Abstract
DNA nonhomologous end-joining (NHEJ) is the major pathway for repairing DNA double-strand breaks in mammalian cells. It also functions to carry out rearrangements at the specialized breaks introduced during V(D)J recombination. Here, we describe a patient with T−B− severe combined immunodeficiency, whose cells have defects closely resembling those of NHEJ-defective rodent cells. Cells derived from this patient show dramatic radiosensitivity, decreased double-strand break rejoining, and reduced fidelity in signal and coding joint formation during V(D)J recombination. Detailed examination indicates that the patient is defective neither in the known factors involved in NHEJ in mammals (Ku70, Ku80, DNA-dependent protein kinase catalytic subunit, Xrcc4, DNA ligase IV, or Artemis) nor in the Mre11/Rad50/Nbs1 complex, whose homologue in Saccharomyces cerevisiae functions in NHEJ. These results provide strong evidence that additional activities are crucial for NHEJ and V(D)J recombination in mammals.
DNA double-strand breaks (DSBs) can be caused by both exogenous DNA-damaging agents, such as ionizing radiation (IR) or reactive oxygen species, and by endogenous cellular processes, such as recombination or replication. Because a DSB can lead to loss or rearrangement of genomic material, pathways for their repair are critically important for genomic stability. Clinical manifestations of defective DNA repair can include increased radiation sensitivity and predisposition to cancer. Because normal development of the immune system requires the introduction of DSBs during antigen receptor gene assembly, defects in the repair of these specialized breaks can also lead to profound immunodeficiencies (1, 2).
During V(D)J recombination, the recombination-activating gene 1 (RAG1) and RAG2 proteins collaborate to introduce a pair of DSBs in the chromosome between recombination signal sequences and the antigen receptor coding segments. The resulting DSBs have two distinct structures, a blunt signal end and a hairpin coding end, which are subsequently resolved to form two new junctions, signal joints (SJ) and coding joints (CJ) (3, 4). Ubiquitously expressed components of the nonhomologous end-joining pathway (NHEJ) are required for the repair of radiation-induced DSBs and the rejoining of the DNA ends specifically generated during V(D)J recombination (1, 5). Several of these factors have been identified to date: the DNA-dependent protein kinase (DNA-PK), which consists of the DNA-PK catalytic subunit (DNA-PKcs) and the Ku70/Ku80 heterodimer that associates with it and binds to DNA ends (for a review, see ref. 5); Artemis (6); Xrcc4; and ligase IV (7, 8). Defects in any of these factors can lead to pronounced radiosensitivity and immunodeficiency.
To determine whether defects in other factors known to be involved in DSB repair could contribute to clinical immunodeficiency and to identify novel genes involved in V(D)J recombination and DSB repair, we analyzed cells derived from patients with a clinical presentation that would be consistent with such a defect. Here we describe the analysis of one such patient. Cells derived from this patient show profound radiosensitivity and a marked defect in DSB repair. V(D)J recombination is also affected, with abnormalities in both SJ and CJ formation. This constellation of defects, in conjunction with the clinical manifestations, is unlike that of any previously described human syndrome. No defects in the factors previously shown to function in the NHEJ pathway in mammalian cells were found, providing strong evidence that the defect lies in an uncharacterized component of the NHEJ pathway.
Materials and Methods
Clinical Assessment of 2BN and 3BN.
Clinical and laboratory investigations for immunodeficiencies were carried out as reported (9, 10).
Cell Culture.
Material was obtained from the patient with informed consent. 2BN represents the primary skin fibroblast cell line derived from the patient and 2BNneo, an SV40-transformed but not immortalized derivative. 2BNhTERT was derived by stable expression of the catalytic subunit of human telomerase. 1BR3 and 1BRneo are normal primary and transformed fibroblast cell lines, respectively, used as controls. 1604hTERT is a normal cell line immortalized by human telomerase. M059J and M059K are glioma cell lines lacking and expressing DNA-PKcs, respectively (11). Cells were cultured in MEM supplemented with 15% FCS, penicillin, and streptomycin, as described (12). Clonogenic assays for radiosensitivity were as described (12).
Measurement of V(D)J Recombination.
V(D)J recombination was measured according to standard methods (13), except that cells were transfected by lipofection with lipofectAMINE (GIBCO/BRL). The fraction of perfect joints was measured by hybridization with a signal junction-specific probe, SJ2 (13). The NHEJ-deficient control cell lines used for the complementation experiments are as follows: Ku70, Ku70-deficient embryonic stem cells (14); Ku80, Xrs-6 (15); DNA-PKcs, SC3T3W (16); Xrcc4, XR-1 cells (17); DNA ligase IV; 411BR (18); Artemis, CJ179. CJ179 is an Artemis-defective fibroblast line derived from a severe combined immunodeficiency patient. The line has no detectable Artemis transcript (unpublished observations).
Measurement of DSB Rejoining.
Two to 3 × 106 cells were labeled with 0.02 μCi/ml [14C]thymidine (Amersham Pharmacia Life Sciences, 50–60 mCi/mmol) for 48–50 h followed by incubation for 2 h in unlabeled medium. Agarose plugs were prepared from the cells, now in stationary phase, irradiated with 40-Gy γ-rays and incubated for repair as described (19). The cells were lysed in the plugs and DNA fragments separated by pulsed-field gel electrophoresis (PFGE) by using a Bio-Rad CHEF DRIII with forward and backward pulse times of 30 min and a total electrophoresis time of 48 h at 1.5 V/cm. After electrophoresis, the gel was placed onto DE18 paper (Whatman), dried for 3 h at 50°C, and analyzed on a PhosphorImager (Storm, Molecular Dynamics) by using imagequant software.
Immunoblotting and Biochemical Assays.
Whole-cell extracts were prepared by the method of Scholer et al. (20). For immunoblotting, whole-cell extracts were boiled in SDS/PAGE loading buffer, separated in 6% SDS/PAGE, and the proteins transferred to nitrocellulose by using a wet-blotting apparatus. The antibodies used were the following: for Ku80, Ku80-4; for Ku70, N3H10; for Xrcc4, SJA4; and an anti-DNA ligase IV antibody as described (21). DNA end-binding (electrophoretic mobility-shift assay), DNA-PK, and DNA ligase IV adenylation assay were as described (22–24). For the in vitro NHEJ assay, whole-cell extracts and DNA-PKcs were prepared and assays carried out as described (25, 26).
Results
Two siblings, patients 2BN and 3BN, were diagnosed with an NK cell predominant form of combined immunodeficiency (details of the immunologic evaluation can be found in Table 4, which is published as supporting information on the PNAS web site, www.pnas.org). The parents of the affected children are of Caucasian background and are not consanguineous, and there was no additional family history of immunodeficiency. Both children received bone marrow transplants before this study. Primary fibroblast cell lines were obtained from both patients, but only the cells from 2BN could be maintained in culture.
Decreased Efficiency and Fidelity of V(D)J Joining in 2BN Cells.
2BN cells were examined for their ability to carry out the V(D)J rearrangement process that assembles antigen receptor genes during lymphoid development (4). No characterization of the endogenous receptor rearrangement in the B and T cells of patient 2BN was possible, because no patient material derived before transplantation was available. V(D)J recombination was, therefore, analyzed in transformed 2BN fibroblast cells (2BNneo) transiently transfected with a recombination substrate and plasmids expressing RAG1 and RAG2 (see Materials and Methods). The frequencies of both SJ and CJ formation were reproducibly lower in 2BNneo cells than in normal 1BRneo control cells, with CJ formation more severely affected than SJ (a 30-fold decrease for CJ vs. 7-fold for SJ; representative data are shown in Table 1).
Table 1.
Analysis of signal and coding joint formation in 2BN cells
| Experiment | Cells | Signal joints
|
Coding joints
|
|||
|---|---|---|---|---|---|---|
| No. of plasmids, ×103 | Rec. freq., % | Perfect joints, % | No. of plasmids, ×103 | Rec. freq., % | ||
| 1 | 1BR3neo | 107 | 4.0 | 100 | 30 | 2.35 |
| 2BNneo | 16 | 0.25 | 54 | 15 | 0.05 | |
| 2 | 1BR3neo | 39 | 4.3 | 100 | 19 | 1.4 |
| 2BNneo | 57 | 0.75 | 55 | 64 | 0.03 | |
| 3 | 1BR3neo | 20 | 3.5 | 100 | 6 | 2.0 |
| 2BNneo | 16 | 0.5 | 59 | 11 | 0.06 | |
| 4 | 1BR3neo | 36 | 1.0 | 100 | 22 | 1.25 |
| 2BNneo | 107 | 0.26 | 48 | 112 | 0.05 | |
The results from four representative transfections are shown. No. of plasmids, number of Amp-resistant plasmids recovered, reflecting the number of plasmids screened; Rec. freq., recombination frequency shown; Perfect joints, percentage of AmpR/CamR colonies that hybridized to the SJ2 probe (see Materials and Methods).
In addition to the decreased frequency of CJ and SJ formation, the junctions themselves were unusual. SJ are normally the result of precise fusions of recombination signal sequences, heptamer to heptamer. However, on average only 54% of the 2BN signal junctions were rejoined precisely, as determined by hybridization to a junction-specific probe. Still lower levels of precise SJ formation were observed in individual subclones of 2BN cells immortalized with human telomerase (see Table 2, experiments 1–5 and 8). Sequence analysis of 35 junctions from 4 independent transfection experiments of 2BN primary cells and 2BNneo revealed nucleotide deletions of 1–23 bases, with no apparent insertions (see Fig. 4A, which is published as supporting information on the PNAS web site). Two major classes of events were found: one, representing 10/35 junctions, was the loss of five nucleotides from the junction (either all five from one heptamer or four from one heptamer and one nucleotide from the other), and the second, observed in 11/35 junctions, was the complete loss of a heptamer with retention of all of the nucleotides in the spacer sequence.
Table 2.
Expression of known NHEJ factors fails to complement the signal-joining defect
| Experiment | cDNA | No. of plasmids, ×103 | Rec. freq., % | Perfect joints, % | |
|---|---|---|---|---|---|
| 1 | A | − | 30 | 0.3 | 11 |
| Ku70 | 18 | 0.6 | 16 | ||
| B | − | 16 | 1.2 | 15 | |
| Ku70 | 9 | 1.0 | 9 | ||
| 2 | A | − | 14 | 1.9 | 9 |
| Ku80 | 14 | 1.5 | 11 | ||
| B | − | 26 | 0.7 | 13 | |
| Ku80 | 12 | 1.2 | 14 | ||
| 3 | A | − | 16 | 0.9 | 25 |
| DNA-PKcs | 23 | 1.3 | 27 | ||
| B | − | 14 | 1.1 | 22 | |
| DNA-PKcs | 11 | 0.2 | 20 | ||
| 4 | A | − | 28 | 0.6 | 15 |
| XRCC4 | 43 | 0.4 | 17 | ||
| B | − | 35 | 0.7 | 10 | |
| XRCC4 | 8 | 1.2 | 9 | ||
| 5 | A | − | 23 | 0.9 | 21 |
| Lig IV | 30 | 0.8 | 19 | ||
| B | − | 16 | 1.1 | 18 | |
| Lig IV | 12 | 1.3 | 24 | ||
| 6 | A | − | 16 | 0.3 | 52 |
| Artemis | 14 | 0.3 | 56 | ||
| B | − | 18 | 0.4 | 46 | |
| Artemis | 15 | 0.3 | 40 | ||
| 7 | − | 136 | 0.35 | 63 | |
| hMre11 | 27 | 0.5 | 54 | ||
| hRad50 | 94 | 0.45 | 48 | ||
| 8 | A | − | 27 | 0.4 | 14 |
| NBS1 | 33 | 0.5 | 11 | ||
| B | − | 7 | 0.7 | 10 | |
| NBS1 | 15 | 0.3 | 13 |
All transfections include RAG1 and RAG2 expression vectors and a signal joint reporter plasmid and either no additional plasmid (−) or a plasmid expressing the indicated cDNA. For experiments 1–6, transfections into cell lines known to be defective in the indicated gene were carried out as a control (see Materials and Methods). In each case, expression of the cDNA served to complement the V(D)J defect in the control cell lines with coding joint formation assayed for the DNA-PKcs and Artemis cell lines and signal joint formation for all others (data not shown). Experiments 1–5 and 8 were carried out in 2BNhTERT cells, whereas experiments 6 and 7 were performed with 2BNneo cells.
Coding junctions derived from 2BN cells were also different from those obtained from their normal counterparts (Fig. 4B). Although the number of bases lost from 1BRneo control cells ranged from 1 to 8 bp, that for 2BN cells ranged from 3 to 37, with 27% of the sequenced junctions having deletions larger than 8 bp. No apparent clustering of break points was detected, but a slightly higher usage of microhomology at junctions in 2BN cells compared with normal cells was noted. Thus, as for the SJ, the CJ of 2BN cells are decreased both in number and fidelity.
2BN Cells Are Dramatically Radiosensitive and Defective in DNA DSB Rejoining.
The abnormal V(D)J recombination observed in 2BN cells expressing wild-type RAG1 and RAG2 indicated that the cells were defective in carrying out the repair phase of the recombination reaction. To further characterize the repair defect in these cells, their response to DNA-damaging agents was assessed. The response of primary (2BN) and SV40-transformed (2BNneo) cell lines was examined in comparison to normal primary (1BR) and transformed (1BRneo) cells and a telomerase-immortalized cell line 1604hTERT. Two additional cell lines, AT1BR and 180BR, were included for comparison where appropriate. AT1BR is derived from a patient with ataxia–telangiectasia (A-T), a syndrome associated with both clinical and cellular radiosensitivity, whereas 180BR cells harbor a defect in DNA ligase IV (21, 27).
2BN primary cells showed dramatic sensitivity to IR in clonogenic survival analysis as did the SV40-transformed derivative (Fig. 1A). 2BNhTERT cells showed the same level of IR sensitivity as 2BN primary cells (data not shown). Elevated chromosomal radiosensitivity as measured by the level of chromosome breaks in mitotic cells 4 h postirradiation was also observed in 2BN cells (see Table 5, which is published as supporting information on the PNAS web site). However, spontaneous chromosome breakage was not significantly elevated (data not shown). 2BN cells have normal sensitivity to UV irradiation and to the DNA crosslinking agent, mitomycin C, demonstrating that their defect is specific to lesions induced by IR (data not shown).
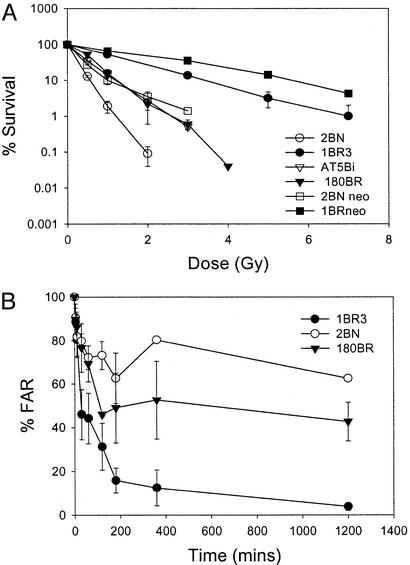
Figure 1.
2BN cells show radiosensitivity and defective DSB repair. (A) Survival of 2BN cells after exposure to IR. Cells were exposed to γ-rays and survival was estimated by colony formation as described (12). The data represent the mean results of a minimum of three experiments for each cell line, and error bars represent the standard deviation of the mean. The survival response of 2BNneo cells represents the mean of two experiments, and error bars have not been included. The survival of 2BNhTERT cells (the mean of two experiments) was identical to that of 2BN primary cells. (B) DSB repair in 2BN cells; estimation of DNA DSB rejoining by PFGE. Cells, radiolabeled with [14C]-TdR, were embedded in agarose, irradiated with 40 Gy γ-rays, incubated for varying times, and subjected to PFGE. The fraction of activity released (FAR) represents the radioactivity that enters the gel, divided by the total activity (well + lane). The results are presented as the ratio of the FAR remaining at the specified time compared with the FAR at time 0. The FAR for unirradiated cells is subtracted before these estimations.
Several assays serve to distinguish radiosensitive cell lines defective in NHEJ from those defective in other damage response mechanisms. A hallmark of cells defective in NHEJ is a reduced ability to rejoin radiation-induced DNA DSBs (5). When DSB rejoining capacity was assessed by using PFGE, 2BN cells were significantly defective compared with control cells (Fig. 1B). This defect was slightly greater than that observed in DNA ligase IV-defective 180BR cells. The cell cycle checkpoint responses of 2BN cells were also assessed. After exposure to IR, normal eukaryotic cells arrest at critical cell cycle checkpoints. Although the operation of the G1/S checkpoint is not impaired in primary cells deficient in the characterized NHEJ proteins (2), radiosensitive cell lines from A-T patients fail to arrest at the G1/S checkpoint (28, 29). The progression of primary 2BN cells from G1 into S phase was monitored by fluorescence activated cell sorting after irradiation (see Supporting Text, which is published as supporting information on the PNAS web site). 2BN cells arrested at the G1/S boundary similarly to normal cells, in marked contrast to the lack of arrest observed in A-T cells (see Fig. 5A, which is published as supporting information on the PNAS web site).
Mammalian cells also exhibit inhibition of DNA synthesis after irradiation, reflecting the operation of an S phase checkpoint. Cell lines defective in this process, including those derived from A-T and Nijmegen Breakage Syndrome patients, display characteristic radioresistant DNA synthesis (30, 31). The arrest of DNA synthesis after irradiation was similar in 2BN and control cells (Fig. 5B). Thus, 2BN cells exhibit the characteristic phenotype of cells defective in NHEJ.
2BN Cell Extracts Fail to Support DNA End-Joining in Vitro.
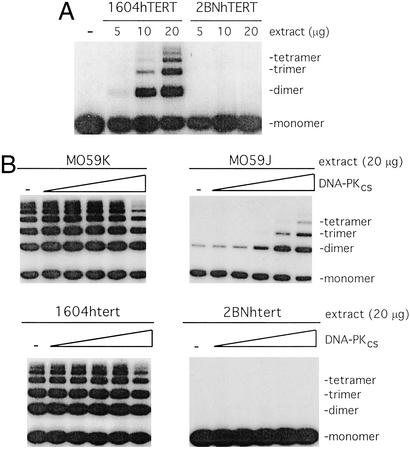
NHEJ activity can also be assessed in vitro where extracts from human cell lines have been shown to promote DNA end joining (25). The joining reaction exhibits a dependence on known components of the NHEJ pathway (Ku70/80, DNA-PKcs, Xrcc4/ligase IV) and is stimulated by IP6 (26). To determine whether cell lines derived from patient 2BN could promote end-joining in vitro, whole-cell extracts were prepared from 2BNhTERT and a control cell line 1604hTERT. Whereas the control line promoted highly efficient end-joining of linearized plasmid DNA, as observed by the formation of multimeric species, the 2BNhTERT extracts were completely devoid of end-joining activity (Fig. 2A). Thus, the NHEJ defect that is observed in vivo is also reflected in the in vitro assay system.
Figure 2.
2BNhTERT cells fail to promote end-joining in vitro. (A) Cell-free extracts prepared from either the control cell line 1604hTERT or 2BNhTERT were incubated with 5′-32P-end-labeled linear plasmid DNA. The products of end-joining were analyzed by gel electrophoresis. (B) Extracts from either 1604hTERT, 2BNhTERT, DNA-PKcs-proficient cells (MO59K), or DNA-PKcs defective cells (MO59J) were incubated with 5′-32P-end-labeled linear plasmid DNA. Where indicated, extracts were complemented by addition of purified DNA-PKcs (0, 0.14, 0.43, 1.3, 3.9, or 11.6 nM). DNA products were analyzed as described in A.
2BN Cells Are Not Defective in Any of the Characterized NHEJ Activities.
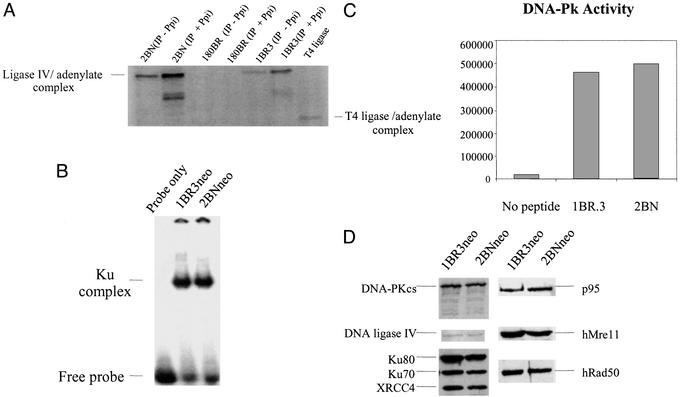
Six mammalian proteins, Ku70, Ku80, DNA-PKcs, DNA ligase IV, Xrcc4, and Artemis, are known to affect both radiosensitivity and V(D)J recombination. To gain insight into the molecular basis underlying the defect in 2BN cells, the expression and function of these factors were examined by a variety of genetic and biochemical methods (see Figs. 2 and 3, Table 2, and summary in Table 3). For each of these proteins except Artemis, standard assays to test for their biochemical activities were performed (see Materials and Methods) and, as detailed below, no defects were found. Similar amounts of DNA ligase IV were immunoprecipitated by using anti-XRCC4 antibodies from control and 2BN cells and equal levels of DNA ligase IV-adenylate complex were generated in an adenylation assay (21) (Fig. 3A). Thus, Xrcc4 and DNA ligase IV can interact efficiently, and a ligase IV adenylate complex, the first step in the ligation reaction, can be formed normally. By electrophoretic mobility-shift assay, 2BN cells had normal levels of a double-stranded DNA end-binding activity, having the mobility expected of Ku (Fig. 3B). DNA-PK activity was normal as assessed by the ability of a DNA-bound fraction of cell proteins to phosphorylate a p53-derived peptide (Fig. 3C), an assay that can detect defects in known DNA-PK-defective cell lines (refs. 11, 24, and 32, and data not shown). We also exploited the in vitro end-joining assay to verify that the defect in 2BN cells is not due to a lack of DNA-PKcs activity. We found that purified DNA-PKcs could complement extracts prepared from the DNA-PKcs-defective cell line M059J, whereas no complementation was observed with 2BNhTERT extracts (Fig. 2D). In addition, Western blotting with specific antibodies to Ku70, Ku80, DNA-PKcs, DNA ligase IV, and Xrcc4 demonstrated that all five proteins were expressed at normal levels (Fig. 3A).
Figure 3.
2BN cells show normal expression and activity of characterized NHEJ proteins. (A) DNA ligase IV adenylation activity. Because DNA ligase IV antibodies are inefficient in IP, anti-Xrcc4 antibodies were used to coimmunoprecipitate DNA ligase IV. DNA ligase IV exists endogenously as an enzyme–adenylate complex; therefore, extracts were or were not treated with 5 mM inorganic pyrophosphate (Ppi) to disrupt preformed AMP–ligase complexes and then analyzed for DNA ligase IV–adenylate complex formation. As expected, the signal is greater after Ppi treatment. DNA ligase IV-defective 180BR cells were included as a control (21). (B) DNA end-binding activity. Whole-cell extracts (5 μg) from 1BR3 and 2BN cells were incubated with [γ-32P]dATP-labeled M1/M2 oligonucleotide probe and separated by PAGE. The position of the free probe and Ku-dependent bands is highlighted. The band was shown to be Ku-dependent, because it is absent in Ku80-defective Xrs-6 cells and is regained in xrs-6 complemented with human Ku80 cDNA (data not shown and ref. 22). (C) DNA-Pk activity. DNA-binding proteins were microfractionated from whole-cell extracts (10 μg) of 1BR3 and 2BN cells using DNA cellulose beads and incubated with a p53-derived peptide in the presence of [γ-32P]dATP. The peptide was separated from other nonspecific phosphorylated proteins by PAGE and quantified by PhosphorImager counting. The results represent arbitrary PhosphorImager units. (D) Western blot analysis of NHEJ proteins in 2BN cells. Whole-cell extracts of transformed 2BN and control cells were analyzed by Western blotting by using antibodies against DNA ligase IV, Ku80 (Ku80-4), Ku70 (N3H10), Xrcc4, and DNA-PKcs (MSAG3). Anti-hRad50, hMre11, and p95 antibodies were a kind gift from J. H. J. Petrini (35).
Table 3.
2BN cells are not defective in factors known to be required for DSB repair and V(D)J joining in mammalian cells
| Factor | Seq. | Western | Activity | Compl. |
|---|---|---|---|---|
| Ku70 | + | + | + | − |
| Ku80 | + | + | + | − |
| DNA-PKcs | ND | + | + | − |
| Xrcc4 | + | + | + | − |
| Ligase IV | + | + | + | − |
| Artemis | + | ND | ND | − |
| hMre11 | ND | + | ND | − |
| hRad50 | ND | + | ND | − |
| Nbs1 | ND | + | ND | − |
Seq., analysis by RT-PCR sequencing (“+” reflects a wild-type sequence); Western, analysis of protein levels by Western immunoblotting (“+” indicates that the protein was present at a size and level indistinguishable from wild type); Activity, examination of the activity associated with the particular protein (see text and Materials and Methods for description of activity tested); Compl., complementation analysis using the defect in signal joint fidelity as an assay (see Table 1). ND, not done.
All of the candidate genes, with the exception of the 12,386-bp DNA-PKcs gene, were examined by RT-PCR sequencing. All genes had wild-type sequences (data not shown). In the case of Artemis, two nucleotide changes were observed compared with the published sequence (a G to T at position 1657 resulting in the conservative amino acid substitution of a leucine for a valine and a silent C to T mutation at position 1788). However, the same nucleotide substitutions were observed in a normal control cell (1BR), suggesting that they are not responsible for the mutant phenotype in 2BN cells.
To further exclude the possibility that defects in the known NHEJ components might underlie the phenotype of 2BN cells, we asked whether expression of functional cDNAs for these candidate genes could complement the V(D)J defect in the frequency and fidelity of SJ formation. For these experiments 2BNhTERT cells were used due to their ease of growth. Analysis of 2BNhTERT cells revealed a slightly more dramatic defect than previously observed with 2BNneo cells in the fidelity of signal rejoining with precise joining ranging from 7.5 to 40% in individual subclones. The frequency of SJ, however, was similar to that observed in 2BNneo cells. Transient transfection of 2BNhTERT cells with plasmids expressing cDNAs for human Ku70, Ku 80, DNA-Pkcs, Artemis, Xrcc4, or DNA ligase IV in conjunction with plasmids expressing the RAG genes and a recombination substrate failed to rescue their V(D)J recombination defect (Table 2). However, when cell lines known to be defective in each of these genes were similarly transfected with the appropriate complementing cDNA, V(D)J recombination was restored to wild-type or near wild-type levels, confirming that the cDNA expression vectors were indeed functional (data not shown, but see Table 2 legend and Materials and Methods for details). Thus, the phenotype observed in 2BN cells cannot result simply from the lack of expression of any of these genes.
Although the six proteins discussed above are the only ones presently known to have an impact on both radiosensitivity and V(D)J recombination, we also examined mammalian homologues of Mre11, Rad50, and Xrs2, because they are required for NHEJ in S. cerevisiae (33, 34). Nbs1, the protein defective in Nijmegen Breakage Syndrome, a disorder also associated with immunodeficiency, appears to represent a functional homologue of Xrs2 (35, 36). 2BN cells expressed normal levels of Mre11, Rad50, and Nbs1 (p95) by Western immunoblotting. Transient transfection of cDNAs expressing hMre11, hRad50, or Nbs1 failed to complement the V(D)J defect (Fig. 3D and Table 2).
Discussion
Here, we have studied cells derived from a patient originally receiving medical attention because of her profound immunodeficiency (and the prior history of immunodeficiency in her sibling). These cells are dramatically radiosensitive, defective in rejoining DSBs, and elevated in their chromosomal radiosensitivity. Moreover, the efficiency and fidelity of V(D)J recombination in these cells is significantly reduced. This combination of features strongly suggests that the cells harbor a defect in a component of the NHEJ machinery.
Extensive examination suggests that the defect in 2BN cells does not reside in any of the characterized factors involved in NHEJ and V(D)J recombination in mammalian cells. As summarized in Table 3, protein expression, biochemical assays, and cDNA sequencing failed to uncover a defect in any of the candidate proteins. Furthermore, complementation studies indicate that expression of the wild-type version of Ku70, Ku80, DNA-PKcs, Artemis, Xrcc4, and ligase IV factors as well as hMre11, hRad50, and Nbs1, does not lead to an increase in SJ fidelity, strongly suggesting that the defect lies in another gene. Although the complementation experiments could be misleading if the defect was a dominant negative, the genetics, with two affected siblings born of unaffected parents, is most consistent with a recessive mutation. Thus, our analysis strongly suggests that NHEJ, the process that functions jointly in V(D)J recombination and the repair of radiation-induced DNA damage in mammalian cells, requires at least one additional component.
Mutations in the different characterized components of the NHEJ pathway lead to a distinct constellation of phenotypes observable in the cultured cell lines, in knock-out mice and in human patients. The severity of the effect on immune system development or V(D)J recombination is not always paralleled by the magnitude of the sensitivity to IR, the deficiency in DSB repair or the severity of clinical features. Furthermore, different aspects of V(D)J joining can be differentially affected by mutations in the different components.
The phenotype that we describe for 2BN cells is distinct from that of previously characterized radiosensitive human cell lines. The level of radiation sensitivity is unparalleled, with sensitivity equal to or greater than cells lacking ataxia telangiectasia mutated, which are among the most exquisitely radiosensitive cell lines described. 2BN cells also show a greater degree of radiation sensitivity than human cell lines deficient in ligase IV (180BR, 411BR, and FB2303) (18). The V(D)J defect is also distinct. Although the LIG4 syndrome cells and 2BN cells both show decreased signal joining fidelity, the effect on CJ (in terms of both frequency and fidelity) is greater for 2BN than for LIG4 syndrome. The LIG4 syndrome patients also show distinct clinical features with more pronounced developmental anomalies than observed in patient 2BN or her brother. The V(D)J defect in 2BN cells is most similar to that of murine severe combined immunodeficiency cells, although 2BN cells show a greater loss of SJ fidelity coupled with a less severe defect in CJ frequency. 2BN cells also differ from the recently described Artemis-deficient cell lines, because the latter, although radiosensitive, are not defective in DSB repair as assessed by PFGE (6, 37). Furthermore, Artemis-deficient cell lines do not display the characteristic effect on SJ fidelity observed for 2BN cells, although they exhibit a more profound deficiency in CJ formation. Finally, 2BN cells are proficient in the cell cycle checkpoint signaling responses defective in both A-T and Nijmegen Breakage Syndrome cell lines, consistent with a specific defect in NHEJ rather than a broader defect in a DSB signaling mechanism (38).
Although the distinctions discussed above could in part reflect the result of hypomorphic non-null mutations and the interaction with different genetic backgrounds, the results serve to underscore the overlapping nature of the pathways involved in the repair of radiation-induced breaks, the broken ends arising from V(D)J joining and restriction enzyme cuts. Our findings provide strong evidence for the existence of an additional factor required for the repair of all three of these types of DSB. They also provide further evidence that defects in the NHEJ components can contribute to human immunodeficiency, and that NHEJ is required for normal human growth.
Supplementary Material
Acknowledgments
We thank David Gossage, M.D., and Richard Harris, M.D., Children's Hospital Medical Center, Cincinnati, for referral of 2BN and 3BN for further evaluation of immune abnormalities; and Drs. P.-M. Girard and B. Singleton from the Jeggo laboratory and Drs. Priya Sudarsanam, Susan Kirch, and Adam Matthews from the Oettinger laboratory for helpful discussions and technical contributions to this work. We thank Drs. J. Petrini (Memorial Sloan–Kettering Cancer Center) and S. Jackson for antibodies; F. Alt (Harvard Medical School) and M. Gellert (National Institutes of Health) for cDNAs; M. Lieber (University of Southern California) for recombination substrates; Y. Gu (University of Washington School of Medicine) for cell lines and cDNAs; and L. McDaniel (University of Texas Southwestern Medical Center) for immortalization of 2BN cells. This work was funded by grants from the Leukaemia Research Foundation (P.A.J.), the Human Frontiers Science Program (P.A.J.), the industry-funded United Kingdom Coordinating Committee on Cancer Research Radiation Research Program (P.A.J.), the Primary Immunodeficiency Association (P.A.J.), the Pew Scholars Program (M.A.O.), the Leukemia and Lymphoma Scholars Program (M.A.O.), National Institutes of Health Grant GM58026 (M.A.O.), and Cancer Research U.K. (S.C.W.).
Abbreviations
- DSB
double-strand break
- RAG
recombination-activating gene
- SJ
signal joint
- CJ
coding joint
- NHEJ
nonhomologous end-joining pathway
- DNA-PK
DNA-dependent protein kinase
- DNA-PKcs
DNA-PK catalytic subunit
- PFGE
pulsed-field gel electrophoresis
- IR
ionizing radiation
- A-T
ataxia–telangiectasia
References
- 1.Taccioli G E, Rathbun G, Oltz E, Stamato T, Jeggo P A, Alt F W. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 2.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 3.Fugmann S D, Lee A I, Shockett P E, Villey I J, Schatz D G. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 4.Gellert M. Adv Immunol. 1997;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 5.Jeggo P A. In: Advances in Genetics. Hall J C, Dunlap J C, Friedmann T, Giannelli F, editors. Vol. 38. San Diego: Academic; 1998. pp. 185–211. [Google Scholar]
- 6.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, et al. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 7.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber M R. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Otevrel T, Gao Y, Cheng H-L, Seed B, Stamato T D, Taccioli G E, Alt F W. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 9.Buckley R H, Schiff S E, Sampson H A, Schiff R I, Markert M L, Knutsen A P, Hershfield M S, Huang A T, Mickey G H, Ward F E. J Immunol. 1986;136:2398–2407. [PubMed] [Google Scholar]
- 10.Schiff R I, Harville T O. In: Allergy, Asthma and Immunology from Infancy to Adulthood. 3rd Ed. Bierman C W, Pearlman D S, Shapiro G G, Busse W W, editors. Philadelphia: Saunders; 1996. pp. 20–54. [Google Scholar]
- 11.Lees-Miller S P, Godbout R, Chan D W, Weinfeld M, Day R, III, Barron G M, Allalunis-Turner J. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 12.Arlett C F, Green M H L, Priestley A, Harcourt S A, Mayne L V. Int J Radiat Biol. 1988;54:911–928. doi: 10.1080/09553008814552321. [DOI] [PubMed] [Google Scholar]
- 13.Oettinger M A, Schatz D G, Gorka C, Baltimore D. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 14.Gu Y, Jin S, Gao Y, Weaver D T, Alt F W. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeggo P A, Kemp L M. Mutat Res. 1983;112:313–327. doi: 10.1016/0167-8817(83)90026-3. [DOI] [PubMed] [Google Scholar]
- 16.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M, Iliakis G, Mehta R, Jackson M. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 17.Stamato T D, Weinstein R, Giaccia A, Mackenzie L. J Somat Cell Genet. 1983;9:165–173. doi: 10.1007/BF01543175. [DOI] [PubMed] [Google Scholar]
- 18.O'Driscoll M, Cerosaletti K M, Girard P-M, Dai Y, Stumm M, Kysela B, Hirsch B, Gennery A, Palmer S E, Seidel J, et al. Mol Cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 19.Kysela B P, Lohrer H, Arrand J E. Radiat Res. 1995;144:276–281. [PubMed] [Google Scholar]
- 20.Scholer H, Haslinger A, Heguy A, Holtgreve H, Karin M. Science. 1986;232:76–80. doi: 10.1126/science.3006253. [DOI] [PubMed] [Google Scholar]
- 21.Riballo E, Doherty A J, Dai Y, Stiff T, Oettinger M A, Jeggo P A, Kysela B. J Biol Chem. 2001;276:31124–31132. doi: 10.1074/jbc.M103866200. [DOI] [PubMed] [Google Scholar]
- 22.Taccioli G E, Gottlieb T M, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann A R, Alt F W, Jackson S P, Jeggo P A. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 23.Gottlieb T M, Jackson S P. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 24.Singleton B K, Torres-Arzayus M I, Rottinghaus S T, Taccioli G E, Jeggo P A. Mol Cell Biol. 1999;19:3267–3277. doi: 10.1128/mcb.19.5.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baumann P, West S C. Proc Natl Acad Sci USA. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanakahi L A, Bartlet-Jones M, Chappell C, Pappin D, West S C. Cell. 2000;102:721–729. doi: 10.1016/s0092-8674(00)00061-1. [DOI] [PubMed] [Google Scholar]
- 27.Taylor A M R, Metcalfe J A, Thick J, Mak Y F. Blood. 1996;87:423–438. [PubMed] [Google Scholar]
- 28.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 29.Lu X, Lane D P. Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 30.Painter R B. Mutat Res. 1981;84:183–190. doi: 10.1016/0027-5107(81)90061-0. [DOI] [PubMed] [Google Scholar]
- 31.Jaspers N G J, Gatti R A, Baan C, Linssen P C M L, Bootsma D. Cytogenet Cell Genet. 1988;49:259–263. doi: 10.1159/000132673. [DOI] [PubMed] [Google Scholar]
- 32.Finnie N J, Gottlieb T M, Blunt T, Jeggo P A, Jackson S P. Proc Natl Acad Sci USA. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore J K, Haber J E. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milne G T, Jin S, Shannon K B, Weaver D T. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carney J P, Maser R S, Olivares H, Davis E M, Le Beau M, Yates J R, III, Hays L, Morgan W F, Petrini J H J. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 36.Varon R, Vissinga C, Platzer M, Cerosaletti K M, Chrzanowska K H, Saar K, Beckmann G, Seemanova E, Cooper P R, Nowak N J, et al. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 37.Nicolas N, Moshous D, Cavazzana-Calvo M, Papadopoulo D, de Chasseval R, Le Deist F, Fischer A, de Villartay J P. J Exp Med. 1998;188:627–634. doi: 10.1084/jem.188.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiloh Y. Annu Rev Genet. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.