1 expression can reverse SGP daughter fates and suppress mutations in upstream Wnt/MAPK components (Kidd et al., 2005; Siegfried et al., 2004). We found that a sys-1 loss-of-function mutation is similarly a strong dominant enhancer of cyd-1(q626) at both permissive and restrictive temperatures, and that ectopic SYS-1 can suppress cyd-1(q626) (Figure 5A). Therefore, cyd-1(q626) appears to influence the Wnt/MAPK pathway.
Figure 5. POP-1 asymmetry in SGP daughters can be abolished in cyd-1 mutants.
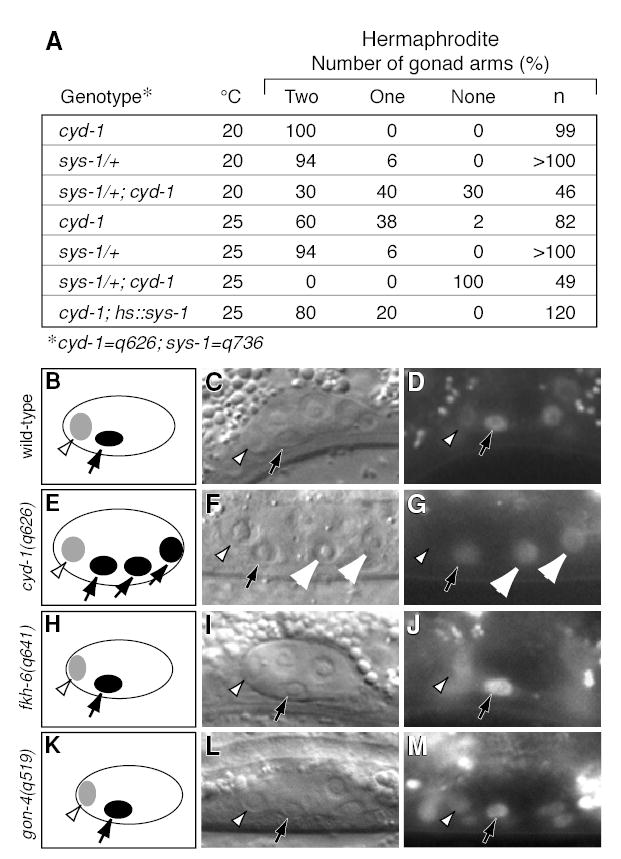
(A) Genetic interactions between cyd-1 and sys-1. (B–J) Left column, diagrams showing POP-1 abundance in SGP daughters. Light grey, low POP-1; dark grey, high POP-1. Middle column, Nomarski micrographs of SGP daughters. Open arrowhead, distal sister with less POP-1; solid arrow, proximal sister with more POP-1; white arrowheads, sisters with equal amounts of POP-1. Right column, same gonads as in middle column, showing fluorescence of GFP::POP-1 reporter. All gonads shown are from XX hermaphrodites. (B–D) Wild-type gonad. GFP::POP-1 is localized asymmetrically in SGP daughters. (E–G) cyd-1(q626) gonad. GFP::POP-1 is localized asymmetrically in anterior SGP daughters (left) and equally in posterior SGP daughters (right). (H–J) fkh-6 null mutant gonad. GFP::POP-1 is localized asymmetrically in SGP daughters. (K–M) gon-4(RNAi) gonad. GFP::POP-1 is localized asymmetrically in SGP daughters.
We next asked if cyd-1(q626) affects POP-1 asymmetry. In wild-type animals, GFP::POP-1 is higher in the nuclei of proximal than distal SGP daughters (Figure 6B–D; Siegfried et al., 2004). By contrast, in cyd-1(q626) hermaphrodites, GFP::POP-1 asymmetry in SGP daughters was sometimes abolished: in 4 of 12 animals, nuclear GFP::POP-1 was present at approximately equal levels in one pair of SGP daughter cells (Figure 5E–G). The frequency of this defect in POP-1 asymmetry was similar to that at which cyd-1(q626) hermaphrodites are missing a gonadal arm. We also examined GFP::POP-1 in fkh-6(0) hermaphrodites, which sometimes lack a DTC (Chang et al., 2004), but found no effect on GFP::POP-1 asymmetry (n=67; Figure 5H–J). GFP::POP-1 localization was also not affected by gon-4(RNAi) (Figure 5K–M). Therefore, cyd-1, but not fkh-6 or gon-4, influences POP-1 asymmetry in SGP daughters.
Figure 6. Gene expression in cyd-1(q626) mutants.
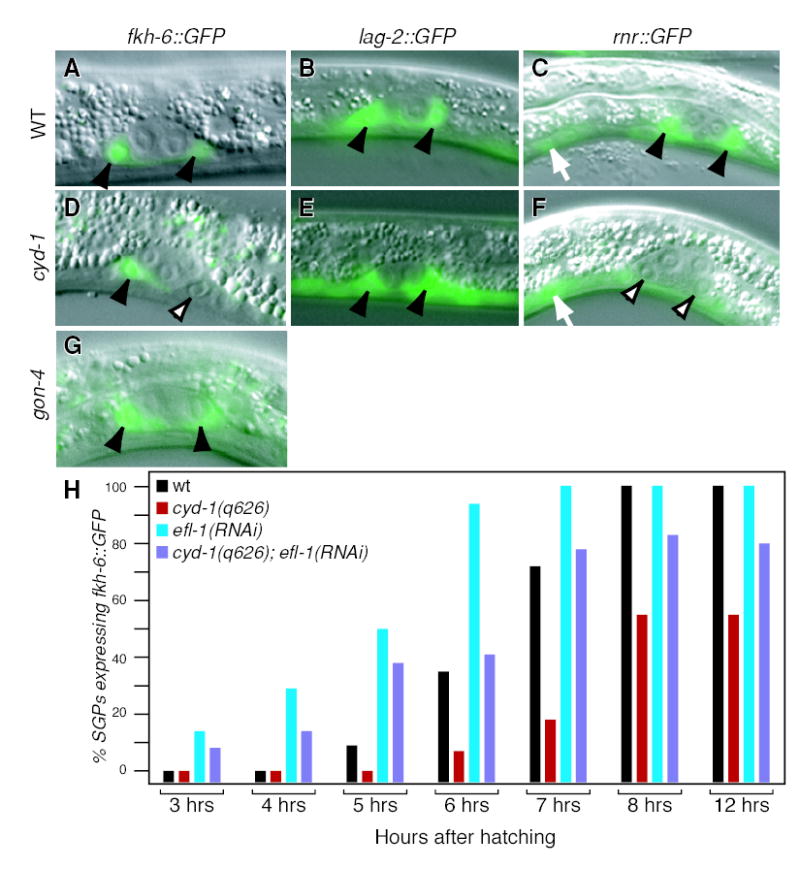
(A–C) Wild-type L1 larvae raised at 25°C; (D–F) cyd-1(q626) L1 larvae raised at 25°C. Solid arrow, SGPs expressing marker; open arrowheads, SGPs that do not express marker; white arrow, non-gonadal cell expressing marker. All images are XX larvae, but the same results are seen in XO larvae. (A–C) Wild-type SGPs express fkh-6::GFP (A), lag-2::GFP (B), and rnr::GFP (C). (D) Some cyd-1(q626) SGPs express fkh-6::GFP (arrow) and others do not (open arrowhead). (E) All cyd-1(q626) SGPs express lag-2::GFP. (F) Most cyd-1(q626) SGPs do not express rnr::GFP (open arrowheads), but non-gonadal cells express rnr::GFP normally (large arrowhead). (G) gon-4(RNAi) gonads express fkh-6::GFP. (F) Time course of fkh-6::gfp expression in WT (black), cyd-1(q626) (red), elf-1(RNAi) (blue), and cyd-1(q626); efl-1(RNAi) (purple) gonads.
To ask if cyd-1(q626) affects POP-1 asymmetry in non-gonadal tissues, we examined T cell and V cell daughters. In wild-type hermaphrodites, nuclear GFP::POP-1 is more abundant in anterior T and V daughters (Herman, 2002). Similarly, POP-1 asymmetry was normal in T and V cell daughters in cyd-1(q626) mutants (n=28). We also looked for T cell lineage defects in cyd-1(q626) animals by their ability to take up the lipophilic dye, DiO (Herman and Horvitz, 1994). In wild-type animals, descendents of the T cell (phasmid neurons in the tail, PHA and PHB) stain with this dye. DiO was taken up normally by phasmid neurons in all cyd-1(q626) mutants (n=84). We conclude that the cyd-1(q626) defect in POP-1 asymmetry is specific to the SGPs.
cyd-1 controls fkh-6 expression in the SGPs
The similarity of the cyd-1(q626) and fkh-6 gonadal defects suggested that cyd-1 might control fkh-6 expression. In wild-type animals, a fkh-6::GFP reporter is expressed in the SGPs of both sexes beginning in L1, prior to the first SGP division. In hermaphrodites, fkh-6::GFP expression ceases just after the SGP division, but in males, expression persists several hours after the SGP division. fkh-6::GFP is also expressed much later (L3-adult) in the hermaphrodite spermatheca (Chang et al., 2004). In wild-type animals of both sexes, fkh-6::GFP expression began in the SGPs at about six hours after hatching (Figure 6A, 6H, black bars). By contrast, fkh-6::GFP expression was either delayed or absent in the SGPs of cyd-1(q626) mutants in both sexes (Figure 6D, 6H, red bars). Specifically, few SGPs expressed fkh-6::GFP six hours after hatching, only about half expressed the reporter after eight hours (males, 50%, n=60; hermaphrodites, 43%, n=124), and even 12 hours after hatching fkh-6::GFP was absent from many SGPs (Figure 6D, 6H, red bars). In those cyd-1(q626) mutants that did express fkh-6::GFP, that expression ceased as normal in both hermaphrodites (just after the SGP division) and males (several hours after the SGP division). Therefore, cyd-1 does not affect the sexually dimorphic nature of fkh-6 expression. Furthermore, fkh-6::GFP was not seen in cyd-1(0) SGPs (n=38 SGPs, data not shown). The hermaphrodite spermathecal expression could not be tested in cyd-1(0) mutants, but it appeared normal in cyd-1(q626) hermaphrodites at restrictive temperature (100%, n=50). We also examined fkh-6::GFP expression in gon-4(RNAi) animals and gon-4(q519) mutants and found no defects (n=26, Figure 6G). Therefore, cyd-1 controls fkh-6 expression specifically in the SGPs of both sexes.
We next examined other SGP markers. In wild-type larvae, tra-1::GFP, pes-1::GFP, and lag-2::GFP are all expressed in both hermaphrodite and male SGPs from hatching (Hope, 1991; Mathies et al., 2004) (Figure 6B, data not shown); similarly, all three reporters were expressed in SGPs of both cyd-1(q626) and cyd-1(he116) mutants in both sexes (Figure 6E; data not shown). Therefore, cyd-1 does not control gene expression in SGPs generally. Finally, we looked at the rnr::GFP S-phase marker (Hong et al., 1998). In wild-type L1 larvae, rnr::GFP expression first begins in the SGPs approximately six hours after hatching, and also occurs in other dividing cells (Figure 6C). By contrast, rnr::GFP was rarely expressed in cyd-1(q626) SGPs (6%, n=32 SGPs), but it was expressed in non-gonadal cells (Figure 6F). We conclude that cyd-1 affects the expression of both fkh-6::GFP and rnr::GFP expression specifically in the SGPs.
Cyclin D appears to relieve E2F repression of fkh-6::GFP
Cyclin D functions, at least in part, to relieve E2F repression of target genes during the G1 to S transition (Coffman, 2004; Koreth and van den Heuvel, 2005). We therefore considered the idea that the efl-1/E2F gene might repress fkh-6 expression. Consistent with that possibility, we sought potential E2F binding sites in the 5′ flanking region of fkh-6, and found one possibility (TTTGCGG) at position −1584 – −1577. As described above, fkh-6::GFP expression in SGPs is delayed or absent in cyd-1 mutants of both sexes compared to wild-type. In efl-1 RNAi animals, fkh-6::GFP was expressed, and it was in fact expressed earlier than in wild-type (Figure 6H, blue bars). Although expression of fkh-6::GFP was earlier than normal in both hermaphrodites and males, hermaphrodite gonadogenesis remained normal and was not masculinized, and male gonadogenesis was also normal and not feminized. Earlier expression of fkh-6::GFP was also seen in cyd-1(q626); efl-1(RNAi) SGPs (Figure 6H, purple bars). Therefore, fkh-6::GFP expression appears to be repressed by E2F, and cyclin D appears to relieve that repression.
Functional relationship of cyd-1 and fkh-6
Loss-of-function mutations in cyd-1 and fkh-6 have similar effects on gonadal sex determination (this work; Chang et al., 2004), and fkh-6 expression is defective in cyd-1 mutants (this work). To ask if cyd-1 and fkh-6 function in the same regulatory pathway, we compared the effects of fkh-6(0) and cyd-1(q626) in double mutants with tra-1. It was previously seen that fkh-6 has a synergistic effect with tra-1: the SGP lineages in fkh-6 tra-1 double mutants arrest after one or at most two divisions (Chang et al., 2004). Similarly, we observed a synthetic phenotype in cyd-1(q626); tra-1 double mutants (Figure 7A). This synthetic effect is less severe than that seen in fkh-6; tra-1 double mutants, but the cyd-1 mutant is not a null allele and its effects on fkh-6 gene expression are partially penetrant. The synthetic effect of both genes with tra-1 supports the idea that cyd-1 and fkh-6 act in the same pathway.
Figure 7. Cyclin D coordinates key regulators of the SGP asymmetric division.
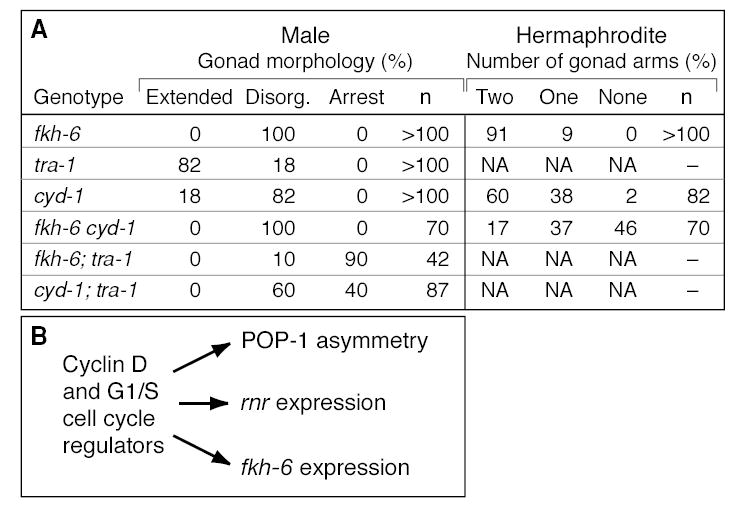
(A) Table comparing effects of cyd-1(q626) and fkh-6(0) mutations on gonadogenesis as single or double mutants. (B) See text for explanation.
We also examined fkh-6(0) cyd-1(q626) double mutants, and found no synergistic effect in males as might be expected since fkh-6 single mutants are fully penetrant (Figure 7A). By contrast, hermaphrodite gonads were affected more severely in fkh-6(0) cyd-1(q626) double mutants than in either single mutant (Figure 7A). It is important to note that the lack of gonadal arms in fkh-6 single mutants results from a defect later in gonadogenesis than seen in cyd-1 mutants (this work; Chang et al., 2004), and, furthermore, that POP-1 asymmetry was not affected in fkh-6 mutants (see above). The simplest interpretation is that cyd-1 and fkh-6 control the generation of hermaphrodite gonadal arms by distinct mechanisms, and that the more severe phenotype typical of fkh-6 cyd-1 double mutant hermaphrodites reflects an additive effect.
DISCUSSION
Cyclin D controls the SGP asymmetric division
Cyclin-dependent kinases (CDKs) and their cyclin binding partners are best known as regulators of the cell cycle (Morgan, 1997). Of particular importance to this work is cyclin D, which has been implicated in control of the G1/S transition (Sherr, 1994). In C. elegans, cyclin D and its CDK-4 partner promote the G1/S transition during most larval cell divisions (Boxem and van den Heuvel, 2001; Park and Krause, 1999). Cyclin D and CDK-4 counteract LIN-35/Rb, EFL-1/E2F, DPL-1/DP and CKI-1, which in turn inhibit Cyclin E and CDK-2 (Koreth and van den Heuvel, 2005). Therefore, the cell cycle machinery governing the G1 to S transition has been conserved in C. elegans.
We have found that cyd-1, the single C. elegans cyclin D gene, controls the asymmetric division of the somatic gonadal precursor (SGP), a highly regulated division that establishes organ axes and embarks on sex-specific programs of gonadogenesis (Kimble and Hirsh, 1979). Specifically, cyclin D affects both POP-1/TCF asymmetry and fkh-6 expression in the early gonad. POP-1/TCF asymmetry is controlled by Wnt/MAPK signaling (Lo et al., 2004; Maduro et al., 2002), and in the SGP daughters, it specifies gonadal axes (Siegfried et al., 2004). The FKH-6 transcription factor is a sex-determining gene that specifies the male gonadal fate (Chang et al., 2004). Therefore, cyd-1 affects two key regulators of early gonadogenesis.
The cyd-1 defects are specific for the SGP division. SGP divisions are delayed, but other larval blast cells divide normally; fkh-6 and rnr expression are affected in SGPs, but other cells express these genes normally; and POP-1 asymmetry is abolished in the SGPs, but it occurs normally in non-gonadal blast cells. Why are the SGPs affected so specifically in cyd-1(q626) mutants? One possibility might have been that cyd-1(q626) identifies a domain that mediates an SGP-specific control. However, gonad defects typical of cyd-1(q626) mutants were also observed after cyd-1 RNAi, which depletes cyclin D but does not eliminate it. We suggest that cyd-1(q626) is a partial loss-of-function allele, and that the SGP division is particularly sensitive to the level of cyclin D activity. Interestingly, the cyd-1(cc600) mutation also acts in a cell-type specific manner, affecting only the division of embryonically-derived coelomocytes (Yanowitz and Fire, 2005). Therefore, individual cell divisions appear to have distinct requirements for cyd-1 activity.
The SGP cell division is delayed in cyd-1 mutants, raising the possibility that the cyclin D effects on POP-1 asymmetry and fkh-6 expression might be non-specific. Cell cycle length has been implicated as a factor in both cell fate specification and proper asymmetric divisions both in C. elegans and Drosophila (Ambros, 2001; Encalada et al., 2000; Fay, 2005; Fay and Han, 2000; Tio et al., 2001). However, delays of the SGP division by other methods did not mimic the cyd-1 effect. Thus, hydroxyurea treatment, which slows S-phase and delays cell divisions, had no apparent effect on gonadal development. Furthermore, gon-4 RNAi, which delays the SGP division specifically (Friedman et al., 2000; this work), did not change the sex-specific asymmetry of the SGP division, POP-1 asymmetry, or fkh-6 expression. Therefore, a simple delay in cell division is not likely to explain the cyclin D effects on the SGP asymmetric division.
Cyclin D coordinates regulators of SGP asymmetric division
We have found that cyclin D regulates three molecular markers in the SGPs or their daughters (Figure 7B). In the SGP itself, cyclin D affects expression of both fkh-6::GFP and rnr::GFP, a marker of S-phase (Hong et al., 1998). Since fkh-6 and rnr reporters are expressed with similar timing in the SGPs, an attractive idea is that cyd-1 coordinates expression of both fkh-6 and rnr as the cell progresses from G1 into S phase. In addition to its control of fkh-6 and rnr in the SGPs, cyclin D affects POP-1 asymmetry in the SGP daughters. The linkage between the cyclin D control of POP-1 asymmetry and the G1/S transition is less evident. POP-1 asymmetry is first observed in SGP daughter cells soon after the SGP division (Siegfried et al., 2004; C. Tilmann, unpublished). One possibility is that CYD-1 controls events in the SGP mother cell to subsequently affect POP-1 asymmetry in her daughters. If this is the case, we note that fkh-6 is not required, because POP-1 asymmetry is not abolished in fkh-6 null mutants. Alternatively, CYD-1 might influence POP-1 asymmetry in the daughters themselves. We attempted to distinguish between these two possibilities with temperature shifts, but the experiment did not have sufficient resolution (C. Tilmann, unpublished). Regardless, the CYD-1 regulation of POP-1 and fkh-6 appear to be distinct, suggesting that cyclin D is a common regulator of both gonadal regulators (Figure 7B).
Cyclin D relieves E2F repression to promote fkh-6 expression
We have found that cyclin D works together with other major regulators of the G1/S transition to control the SGP asymmetric division (Figure 4). Most importantly, RNAi depletion of efl-1, which encodes an E2F-related transcription factor (Ceol and Horvitz, 2001) suppressed the cyd-1 defects. This suppression was observed in both sexes, suggesting an effect of E2F on both POP-1 asymmetry and fkh-6 expression. Although we do not know the specific Wnt/MAPK signaling component regulated, several lines of evidence suggest that fkh-6 is controlled antagonistically by cyclin D and E2F: an fkh-6 reporter was either not expressed at all in cyd-1 mutant SGPs or expressed late; by contrast, fkh-6 expression was restored in cyd-1; efl-1(RNAi) SGPs, and was seen earlier than normal in efl-1(RNAi) SGPs. Therefore, cyclin D and E2F have opposite effects on fkh-6 expression. Given the gonadal feminization by depletion of cyclin D, one might think a priori that depletion of E2F might masculinize the hermaphrodite gonad. However, in wild-type L1s, fkh-6 is typically expressed in both hermaphrodite and male SGPs, and E2F depletion did not masculinize the hermaphrodite gonad. The simplest explanation is that E2F normally represses fkh-6 expression, and that cyclin D relieves that repression. Although we identified a canonical E2F binding sequence in the fkh-6 promoter, biochemical experiments will be required to learn whether fkh-6 is a direct target of E2F repression. We conclude that the G1/S regulatory machinery controls fkh-6 expression.
Cyclin D as a key regulator of metazoan development
Cyclin D is not an essential regulator of all metazoan cell cycles, a conclusion based on RNAi, deletion and nonsense mutants of the single cyclin D gene in C. elegans (Boxem and van den Heuvel, 2001; Park and Krause, 1999), deletion mutants of the single cyclin D gene in Drosophila (Emmerich et al., 2004), and a triple knockout of the three cyclin D genes in mice (Kozar et al., 2004). Instead, cyclin D appears to be essential for a variety of developmental functions: regulation of the G1/S transition in most larval cell divisions in C. elegans (Boxem and van den Heuvel, 2001; Park and Krause, 1999), control of cell growth and cell number in Drosophila (Datar et al., 2000; Emmerich et al., 2004; Meyer et al., 2002), and regulation of heart development and expansion of hematopoietic stem cells in mice (Kozar et al., 2004). A caveat to interpreting these deletion studies is that other cyclins may fill in for cyclin D, an idea supported by genetic interactions with CDK-2 in Drosophila (Emmerich et al., 2004). Therefore, the scope of biological functions mediated by cyclin D and the G1/S cell cycle machinery more generally are poorly understood in metazoans.
Over the past ten years, it has become clear that G1/S cell cycle regulators are intimately linked with controls of cell fate. An early example was myoD, which drives muscle myoblasts out of the cell cycle by induction of p21(Waf1/Cip) (Halevy et al., 1995). In addition, myoD and cyclin D1 physically interact with each other and antagonize each other’s activities (Zhang et al., 1999). Furthermore, Drosophila Cyclin D/CDK-4 binds and stabilizes STAT92E, a control critical for embryonic segmentation (Chen et al., 2003), and mammalian cyclin E-CDK2 binds and phosphorylates cytosolic β-catenin, leading to its rapid degradation during G1 (Park et al., 2004). An effect of G1/S regulators on asymmetric divisions has also been seen in Drosophila (Prokopenko and Chia, 2005). Most strikingly, cyclin E controls the asymmetric division of an NB6-4 neuronal precursor (Berger et al., 2005). However, other components of the cell cycle machinery were not involved in this asymmetric division, and therefore this Drosophila control may be distinct from that reported here. While these previous results have forged a link between the cell cycle machinery and controls of cell fate, our findings extend this idea and suggest that cyclin D and other regulators of the G1 to S transition coordinate the activity of multiple cell fate determinants during a single asymmetric cell division.
Experimental Procedures
Strains and genetics
Standard protocols were used for culturing C. elegans strains (Sulston and Hodgkin, 1988). Strains were derived from the Bristol strain N2 and maintained at 20°C unless otherwise noted (Sulston and Horvitz, 1977). The high incidence of male mutation, him-5(e1490), was included in strains to generate males (Hodgkin, 1983). The following mutations were used for this work: LGI, lin-35(n745); pop-1(q645); sys-1(q736); LGII, fkh-6(q641); cyd-1(he116); cyd-1(he112); unc-52(e444); dpy-10(e128); unc-4(e120); rol-1(e91); bli-1(e769); LGIII, tra-1(e1099); LGIV, gon-4(q518); LGV, him-5(e1490). The following integrated transgenes were used: arIs51 [cdh-3::GFP] (Pettitt et al., 1996); ezIs1 [K09C8.2::GFP] and ezIs2 [fkh-6::GFP] (Chang et al., 2004); tnIs6 [lim-7::GFP] (Hall et al., 1999); maIs103 [rnr::GFP] (Hong et al., 1998); qIs74 [GFP::POP-1] (Siegfried et al., 2004); qIs76 [tra-1::GFP] (Mathies et al., 2004); qIs56 [lag-2::GFP] (Blelloch et al., 1999); qIs61 [pes-1::GFP] (Molin et al., 2000); and qIs77 [unc-122::GFP] (Mathies et al., 2003). Extrachromosomal arrays used include: leEx780 [ZK813.3::GFP, pRF4] (a gift of Ian Hope); qEx500 [hs::sys-1] (Kidd et al., 2005). Balancers used include: LGI and LGIII, hT2[qIs48]; LGII, mIn1[mIs14 dpy-10(e128)]; LG IV and V, nT1[qIs51]. qIs48 and mIs14 carry the integrated array of ccEx9747 which is an extrachromosomal array of three GFP constructs: the myo-2 and pes-10 promoters and one driven by a gut-specific enhancer (Edgley and Riddle, 2001).
Isolation, mapping, and molecular analysis of cyd-1(q626)
cyd-1(q626) was isolated in an F2 mutagenesis screen of him-5(e1490) animals for gonadogenesis defects. The q626 mutation was then mapped to a region on LGII between ptr-18 and zyx-1 using a combination of three-factor mapping and SNP mapping as described by Wicks et al. (2001). For SNP mapping, q626, non–unc-52(e444) mutant males were isolated and the following primer sets were used to narrow the q626 region: corresponding to C50E10:22022, 5′-TCGGGTCTCTCCAAAAACTC and 3′-CTCCTTCAGTACCATATGGCTC, cut with Eco R I; corresponding to Y38F1A:41752, 5′-TAGGAAAGTTGTGTCCACCTGG and 3′-TGATGACTCCTTCTTCAGCTGC, cut with Ban II; corresponding to F15D4;22810, 5′-TTCCCATTTTCCTCCCAG and 3′-TCAAAAACCCAGACACTGG, cut with Dra I; corresponding to F37B1:13109, 5′-TCTCCAAGATGAGAGAGAACCACTG and 3′-TCGCCGATTTGCTGGTACAG, cut with Ase I; corresponding to C31C9:11976, 5′-CAGTGACTAACGCCCCAAAACTC and 3′-GCTTCGTGTGATTCTTCTTTTGGG, cut with Nla III; corresponding to Y51H1A:30057, 5′-GATTCGGAATGGGTGTTG and 3′-TCTTGAATGCGTGGTGTG, cut with Taq I; corresponding to Y48B6A:98880, 5′-GGTTTCCAGGTGATTCATAGCG and 3′-TATAGGACGGTTCGGTGAGAAG, cut with Nla III. q626 was placed to the right of the SNP on Y38F1A and to the left of the SNP on F15D4. q626 failed to complement two null alleles of cyd-1, he112 and he116. A PCR fragment containing 1000 bp upstream of the predicted start codon through the predicted poly(A) signal rescued cyd-1(q626) mutants. cyd-1 RNAi was performed as described below. Template for sequencing of cyd-1(q626) was made by amplifying genomic DNA from N2 or cyd-1(q626) animals using Roche PCR reagents. Three independent PCR reactions were sequenced for each cyd-1 fragment amplified using Big-Dye Terminator Ready Reaction Mix (PE/Applied Biosystems). Sequencing of q626 DNA identified an A to G change in the third exon of the coding region.
DAPI staining, lineage analysis, and imaging
DAPI staining was performed as previously described by (Kadyk and Kimble, 1998). Briefly, animals were collected from plates in Eppendorf tubes and washed once in M9, fixed 5 minutes in −20°C methanol, washed twice in M9, stained with 0.1 ug/ml DAPI in M9 for 20 minutes, washed twice in M9 and mounted for fluorescence microscopy. Fluorescent images were captured with a Zeiss Axioskop using Openlab software and processed using Adobe Photoshop and Illustrator. Lineage analysis was performed by following cell divisions with standard DIC microscopy methods on a Zeiss Axioskop (Sulston and Horvitz, 1977). Males were identified by the presence of the large B blast cell in the tail.
RNAi and hydroxyurea experiments
All RNAi experiments were performed by growing synchronized larvae on bacteria expressing dsRNA as previously described (Ashrafi et al., 2003; Kamath et al., 2001) with the exception of RNAi corresponding to cki-1/cki-2, which was injected. For hydroxyurea treatment (HU), synchronized larvae were grown at 20°C for 6 hours then transferred to plates containing 40 mM HU for 4 to 6 hours (Ambros, 1999). Larvae were then transferred back to normal plates. SGP cell division was assayed at the time larvae were transferred back to normal plates and at one-hour intervals until division was observed. In all gon-4(RNAi) experiments, SGPs were delayed by approximately three hours compared to wild-type controls.
Acknowledgments
We gratefully acknowledge Victor Ambros, Ian Hope, Laura Mathies, Jonathan Pettitt and Dave Zarkower for sharing GFP reporter strains, and members of the Kimble laboratory for discussions during the course of this work. We thank Anne Helsley-Marchbanks and Laura Vanderploeg for help preparing the manuscript and figures. J.K. is an investigator with the Howard Hughes Medical Institute (HHMI). C.T. was supported by a National Institutes of Health postdoctoral fellowship (F32 GM069716).
References
- Ahringer J. Control of cell polarity and mitotic spindle positioning in animal cells. Curr Opin Cell Biol. 2003;15:73–81. doi: 10.1016/s0955-0674(02)00018-2. [DOI] [PubMed] [Google Scholar]
- Ambros V. Cell cycle-dependent sequencing of cell fate decisions in Caenorhabditis elegans vulva precursor cells. Development. 1999;126:1947–1956. doi: 10.1242/dev.126.9.1947. [DOI] [PubMed] [Google Scholar]
- Ambros V. The temporal control of cell cycle and cell fate in Caenorhabditis elegans. Novartis Found Symp. 2001;237:203–214. doi: 10.1002/0470846666.ch16. discussion 214–220. [DOI] [PubMed] [Google Scholar]
- Ashrafi K, Chang F, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory cells. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Bei Y, Hogan J, Berkowitz LA, Soto M, Rocheleau CE, Pang KM, Collins J, Mello CC. SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev Cell. 2002;3:113–125. doi: 10.1016/s1534-5807(02)00185-5. [DOI] [PubMed] [Google Scholar]
- Berger C, Pallavi SK, Prasad M, Shashidhara LS, Technau GM. A critical role for Cyclin E in cell fate determination in the central nervous system of Drosophila melanogaster. Nat Cell Biol. 2005;7:56–62. doi: 10.1038/ncb1203. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Knoblich JA. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr Biol. 2004;14:R674–685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Blelloch R, Santa Anna-Arriola S, Gao D, Li Y, Hodgkin J, Kimble J. The gon-1 gene is required for gonadal morphogenesis in Caenorhabditis elegans. Dev Biol. 1999;216:382–393. doi: 10.1006/dbio.1999.9491. [DOI] [PubMed] [Google Scholar]
- Boxem M, van den Heuvel S. lin-35 Rb and cki-1 Cip/Kip cooperate in developmental regulation of G1 progression in C. elegans. Development. 2001;128:4349–4359. doi: 10.1242/dev.128.21.4349. [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Horvitz HR. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol Cell. 2001;7:461–473. doi: 10.1016/s1097-2765(01)00194-0. [DOI] [PubMed] [Google Scholar]
- Chang W, Tilmann C, Thoemke K, Markussen FH, Mathies LD, Kimble J, Zarkower D. A forkhead protein controls sexual identity of the C. elegans male somatic gonad. Development. 2004;131:1425–1436. doi: 10.1242/dev.01012. [DOI] [PubMed] [Google Scholar]
- Chen X, Oh SW, Zheng Z, Chen HW, Shin H-h, Hou SX. Cyclin D-Cdk4 and cyclin E-Cdk2 regulate the Jak/STAT signal transduction pathway in Drosophila. Dev Cell. 2003;4:179–190. doi: 10.1016/s1534-5807(03)00024-8. [DOI] [PubMed] [Google Scholar]
- Coffman JA. Cell cycle development. Dev Cell. 2004;6:321–327. doi: 10.1016/s1534-5807(04)00067-x. [DOI] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu Rev Cell Dev Biol. 2004;20:427–453. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- Datar SA, Jacobs HW, de la Cruz AF, Lehner CF, Edgar BA. The Drosophila cyclin D-Cdk4 complex promotes cellular growth. EMBO J. 2000;19:4543–4554. doi: 10.1093/emboj/19.17.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ, Bowerman B. Asymmetric cell division: fly neuroblast meets worm zygote. Curr Opin Cell Biol. 2001;13:68–75. doi: 10.1016/s0955-0674(00)00176-9. [DOI] [PubMed] [Google Scholar]
- Edgley ML, Riddle DL. LG II balancer chromosomes in Caenorhabditis elegans: mT1(II;III) and the mIn1 set of dominantly and recessively marked inversions. Mol Genet Genomics. 2001;266:385–395. doi: 10.1007/s004380100523. [DOI] [PubMed] [Google Scholar]
- Emmerich J, Meyer CA, de la Cruz AF, Edgar BA, Lehner CF. Cyclin D does not provide essential Cdk4-independent functions in Drosophila. Genetics. 2004;168:867–875. doi: 10.1534/genetics.104.027417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encalada SE, Martin PR, Phillips JB, Lyczak R, Hamill DR, Swan KA, Bowerman B. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev Biol. 2000;228:225–238. doi: 10.1006/dbio.2000.9965. [DOI] [PubMed] [Google Scholar]
- Fay DS. The cell cycle and development: lessons from C. elegans. Semin Cell Dev Biol. 2005;16:397–406. doi: 10.1016/j.semcdb.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Fay DS, Han M. Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell-cycle control and vulval development. Development. 2000;127:4049–4060. doi: 10.1242/dev.127.18.4049. [DOI] [PubMed] [Google Scholar]
- Friedman L, Santa Anna-Arriola S, Hodgkin J, Kimble J. gon-4, a cell lineage regulator required for gonadogenesis in Caenorhabditis elegans. Dev Biol. 2000;228:350–362. doi: 10.1006/dbio.2000.9944. [DOI] [PubMed] [Google Scholar]
- Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, Lassar AB. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- Hall DH, Winfrey VP, Blaeuer G, Hoffman LH, Furuta T, Rose KL, Hobert O, Greenstein D. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: Relations between the germ line and soma. Dev Biol. 1999;212:101–123. doi: 10.1006/dbio.1999.9356. [DOI] [PubMed] [Google Scholar]
- Herman MA. Control of cell polarity by noncanonical Wnt signaling in C. elegans. Semin Cell Dev Biol. 2002;13:233–241. doi: 10.1016/s1084-9521(02)00051-4. [DOI] [PubMed] [Google Scholar]
- Herman MA, Horvitz HR. The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development. 1994;120:1035–1047. doi: 10.1242/dev.120.5.1035. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. Male phenotypes and mating efficiency in Caenorhabditis elegans. Genetics. 1983;103:43–64. doi: 10.1093/genetics/103.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin JA, Brenner S. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics. 1977;86:275–287. [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Roy R, Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 1998;125:3585–3597. doi: 10.1242/dev.125.18.3585. [DOI] [PubMed] [Google Scholar]
- Hope IA. ‘Promoter trapping’ in Caenorhabditis elegans. Development. 1991;113:399–408. doi: 10.1242/dev.113.2.399. [DOI] [PubMed] [Google Scholar]
- Kadyk LC, Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biology. 2001;2:research0002.0001–0002.0010. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd AR, III, Miskowski JA, Siegfried KR, Sawa H, Kimble J. A β-catenin identified by functional rather than sequence criteria and its role in Wnt/MAPK signaling. Cell. 2005;121:761–772. doi: 10.1016/j.cell.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Kimble J. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev Biol. 1981;87:286–300. doi: 10.1016/0012-1606(81)90152-4. [DOI] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kimble JE, White JG. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 1981;81:208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- Koreth J, van den Heuvel S. Cell-cycle control in Caenorhabditis elegans: how the worm moves from G1 to S. Oncogene. 2005;24:2756–2764. doi: 10.1038/sj.onc.1208607. [DOI] [PubMed] [Google Scholar]
- Kozar K, Ciemerych MA, Rebel VI, Shigematsu H, Zagozdzon A, Sicinska E, Geng Y, Yu Q, Bhattacharya S, Bronson RT, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Lin R, Thompson S, Priess JR. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- Lo MC, Gay F, Odom R, Shi Y, Lin R. Phosphorylation by the β-catenin/MAPK complex promotes 14-3-3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C. elegans. Cell. 2004;117:95–106. doi: 10.1016/s0092-8674(04)00203-x. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Lin R, Rothman JH. Dynamics of a developmental switch: recursive intracellular and intranuclear redistribution of Caenorhabditis elegans POP-1 parallels Wnt-inhibited transcriptional repression. Dev Biol. 2002;248:128–142. doi: 10.1006/dbio.2002.0721. [DOI] [PubMed] [Google Scholar]
- Mathies LD, Henderson ST, Kimble J. The C. elegans Hand gene controls embryogenesis and early gonadogenesis. Development. 2003;130:2881–2892. doi: 10.1242/dev.00483. [DOI] [PubMed] [Google Scholar]
- Mathies LD, Schvarzstein M, Morphy KM, Blelloch R, Spence AM, Kimble J. TRA-1/GLI controls development of somatic gonadal precursors in C. elegans. Development. 2004;131:4333–4343. doi: 10.1242/dev.01288. [DOI] [PubMed] [Google Scholar]
- Meyer CA, Jacobs HW, Lehner CF. Cyclin D-cdk4 is not a master regulator of cell multiplication in Drosophila embryos. Curr Biol. 2002;12:661–666. doi: 10.1016/s0960-9822(02)00770-4. [DOI] [PubMed] [Google Scholar]
- Miskowski J, Li Y, Kimble J. The sys-1 gene and sexual dimorphism during gonadogenesis in Caenorhabditis elegans. Dev Biol. 2001;230:61–73. doi: 10.1006/dbio.2000.9998. [DOI] [PubMed] [Google Scholar]
- Molin L, Mounsey A, Aslam S, Bauer P, Young J, James M, Sharma-Oates A, Hope IA. Evolutionary conservation of redundancy between a diverged pair of forkhead transcription factor homologues. Development. 2000;127:4825–4835. doi: 10.1242/dev.127.22.4825. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Park CS, Kim SI, Lee MS, Youn C-y, Kim DJ, Jho E-h, Song WK. Modulation of β-catenin phosphorylation/degradation by cyclin-dependent kinase 2. J Biol Chem. 2004;279:19592–19599. doi: 10.1074/jbc.M314208200. [DOI] [PubMed] [Google Scholar]
- Park M, Krause MW. Regulation of postembryonic G1 cell cycle progression in Caenorhabditis elegans by a cyclin D/CDK-like complex. Development. 1999;126:4849–4860. doi: 10.1242/dev.126.21.4849. [DOI] [PubMed] [Google Scholar]
- Pettitt J, Wood WB, Plasterk RHA. cdh-3, a gene encoding a member of the cadherin superfamily, functions in epithelial cell morphogenesis in Caenorhabditis elegans. Development. 1996;122:4149–4157. doi: 10.1242/dev.122.12.4149. [DOI] [PubMed] [Google Scholar]
- Prokopenko SN, Chia W. When timing is everything: role of cell cycle regulation in asymmetric division. Semin Cell Dev Biol 16, 423–437. Roegiers, F., and Jan, Y. N. (2004). Asymmetric cell division. Curr Opin Cell Biol. 2005;16:195–205. doi: 10.1016/j.semcdb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- Siegfried K, Kimble J. POP-1 controls axis formation during early gonadogenesis in C. elegans. Development. 2002;129:443–453. doi: 10.1242/dev.129.2.443. [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Kidd AR, III, Chesney MA, Kimble J. The sys-1 and sys-3 genes cooperate with Wnt signaling to establish the proximal-distal axis of the C. elegans gonad. Genetics. 2004;166:171–186. doi: 10.1534/genetics.166.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg PW, Horvitz HR. lin-17 mutations of Caenorhabditis elegans disrupt certain asymmetric cell divisions. Dev Biol. 1988;130:67–73. doi: 10.1016/0012-1606(88)90414-9. [DOI] [PubMed] [Google Scholar]
- Sulston, J., and Hodgkin, J. (1988). Methods, In The nematode Caenorhabditis elegans, W. B. Wood, ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp 587–606.
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tio M, Udolph G, Yang X, Chia W. cdc2 links the Drosophila cell cycle and asymmetric division machineries. Nature. 2001;409:1063–1067. doi: 10.1038/35059124. [DOI] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RHA. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Yanowitz J, Fire A. Cyclin D involvement demarcates a late transition in C. elegans embryogenesis. Dev Biol. 2005;279:244–251. doi: 10.1016/j.ydbio.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Zarkower D, Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell. 1992;70:237–249. doi: 10.1016/0092-8674(92)90099-x. [DOI] [PubMed] [Google Scholar]
- Zhang JM, Chen L, Krause M, Fire A, Paterson BM. Evolutionary conservation of MyoD function and differential utilization of E proteins. Dev Biol. 1999;208:465–472. doi: 10.1006/dbio.1999.9218. [DOI] [PubMed] [Google Scholar]


