Abstract
Diverse data sets have become key building blocks of translational biomedical research. Data types captured and referenced by sophisticated research studies include high throughput genomic and proteomic data, laboratory data, data from imagery, and outcome data. In this paper, the authors present the application of an XML-based data management system to support integration of data from disparate data sources and large data sets. This system facilitates management of XML schemas and on-demand creation and management of XML databases that conform to these schemas. They illustrate the use of this system in an application for genotype–phenotype correlation analyses. This application implements a method of phenotype–genotype correlation based on phylogenetic optimization of large data sets of mouse SNPs and phenotypic data. The application workflow requires the management and integration of genomic information and phenotypic data from external data repositories and from the results of phenotype–genotype correlation analyses. Our implementation supports the process of carrying out a complex workflow that includes large-scale phylogenetic tree optimizations and application of Maddison's concentrated changes test to large phylogenetic tree data sets. The data management system also allows collaborators to share data in a uniform way and supports complex queries that target data sets.
A primary goal of biomedical research is to arrive at a better understanding of biological systems and underlying mechanisms of complex diseases and to translate this knowledge into timely disease diagnosis and effective treatment. There is widespread recognition1 of the value of integrating information from locally generated data sets and from external data sources such as public databases (e.g., GenBank,2 The Jackson Laboratory,3 and the Mouse Phenome database [MPD]).4 The ability to reference data from diverse sources is an invaluable mechanism for researchers to better analyze information, statistically evaluate results, and prioritize disease candidate genes, transcripts, and proteins for further study.
Over the past few years, widespread adoption of high throughput molecular methods in biomedical research has led to substantial increases in the size and complexity of data used in research studies. Genotype–phenotype correlation analyses of the type described here have benefited from a sustained growth in the number of validated single nucleotide polymorphisms (SNPs) for many model organisms and patient populations. These studies have also benefited from the considerable effort now under way to make phenotypic variation among inbred mice available in digital formats.5 However, researchers are often exasperated by the vastness, complexity, and lack of interoperability in data resources. In practical terms, this means that researchers spend more and more of their time assembling, documenting, and updating data sets.
We have developed a data management and integration framework for developing applications to query and examine information from multiple data types and large data sets and to share data generated in a research project in a distributed environment such as the Grid. The salient features of the framework include support for (1) distributed and coordinated management of data definitions as XML schemas, (2) querying of distributed data sources as XML databases, and (3) on-demand creation of databases that conform to well-defined and published schemas; this ability allows for creating and managing medium- and long-term caches of external public data sources on a storage system. This framework has been used in several applications, including a Grid-enabled biomedical image management and analysis application and an application to manage data from SNPlex analyses. The metadata service component (Mobius Global Model Exchange) of the framework is also being employed in the caGrid software infrastructure of the cancer Biomedical Informatics Grid (http://cabig.nci.nih.gov) (caBIG) effort.
We presented a brief overview of the framework in an earlier work.6 In this work, we implement an application using the framework to support genotype–phenotype studies. This application is part of a larger project that studies the genetic bases of coronary artery disease. The overall application workflow incorporates novel methods to discover candidate genes, sequence similarity searches, and data integration to correlate genotypes and phenotypes. A key step in the workflow is the management and integration of genotypic and phenotypic data from multiple sources. In this paper, we propose and implement a schema for storing and managing output from phylogenetic tree optimization operations. We also describe how the framework is used to create and manage local caches of external data sources for data integration and analysis. The key contribution of this research study is to elucidate a middleware framework designed to provide investigators with a uniform way of interacting with a heterogeneous collection of data sources. Investigators can use these tools to invoke complex queries and design workflows. To demonstrate the value of this approach, we describe our implementation of a complex workflow that includes large-scale phylogenetic tree optimizations and application of Maddison's concentrated changes test to large phylogenetic tree data sets. We should note that this article focuses on the design and implementation of the software system to support the application. The scientific findings from synthesis of information from databases supported by our implementation will be presented in a future publication.
Background
Federated and parallel database technologies have been developed by the database management systems community to enable efficient access to distributed data sources.7,8,9,10 Two main types of virtualization are offered by a federated database management system: (1) transparency in the heterogeneity of attribute names, data schemas, and query languages of data sources and (2) masking the distributed nature of the sources, that is, the client sees a unified, virtually centralized system. Our approach draws from the notion of database federation to provide a unified view of disparate data sets. However, the system described in this paper not only allows federation of existing databases, but also enables on-demand creation of distributed databases and integration of user-defined data processing with data storage.
Model-management tools11,12 and mediator-based systems have also been employed for integration of semantic information and data across heterogeneous data sets.13 The mediator-based architecture developed by Ludäscher et al.14 provides support for creation, management, and querying of integrated view definitions. Their system enables linking of data at the semantic level that encodes the domain specific knowledge about data elements and their relationships. Mediation-based knowledge integration layers need to interact with a data management layer to access data sources; that is, semantic queries submitted by clients through a portal interface are translated into queries against databases controlled by the data management layer. Our system addresses the requirements of the data management layer.
Grid computing has emerged as a widely accepted mechanism to harness computing, storage, and data resources that are hosted at different locations and connected over wide area networks (e.g., the Internet).15,16 As Grid computing has become more prevalent, a service-oriented view of the Grid has been proposed. The Open Grid Services Architecture (OGSA),17,18 the core set of standards developed by the Global Grid Forum (GGF),19 builds on and extends the Web Services technologies20 to address standard mechanisms and definitions for creating, naming, and integrating Grid services. There are some recent efforts to develop database technologies on Grid and Web services. Bell et al.21 develop interfaces, data models, and security support for relational databases using Web services. Smith et al.22 address the problems associated with distributed execution of queries in a Grid environment. They describe an object-oriented database prototype running on Globus.23 Narayanan et al.24 implement a Grid-enabled infrastructure to support management and querying of scientific data sets stored in distributed collections of flat files. The OGSA Data Access and Integration Services25 (DAIS) working group of the GGF is a focused effort that has been developing the service definitions and standards, drawing from the core OGSA standards, for data access and integration in the Grid. Our software system has been designed as a Grid-aware system and can take advantage of emerging standards. In fact, the XML-based data management system employed in this paper builds on the evolving OGSA-DAIS standards.
A number of large projects have been driven by the need to access distributed repositories. The Biomedical Informatics Research Network (BIRN)26 project, funded by the National Institutes of Health (NIH), targets shared access to medical data in a wide-area environment. The BIRN focuses on support for data sets generated by neuroimaging studies. The Shared Pathology Informatics Network (SPIN)27 initiative is intended to provide a virtual database of human tissue specimens. It develops an Internet-based software infrastructure to support a network of tissue specimen data sets and the clinical information associated with these data sets. It allows searches and requests from approved researchers into these data sets while making sure that patient confidentiality requirements are met. Several multi-institutional research projects have been funded by the European Union to investigate the application of Grid technologies for manipulating large medical image databases.28,29,30 The caBIG is an initiative supported by the National Cancer Institute (NCI). The goal of this initiative is to develop applications and the underlying systems architecture for a nationwide cancer research network. The caBIG Grid software infrastructure (called caGrid) will facilitate distributed management and sharing of a vast array of data sources and analytical tools hosted at multiple institutions. The Mobius Global Model Exchange (GME) component of our framework is used in the caGrid infrastructure to manage XML schemas, which represent the structure of data objects that are managed and shared in the caBIG environment.
Design Objectives
Our main objective is to support a set of core functions for management and integration of data from disparate data sources and different data types. We implement this support in the form of a framework consisting of a set of tools and loosely coupled services, which can be directly used or extended and customized for a particular application. In this section, we present the set of functions that have motivated the design of our framework.
Virtualization
One of the challenges that an application developer has to overcome is the fact that different storage formats and database management technologies may be employed by each data source. Querying and accessing data in such a setting become challenging, as applications need to interact with different types of systems. When a new data source is incorporated into the environment, application developers have to update their applications to interact with the new data source correctly. The goal of virtualization is to hide the heterogeneity of software systems used by different data sources by enabling access to the sources with well-defined interfaces and common information exchange protocols. In our framework, this is achieved by services and communication protocols that expose data sources as XML data sources.
Management of Data Types
In a setting where data can be consumed by disparate clients, it is important to be able to define the structure of a data type and to publish and manage this definition. A schema provides a formal and complete representation of the structure of a data type and can act as a contract of data representation between data producers and data consumers. In this way, a client can correctly interpret a data object served by a data source and client programs can interact with the data source programmatically. Our framework provides support for creating, publishing, and managing data types as XML schemas in a distributed environment.
On-demand Creation of Local Caches of External Data
Virtualization provides a mechanism to hide the complexity and heterogeneity of external data sources. However, there are cases in which it is desirable to create local caches of data from multiple data sources.
Snapshots: In some applications it is important to be able to maintain a local snapshot of the data source. For instance, a community of researchers may want to compare the performance of various statistical or data mining algorithms directed at a static data set. Such studies often involve repeated requests to the database. By storing a snapshot of a data source, the query load on the original data source can be reduced.
Interface stability: Both the data attributes and the interface of a data source may change over time. In such cases, it is useful to maintain a data source view on which analysis operations can continue to function.
Query optimization: A data source may provide suboptimal infrastructure for associative queries. For example, the data source might allow a client to only download or upload files, despite the fact that these files can be structured documents and queried. By creating an optimized database on the contents of such files, complex queries can be executed efficiently.
Distributed Execution of User-defined Operations
Processing of data by user-defined operations is a common aspect of data analysis in almost every application domain. In the context of data management and integration, user-defined functions can be implemented to extract the data of interest from one or more data sources and transform them into a format that complies with published schemas and is more efficient to manage and query. It is also desirable (and necessary in some cases) to be able to compose multiple user-defined operations into a data flow network and take advantage of distributed and parallel computing clusters for more efficient processing.
System Description
Mobius Framework
Our implementation builds on the Mobius framework.31,32 Mobius supports distributed creation, versioning, management, and semantic discovery of data models and data instances, on-demand creation of databases, federation of existing databases, and querying of data in a distributed environment. Mobius services employ XML schemas to represent data definitions and XML documents to represent, manage, and exchange data instances. In the context of data integration, the management of data definitions and data adhering to those definitions is of particular importance. We present the two Mobius services, GME and Mako, which provide the required functionality.
Mobius Global Model Exchange
Mobius adopts the philosophy that data standards should be allowed to evolve organically, but they should be managed under a common infrastructure. In this vein, Mobius implements a distributed service, the GME, for schema management. The GME provides a well-defined protocol and service implementation for publishing, versioning, and discovering XML schemas in a distributed environment. A schema can be a completely stand-alone description of a particular data set or it can be an elaborate composition of new attributes and references to multiple existing schemas. By referencing other schemas and entities in other schemas, a user can compose more complex data types from simple data types. Since references from a schema to other schemas are allowed, it is essential to ensure referential integrity. Once a schema is published to a GME instance, it is assigned a version and made available for use by other clients. Once published, a schema cannot be modified; however, a new version of the schema can be published.
The architecture of GME is similar to the Domain Name Server (DNS). A schema published through a GME instance has to be registered under a namespace. Namespaces enable publishing and management of schemas in a controlled way. A hierarchy of namespaces can be created (like domain names in DNS) and managed by multiple GMEs, each of which is an authority for a set of namespaces and delegates responsibility of subnamespaces to subordinate GMEs. Any published schema can be discovered or resolved through any given GME instance, as the authority hierarchy can be navigated, while the storage of schemas can be distributed across a collection of GMEs.
It is reasonable to expect that data types captured in a study will evolve over time; new data attributes may be added or existing attributes may be modified or deleted. It is therefore necessary to support evolution of schemas in a controlled way so that existing programs do not break when a schema goes through changes. The GME architecture formalizes the concept of versions. Any changes made to a data schema reflect either a new version of that schema or a completely new schema under a different name or namespace. This restriction is an important one because it facilitates the evolution of data types while enabling clients and services to still make use of the older versions of the schema.
Mobius Mako: Data Storage and Retrieval
A critical component in supporting access to heterogeneous data sources is the mechanism by which the data sources are exposed (or virtualized). Mako is a strongly typed data storage service that provides a set of well-defined interfaces that expose a data source as an XML data source; since Mako is a distributed service, its interfaces are motivated by the work of the Data Access and Integration Services working group in the Global Grid Forum.33 The Mako client interfaces are similar to those of an XML database. For example, a relational or object database, once exposed through Mako service interfaces, could be queried using XPath as opposed to SQL or OQL (Object Query Language). When an XPath query is received by Mako, it is translated into the query language of the back-end database system; our current implementation of Mako provides support for accessing XML views of existing relational databases via the XQuark Bridge tool (www.xquark.org) and native XML databases via the XMLDB API standard.
Virtualization of data sources as XML data sources trades off some amount of native optimization and expressivity of the underlying data resource in favor of uniformity of data access. This trade-off is acceptable in most cases, since in the process of virtualization, the salient features of the individual data sets of interest have been made prominently available and accessible in the data model being exposed to the client. Often underlying data are aggregated, projected, and denormalized into the corresponding XML view, making attributes of interest easily accessible via XPath queries. While this approach has been very successful, we have identified that there are still occasions when a higher level of expressivity is required by some clients and data aggregation scenarios. The root of this limitation is that XPath does not support expression of joins between documents and document collections. It is solely a data access or location language. Queries, therefore, are limited to accessing the data model as designed by the virtualization process. To support more complex user-defined filters, joins, and more complex queries, we are in the process of developing XQuery support in Mobius.
Virtualization, using Mako, of a data source to a common access model makes it easy and uniform for applications, services, and clients to interact with the data source. Nevertheless, for the service provider, the problem of accessing the native data source remains. Mako abates this problem by providing implementations for common data storage mechanisms (e.g., support for relational databases using the XQuark Bridge tool) and a simple method for extension to new storage and data management systems. The implementation of Mako also provides a custom back end named MakoDB, which is an XML database layered on top of MySQL and optimized for data interactions within the Mako framework. MakoDB provides many advanced features such as (1) the ability to uniquely reference and retrieve individual elements, (2) support for on-demand creation of optimized databases from XML schemas (published in GME), and (3) efficient storage and retrieval of binary data.
The structure of a database in Mako is required to comply with a schema registered in the GME. In this way, Mako enforces all data sets stored and exposed to the Grid to conform to strongly typed, published, and discoverable data models. Each data collection in Mako can be restricted to only accept XML documents from a set of certain schemas. When an XML document conforming to a schema is submitted to a Mako server that accepts said schema, the document is stored and indexed so that the instance data in the document can be queried and subsets of the data can be retrieved.
System Implementation
Our implementation consists of three main services: (1) a metadata service, (2) a data service, and (3) a distributed execution service. The metadata service is built on the GME and provides tools to support management of data models in a distributed environment. An application developer can create, register, and modify schemas that define the structure of data sets managed by the data service. The data service implements tools for efficient storage, management, and querying of distributed data sets. The data service is layered on the Mobius Mako service and designed to take advantage of aggregate storage capacity of distributed disk-based storage platforms and clusters. Using the on-demand database creation capabilities of Mako, local caches of data stored in external data repositories can be created and managed in this service. The distributed execution service is designed to allow execution of user-defined data processing procedures on compute clusters. A more detailed description of the distributed execution service and the underlying runtime system can be found in our earlier work.34,35 The distributed execution service works in tandem with the metadata and data services. The input and output of a data processing component can be described using XML schemas to enable better type checking. Moreover, data output from an application component can be stored in databases maintained in the data service.
An application developer using our system is expected to implement application-specific schemas for data types and user-defined operations for data filtering and format transformations. As an example, consider integration of information from protein databases from SwisProt, gene data from the National Center for Biotechnology Information (NCBI) Gene, genome annotations from the University of Southern California (USC), and pathway information from the Kyoto Encyclopedia of Genes and Geonomes (KEGG). In this case, the developer will need to create XML schemas that define each data type (i.e., protein, gene, genome annotations, and pathways) and define join attributes that will be common across these data types. These schemas can be registered in the metadata service. The next step would be to develop customized Mako services that will expose each data source (SwisProt, NCBI Gene, USC, KEGG data servers) as XML data sources that conform to the schemas registered in the metadata service. Alternatively, the application developer can implement data extraction and translation components, which will interact with the individual data sources and create XML documents that conform to the schemas registered in the metadata service. Using the distributed execution service, the data extraction and translation components can be executed on a cluster system and the local caches of the remote sources can be created. User-defined operations are not limited to format translations only; more complex operations (e.g., translation of mass spectral data to predicted peptide sequence) can also be implemented, registered, and executed in the system.
Clearly, there are several steps and application-specific implementations that an application developer has to do before being able to implement support for data integration and management in the application. However, once the data sources have been exposed using common protocols and service interfaces and/or the local caches of the remote data sets have been created, many client programs can be developed without needing to customize each program for each data source. In addition, since the data types are published in the metadata service and data sources are accessed via unified protocols and service interfaces, other researchers also can develop applications that can access the same data. These are the main strengths of our framework. Using our framework, application developers, data providers, and users in a distributed environment can define, publish, and share common data types and implement uniform mechanisms for data source access. This enables data integration, and client programs can operate, even if the back-end data server technologies are different and changed over time.
Status Report
In this section, we describe an application that we have developed for phenotype–genotype correlation studies as part of an ongoing research project on coronary artery disease (CAD). These studies involve collaboration between biomedical researchers (Dan Janies, Andrew Johnson, and Farhat Habib) with different expertise and analytical skills. They require support for the management, sharing, and integration of data generated by each researcher and gathered from external resources. This support was originally realized using flat files and awk scripts. This approach was labor intensive, prone to errors, and inefficient when the researchers wanted to work with numerous large data sets. By implementing the application using the framework described in this paper, we have facilitated access to different data sets via a uniform interface, allowed more effective sharing of data among the collaborators, and enabled execution of complex queries on large data sets.
The application presents a workflow for combined analysis of genome wide DNA polymorphisms (SNPs) and phenotype data for cardiac function, blood composition, and lipid metabolism among a variety of mouse models. Given the high labor and reagent costs necessary to evaluate candidate genes in a wet laboratory, it is entirely reasonable to expect that candidate genes can be prioritized for empirical studies based on initial discovery and characterization via in silico association studies. The emerging field of in silico association of genetic variants to disease traits has shown some success.36,37,38 A major limiting factor in the field remains the availability of densely sampled DNA data across diverse strains of mice. This limitation should be substantially reduced because sequencing of whole genomes for 15 diverse mouse strains is underway (http://mouse.perlegen.com/mouse). In silico methods have been controversial as they appear quite powerful for penetrant (the likelihood that a given gene will result in disease) traits but less powerful for incompletely penetrant traits such as those that have quantitative variation or are controlled by multiple genes.39 However, the labor and expense necessary for mapping of quantitative and polygenic traits in mouse breeding regimens are considerable.40 Another limitation of mouse studies in general is that genetic components of complex traits in mice are not necessarily relevant to human etiologies. For instance, the human gene CETP plays a role in cholesterol processing, but no mouse CETP gene has been discovered.41 Thus, our workflow includes means to rapidly evaluate the applicability to human disease of a candidate gene discovered in mice.
Application Overview
We have implemented a method of phenotype–genotype correlation based on phylogenetic optimization of large data sets of mouse SNPs and phenotypic data. We have adopted a phylogenetic approach because the hereditary relationships of laboratory mice strains mean that they are not statistically independent entities, thus rendering standard statistical tests inappropriate.42 Our method takes a global approach by optimizing the SNP data into phylogenetic trees to minimize the mutations necessary to explain the observed variation. Phylogenetics sorts variation into attributes that result from common ancestry and those that have evolved independently. In doing so, it can identify mutations that occur in step with the phenotypic changes of interest. Once a set of putative phenotype–genotype correlations has been established, we assess the statistical strength of the correlation and the human biomedical relevance of candidate genes. This filtering process needs to take into account both phylogenetic tree–related statistics, merged with preexisting information contained in mouse and human gene annotations. The results can be used to (1) identify candidate genes that are already known to be relevant to the human disease under study, (2) detect novel candidate genes that are then prioritized for empirical follow-up, and (3) identify false positives. While some results are well-known candidate genes for CAD, other genes have not previously been described as candidates. Such novel candidates are prioritized for empirical follow-up.
Our application provides the researcher with a mechanism to systematically support the integration of locally generated experimental information, with information obtained from diverse sources. The application consists of two main parts. In the first part, a researcher calculates one or more phylogenetic trees from SNPs using a phylogenetic tree optimization program called POY.43 The researcher may incorporate phenotypic data into the construction of the trees or optimize the phenotypic data onto a tree based only on genotypic data. Once a satisfactory tree has been found, the researcher creates, using POY and the phylogenetic tree, a table of inferred mutations and phenotypic changes among strains of mice (an example of the table is provided in ▶). The inferred mutations and the phenotypic changes are then examined using Maddison's44 concentrated changes test to score putative correlations among the mutations and the phenotypic changes. Any highly correlated SNPs are output for each phenotype. The actual implementation of these steps is explained for our use cases in the section “Finding Single Nucleotide Polymorphisms of Interest in Specific Use Cases.” The second part of the application carries out an evaluation of the biomedical relevance of the candidate SNPs. Using a graphical user interface, the researcher selects the list of well-correlated SNPs to guide the search for haplotype blocks. The researcher then gathers annotation data on the biomedical relevance of genes contained in the haplotype blocks from multiple sources, including Mouse Genome Informatics (MGI).45 Finally, the researcher assesses the relevance of the candidate gene(s) based on the annotation data, primary biomedical literature, and the presence of human orthologs. These steps are described in detail in the section “Estimating Haplotypes in Strains of Interest and Using Annotation Data to Assess Biomedical Relevance of Candidate Genes.” Execution of all the steps in the application requires sharing data among the researchers responsible for various analyses and the integration of the results with public data sources.
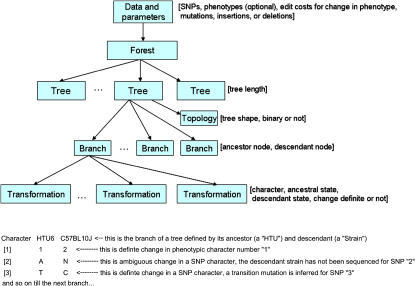
Figure 1.
(Top) The data model for the output from POY, a program for phylogenetic tree optimization. The rectangles and arrows represent the element types and parent-child relationships. The contents of the square brackets denote the attributes associated with a node type. For example, a branch element type has attributes for the ancestor node, the descendant node, and the minimum and maximum length as computed by the POY program. (Bottom) An annotated example of section of tabular POY output depicting inferred transformations in three characters on a branch of a phylogenetic tree. Character 1 is a phenotype that changed from state 1 in the ancestor (HTU6) to state 2 in the descendant (strain C57BL10J). This change could be from normal to elevated non–high-density lipoprotein cholesterol plasma levels. Character 2 is a single nucleotide polymorphism (SNP), in which change could have occurred along this branch but is ambiguous due to missing data. Character 3 is an SNP, in which a transition mutation occurs. This could be SNP rs3023213 as depicted in ▶.
Finding Single Nucleotide Polymorphisms of Interest in Specific Use Cases
The first part of our application searches for mutations that change in step with phenotypes among diverse strains of mice. In this paper, we used data sets from the Mouse Phenome Database (MPD) and GNF2. To facilitate the study of complex genetic diseases in mouse models, the Jackson Laboratory has compiled an extensive MPD.4 Phenotype data from many mouse strains are collected from the literature and via collaboration with experts through consistently applied protocols. The data are compiled into a database and flat files for download. In addition, a large number of SNP data sets are now publicly available in the MPD. The mpd146 data set available in the MPD is a compilation of 439,942 single and multiple nucleotide polymorphisms genotyped by many research groups for 17 mouse strains. We analyzed SNP data for 15 strains represented in mpd146, for which there were accompanying phenotype data in the MPD. The phenotype data were selected from a lipid study data set titled “Paigen2”46 in the MPD. The GNF2 database contains 8,944 SNPs.38 Although the GNF2 data have fewer SNPs, the data sets are more uniform in strain coverage, include a larger number of strains (a total of 48 strains), and are more evenly distributed along the genome. Phenotype data were available in the MPD for 39 of the strains represented in the GNF2.
In the application, we use the commands -replicate -tbr -spr of POY43 to simultaneously align the sequence data for characters longer than 1 as various trees are constructed and refined by POY in searching for an optimal tree and alignment. The need for an algorithm such as POY to coordinate alignment and tree construction arises from length differences in multiple nucleotide polymorphisms (MNP) that are part of mpd146. In the case of the GNF2 data, the alignment is fixed because each polymorphism is of length 1 (i.e., an SNP). As a result of the organized nature of the GNF2 data set, data prealigned by any tree search program (such as TNT47) could be used as input. To efficiently store and query the POY output, we developed a data model (▶) and the corresponding XML schema. This model represents a view of the POY output that accommodates a forest, containing one or more trees, implied by the data and analytical parameters (e.g., the number of replicates examined and edit costs for transformations imposed). The data model also includes information on the branches of each tree that represent inferred ancestor–descendant relationships. For each branch, transformations in the nucleotide and phenotype data from the inferred ancestral states to descendant states are assigned and output by POY. The transformations considered in the use cases include mutations, insertion events, or deletion events in nucleotide data and changes in phenotypes (e.g., from normal to elevated non–high-density lipoprotein (HDL) cholesterol plasma levels). A branch may contain multiple transformations (▶).
Our phylogenetic tree schema complements the schema being developed in the TreeBase component of the CIPRes project (http://www.phylo.org/). Our schema supports transformations assigned to various branches that occur in trees and their implied alignments. The TreeBaseII schema, on the other hand, expresses data types and attributes for studies and related publication references, analyses, matrices, taxon labels, and trees. To have a more comprehensive data model for phylogenetic trees, our data model could be combined with the TreeBaseII schema to capture transformations, implied alignments, and information on studies and publications.
Once phylogenetic trees are constructed, we need to assess the potential that a phenotypic trait is correlated with a DNA polymorphism by chance. To assess the significance of a given SNP, we use Maddison's44 concentrated changes test (CCT). The CCT is a measure of correlation between two binary characters on a phylogenetic tree. It produces a p-value for the type one error on the null hypothesis that the DNA polymorphism is associated with the trait by chance. Since the CCT works on binary data values, the phenotype data are made suitably binary by using a threshold value such as exceeding a standard deviation above or below the mean (e.g., the mean value and standard deviation value of non-HDL cholesterol levels) (▶). The SNPs are often naturally biallelic and thus binary. The CCT determines whether the observed changes in a binary character are concentrated on the branches of a tree that has a particular state of the second character. A step in computing the CCT is to extract the sets of transformations from the branches of interest from the phylogenetic tree and compute intersections and differences of these sets. For instance, one can look for transformations that occur in a group of branches but do not occur in another branch. In our implementation, the phylogenetic tree output from POY can be queried using XPath to search for and extract the branches of interest. The CCT calculations in our case determine whether changes in phenotype are concentrated on the branches that have a derived allele for an SNP of interest (i.e., a recent mutation). We count the number of gains and losses of the phenotypic character over the whole tree and the number of these gains and losses that fall on branches reconstructed to have the derived allele for an SNP of interest.
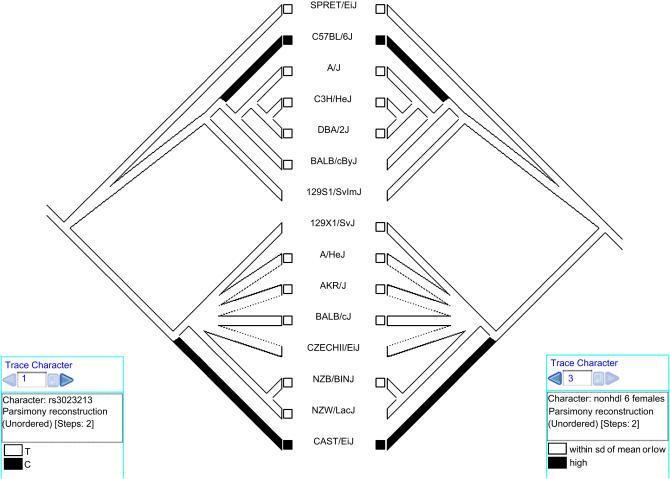
Figure 2.
Two views of the same phylogenetic tree of females of mouse strains displaying correlated changes of a phenotype and a genotype across 15 mouse strains. The right tree depicts phenotypic change in non–high-density lipoprotein (non-HDL) cholesterol plasma levels in female mice after six weeks of atherogenic diet. Black branches indicate strains (C57BL/6J and CAST/EiJ) with non-HDL levels greater than one standard deviation (sd) above the mean after treatment. Genotype observations for each strain for the SNP of interest (rs3023213; T or C) are indicated on the left tree. Boxes at the terminal branches of the trees indicate genotype or phenotype observations in databases for those strains. Concentrated changes test results for this phenotype–genotype correlation differ for females (p = 0.004) and males (p = 0.088) (not shown).
Although the CCT is implemented in Maddison et al.,48 a user can only calculate the CCT for one pair of characters at a time. In addition, counting the gains and losses of the dependent character has to be done manually. In the research we describe here, we calculate CCTs for very large numbers of SNPs and phenotypes using the macro language in a command line–driven version of Tree analysis using New Technology (TNT).47 Given that there were often several most parsimonious optimizations of an SNP, we used the delayed transformation (DELTRAN)48 optimization to obtain a unique reconstruction of each SNP. Once compiled, the CCT values are stored in the data management and integration system. We have developed a simple schema that consists of the following attributes: cct_value, character, position, trait, and SNP id. The SNPs can be filtered using XPath queries into the CCT data stored in the system (e.g., /snp[(traits/trait/@cct_value < “.05”)] will return SNPs that have significant p-values for the concentrated changes test).
Estimating Haplotypes in Strains of Interest and Using Annotation Data to Assess Biomedical Relevance of Candidate Genes
Portions of the genomes of inbred mouse strains are often similar over large distances, complicating attempts to finely map the genetic basis of traits.49 Thus, in silico approaches that identify correlated markers must also consider the genomic context of those markers.37,38 Although SNPs vary in density throughout the mouse genome, they lie within genes or in intergenic regions, thus providing the potential for resolution at the locus level (defined by the SNP and flanking DNA). However, the genomic context of the SNP of interest is very important because an SNP may vary with a phenotype, yet may only be associated with the causal gene via linkage disequilibrium. We have implemented a simple algorithm that takes an SNP of interest from the previously described steps as input (e.g., rs3023213 from ▶) and returns a range on the genome containing 1 or more markers, thus defining a region of similarity between the query strain pair (e.g., C57BL/6J and CAST/EiJ from ▶). Because this process is repeated many times for different regions resulting in many potential candidate genes, an integrated database support was designed.
To evaluate the potential biomedical relevance of candidate genes that fall within the region of similarity for the phenotype of interest, a researcher needs to retrieve and organize knowledge from various data sources. A challenging issue is the need for querying and integrating data from several public data sets. Internet-based biomedical and genomic data repositories differ, in both data formats and mechanisms by which clients can query and retrieve data. A data source might be accessed via a Web service interface or, alternately, the data source might be a Web or ftp site from which the data set of interest needs to be downloaded. Our approach to supporting integration of data from multiple resources and complex queries is to create caches of subsets of external data sources in our data management and integration system.
Data collection from external data sources can be done manually or can be automated in our system. In the manual mode, the user can download a data set from an external data source and load it into the system using one of the available data extraction and loading programs. The data loading programs identify the input data set format and generate a set of XML documents, which are then stored in Mako servers. In the automated mode, our system can support processes, referred to as spiders (or data extractors), that scrape the individual data resources for data. A data source catalog maintains the data access method or the Web site wrapping service to retrieve data files from an Internet repository and the extractor method to parse data attributes and attribute values from the retrieved data files. The distributed execution service can be used to execute spiders and data extraction and loading programs on a PC cluster. In this way, multiple data sources can be accessed and processed concurrently. A spider for each data resource can be executed to obtain the relevant data. The results, returned by each spider, are mapped onto the corresponding data model using the data extraction and loading methods, and then are stored in the system.
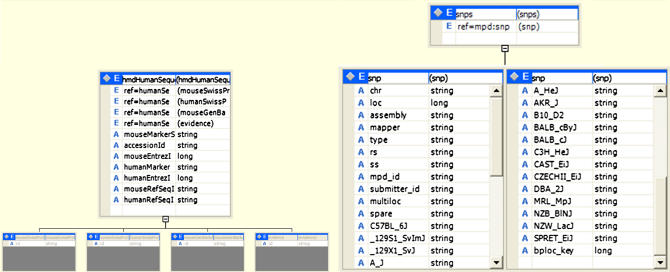
We have implemented data models for data subsets from different sources so that the contents of data files downloaded from a data source can be stored in the system and queried along with other information. These data models are described as XML schemas, which are stored and managed by the metadata service. Once a set of files from an external data source are downloaded, they are parsed using the corresponding extractor method and XML documents, which conform to the corresponding data model (XML schema) and are created and stored on Mako servers. In our current implementation, we have parsers and data models for a number of databases that can be downloaded from the Jackson Laboratory and for the GNF2 data set. These data sets include MPD-merged mouse strain SNPs (mpd146), Gene Ontology terms data set, Human and Mouse Orthology with Human Online Mendelian Inheritance in Man (OMIM) IDs, MGI Sequence Coordinates, and Human and Mouse Orthology with Sequence information (▶). The data models of the mpd146 and HMD Human Sequence data sets are shown in ▶ as examples. Clients can access and query the databases through a graphical user interface that allows execution of a number of query templates (▶).
Table 1.
List of Data Sets Currently Managed by the Data Management and Integration System
| Data Set | Explanation |
|---|---|
| Mpd146, Paigen 2, GNF2 | Mouse phenome database |
| MGI_Coordinate.rpt | MGI sequence coordinates |
| Gene_association.mgi | Gene ontology (GO) annotations of mouse markers |
| Go_terms.mgi | GO terms and GO IDs |
| HMD_HGNC_Accession.rpt | Human and mouse orthology |
| HMD_HumanSequence.rpt | Human and mouse orthology with sequence information |
| HMD_OMIM.rpt | Human and mouse orthology with human OMIM IDs |
Figure 3.
Data models for the HMD Human Sequence (left) and mpd146 (right) data sets.
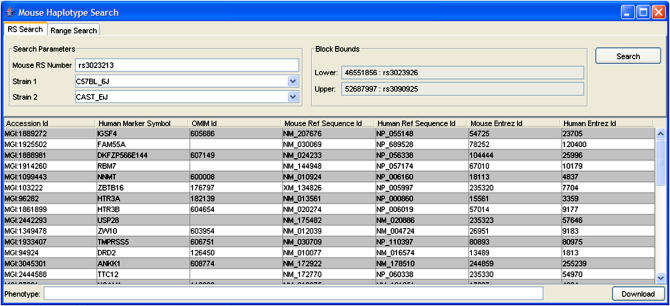
Figure 4.
A view of the Client Interface after a query has been executed for a single nucleotide polymorphism (rs3023213) correlated with higher non–high-density lipoprotein cholesterol levels (see also ▶). Within this block of genetic similarity between the query strains (C57BL/6J and CAST/EiJ), a candidate gene, NNMT (nicotinamide N-methyltransferase), for the trait was noted.
Detailed results obtained through this application will be presented elsewhere (Habib et al., in preparation). Here we present an example of the types of results obtained. An example query (rs3023213) identified NNMT (nicotinamide N-methyltransferase) as a candidate gene for high non-HDL levels in female mice of strains C57BL/6J and CAST/EiJ, within a block on mouse chromosome 9. NNMT is highly expressed in liver tissue and is known to exhibit large differences, in level and activity, between mouse strains and genders51 and among humans.52 N-methyltransferases (e.g., NNMT) are involved in the biochemical synthesis of homocysteine, a cardiovascular disease risk factor. NNMT was recently implicated as a genetic factor for plasma homocysteine levels, in a genome-wide linkage study in humans.53 The potential link between N-methyltransferases, homocysteine, and cholesterol levels is supported by findings in a knockout mouse model.54
Performance: Creation of Local Caches and Data Querying
In the first set of experiments, the performance of the system in the format translation and on-demand database creation (data loading) step is examined. ▶ shows the execution time for data sets extracted from the mpd146, MGI Gene Association, and MGI coordinate databases from the Jackson Laboratory. In these experiments, the data extractor was run on a machine with a Pentium IV 3-GHz central processing unit (CPU) and 1,024 MB of memory. The data server machine was an SMP node with dual Xeon 2.4-GHz CPUs and 2,048 MB of memory. The two machines were connected to each other over a Gigabit switch. All timings are in seconds and represent the execution time from a single run for each data set. In our implementation, the extractor reads one or more data rows from the data set file and transforms them into XML documents, conforming to the corresponding schemas. When an XML document is created, the extractor submits the document to a Mako server over the network for storage. It can be expected that running the data extractors and the data servers on two separate machines will incur communication overhead. To reduce this overhead, these two components could be colocated. We should note that our implementation allows for placement of a data extractor on the machine where the data server is. However, in many cases, machines hosting data servers may not be configured to run user-defined operations because of security and performance concerns. Our experiments emulate such a configuration.
Figure 5.
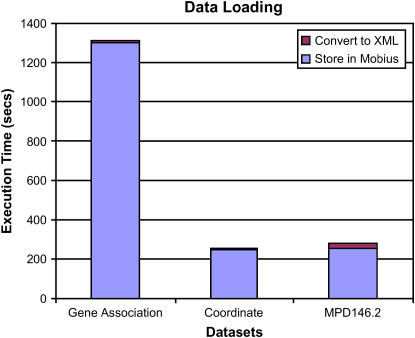
The execution time of the data set extraction and loading step for the gene association, mpd146, and coordinate data sets.
As is seen in ▶, the bulk of the execution time is spent submitting XML documents to the data server; the time to parse a data file and create XML documents takes less than 5% of the overall execution time on average. In this experiment, the data extraction and loading time for the Gene Association data set was about 20 minutes. The reason for this is that the data set used in the experiment contained about 90,000 rows. The XML schema for this data set generates an XML document for each row, containing all the attributes as XML node elements. Hence, the extractor had to submit 90,000 documents to the data server, thus incurred a large network communication overhead. The data loading time for the coordinate data set was much less because there were about 18,000 rows in total. We observe that the execution time increases linearly as the number of rows increases. This is a performance bottleneck in our current implementation. The network overheads would be more pronounced in a more distributed environment (e.g., the Grid). To reduce the network overhead, multiple documents could be combined into one buffer by the runtime system, and this buffer could be submitted to the server. This requires that the server can handle buffers containing more than one XML document. We are in the process of adding this functionality to the data service component.
The number of data rows for the mpd146 data set was 439,942. If a single XML document were created for each row of this data set, the execution time would be around 105 minutes. Since our current implementation does not support concatenation of multiple documents into one buffer and processing of such buffers, we developed an XML schema that allowed us to combine multiple rows of the data set into one XML document. In our experiments, we stored 10,000 rows of the data set in one XML document, thus submitting only 44 documents to the data server. As seen in the figure, the execution time of the data extraction and loading step for mpd146 is less than that of the Gene Association data set because of this optimization.
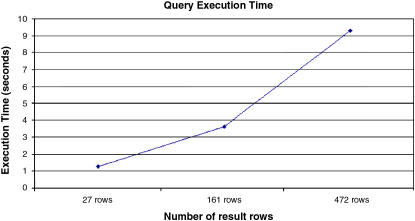
In the next set of experiments, we look at the query execution performance. The current client interface supports two types of queries. The first query type specifies an SNP identification (rsID) and two strains. In the second type of query, the client inputs a chromosome and a genomic location range. Our experimental results show that the first type of query takes on average about eight times longer to execute than the second type of query. We tried three different queries for each query type; the first query returned one result row, the second 33 result rows, and the last one 241 result rows. The average execution time of the first query type was 76 seconds, whereas the second type of query took 9 seconds on average. When the first type of query is submitted, the mpd146 data set is first searched to find the chromosome corresponding to the rsID and a corresponding genomic location. Next, a region of genetic similarity between the two query strains is computed as follows: multiple accesses to the mpd146 data set are made to find the third mismatch in alleles between the strains in regions downstream and upstream from the location identified by the rsID. On the other hand, the second type of query requires only two accesses to the mpd146 data set to extract the corresponding SNP accession numbers and other information required. In either query, once the range values are determined, the upper and lower bound values are used to find the list of marker accession IDs (see ▶, the left-most column). The marker accession IDs are used as join attributes to extract information from other databases, where the marker accession ID of a data element matches one of the marker accession IDs in the list obtained from the coordinate data set. ▶ shows the execution time of the second type of query as the query size is scaled up. We executed three different queries in this experiment; the number of rows returned was 27, 161, and 472 rows. The timing numbers are the average execution time over four runs of each query size. As seen from the figure, the execution time increases in proportion to the size of the query results. The standard deviations were 0.15 (27 rows), 0.04 (161 rows), and 0.73 (472 rows) seconds in these experiments.
Figure 6.
Query execution time as the query size is scaled.
Limitations
There are several issues that have not been addressed in our current implementation. The first issue stems from the use of XPath as our querying language. Although our system can support XPath queries of any complexity, the XPath language does not support joins on XML databases; it provides the basic data-subsetting capabilities. In our implementation, queries involving joins between different data sources have been implemented as client side operations; that is, individual data sets are accessed using XPath queries and any joins between the data subsets are performed in the client application. We are currently working on integrating a subset of XQuery functionality (i.e., support for XQuery FLWR expressions) in our framework. This will make it possible to support more complex queries without implementing client side extensions.
Another limitation of our current implementation is that it does not support integration at the semantic level. It provides the core support needed to enable management of data types and data instances. A semantic integration component can be layered on this core support. However, we recognize that establishing semantically meaningful relationships between data elements from different resources is a complex issue that requires innovative solutions in the semantic layer. An approach to address semantic interoperability is to standardize common data elements, controlled vocabularies, and taxonomies and implement these standards in a framework. There are a number of standardization efforts such as SNOMED (Systematized Nomenclature of Medicine), LOINC (Logical Observation Identifiers Names and Codes), and DICOM (Digital Imaging and Communication in Medicine) for naming and description of data attributes and data. In cancer research, the NCI Center for Bioinformatics has developed the Cancer Data Standards Repository and the Enterprise Vocabulary Services to serve as a controlled vocabulary source in order to enable common semantics across related databases. There is also an ongoing effort in the caBIG initiative to define common data elements and controlled vocabularies to support multi-institutional basic and clinical cancer research projects. We have implemented a prototype service in Mobius to support management of semantic information using resource description framework (RDF). We plan to extend this prototype and integrate it in our software system. We also plan to use the caBIG technologies as they become available to provide support for management and use of controlled vocabularies as well as common data elements.
A performance bottleneck in our current implementation is the data-loading overhead. A network overhead is incurred if data extractors and Mako servers are located on different machines. The data-loading overhead also stems from the fact that during data loading, each row in the source database is encoded in an XML document and submitted to a Mako server. This overhead could be reduced by combining multiple documents into a message buffer that is submitted to the server. Our current implementation does not support multiple documents in a single buffer. We plan to incorporate this performance optimization into our implementation in future. Another approach to reduce the data loading overhead would be to colocate extractors and Mako-servers. We also observed that the cost of executing the first type of query (as explained in the section “Performance: Creation of Local Caches and Data Querying” is expensive. This is because of the need for multiple accesses to the database to find the bounds for the region of genetic similarity. This overhead could be reduced by implementing data prefetching and in-memory caching. In this strategy, the entire set of attributes required for computing the bounds would be prefetched from the database and cached in an in-memory data structure (e.g., an array) in the application client when the client is started. This would reduce the overhead of querying the database multiple times. However, a caching mechanism would need to be implemented in the client to manage the in-core data structure if the set of required attributes did not fit in memory.
Conclusions
In this paper, we presented the application of an XML-based generic metadata and data management system for management and integration of data in a biomedical application. XML has become a de facto standard for representing structured and semistructured data sets in heterogeneous and distributed environments. The software infrastructure presented in this paper provides core services and common protocols for (1) distributed, but coordinated, managing of metadata definitions, (2) exposure of subsets of data stored in ad hoc data warehouses and enterprise information systems, and (3) on-demand creation of databases along with management of complex data analysis workflows. These capabilities make it possible to support the management and querying of distributed collections of heterogeneous biomedical data sets and integration of such data sources in a unified framework. This type of common distributed data environment, with strongly typed data, can enable implementation of applications that can remove barriers to better synthesis and analysis of information in translational research.
In this work, the data management and integration framework allowed us to manage and query much larger data sets from phylogenetic tree optimizations, apply CCT on large data sets, and integrate data from external repositories efficiently. Without the ability to create and manage local caches of public databases, it would have required the application to submit large numbers of queries to the respective database servers, thus incurring a lot of network overhead and load on those servers. Moreover, the software system enabled the researchers to share data in a more uniform way and to execute complex queries on large data sets. This was a major step forward from using simple shell and awk scripts to manage, filter, and integrate data generated and referenced in the application.
The development of the application involved close collaboration between a group of biomedical researchers and a group of information technology developers. In this collaboration, we observed that the analysis of the requirements was the most challenging part. This was mainly because it required a good understanding of the scientific application as well as the capabilities of the software system by both groups. This process proved that the development of good software applications to support biomedical research is only possible through close collaboration of application domain experts and information technology developers.
Although we focused on one application in this paper, our system is extensible and customizable for other applications. In another collaborative project, we have developed an application to support data sets from SNPlex analyses. In a follow-up to that implementation, we are developing a quality control application that will enable integration of data obtained from multiple experimental and bioinformatics analysis techniques done on the same or overlapping groups of samples. The system will allow a user to query data based on individual or groups of data collection/analysis techniques, perform joins between different data subsets, and carry out statistical analyses on the selected data. For example, a user will be able to query for all samples, for which at least one data collection/analysis technique has generated a different result. Using our framework, we are also implementing an application similar to the application in this paper for study of coronaviruses and influenza A viruses. The application will support management and analysis of data from phylogenetic tree optimizations, results from bioinformatics analysis methods, and integration of data obtained from public repositories to the overall workflow. We plan to report on the status of these applications in a future paper.
The Mobius framework presented in the paper is available for download from our Web site (www.bmi.osu.edu). We plan to make the application implementation available for download in future after further performance testing and improvement are carried out and a user guide for the application has been written. In the meantime, interested readers are encouraged to contact the authors of this paper.
This research was supported in part by the National Science Foundation under grants ACI-9619020 (UC Subcontract 10152408), EIA-0121177, ACI-0203846, ACI-0130437, ANI-0330612, ACI-9982087, Lawrence Livermore National Laboratory under grant B517095 (UC subcontract 10184497), NIH NIBIB BISTI P20EB000591, Ohio Board of Regents BRTTC BRTT02-0003.
References
- 1.Leppert M, Singh NA. Nonsyndromic seizure disorders: epilepsy and the use of the Internet to advance research. Annu Rev Genomics Hum Genet. 2003;4:437–57. [DOI] [PubMed] [Google Scholar]
- 2.GenBank. 2005. Available at: http://www.ncbi.nlm.nih.gov/Genbank/. Accessed March 2006.
- 3.The Jackson Laboratory. 2005. Available at: http://www.jax.org/. Accessed March 2006.
- 4.The Mouse Phenome Databases. 2005. Available at: http://www.jax.org/phenome/. Accessed March 2006.
- 5.Bogue M. Mouse Phenome Project: understanding human biology through mouse genetics and genomics. J Appl Physiol. 2003;95:1335–7. [DOI] [PubMed] [Google Scholar]
- 6.Hastings S, Langella S, Oster S, Kurc T, Pan T, Catalyurek U, et al. Grid-based Management of Biomedical Data using an XML-based Distributed Data Management System. In: Proceedings of the 20th ACM Symposium on Applied Computing (SAC 2005), Bioinformatics Track. Santa Fe, NM: ACM Press, 2005.
- 7.Conrad S, Eaglestone B, Hasselbring W, Roantree M, Saltor F, Schonhoff M, et al. Research issues in federated database systems: report of EFDBS '97 Workshop. SIGMOD Rec. 1997;26:54–6. [Google Scholar]
- 8.DeWitt D, Gray J. Parallel database systems: the future of high performance database systems. communications of the ACM. Communications of the ACM. 1992;35:85–98. [Google Scholar]
- 9.Kossman D. The state of the art in distributed query processing. ACM Comput Surv. 2000;32:422–69. [Google Scholar]
- 10.Ozsu MT, Valduriez P. Distributed and parallel database systems. ACM Comput Surv. 1996;28:125–8. [Google Scholar]
- 11.Alagic S, Bernstein PA. A model theory for generic schema management. In: Proceedings of the 8th Biennial Workshop on Data Bases and Programming Languages (DBPL '01). Frascati, Rome; 2001.
- 12.Melnik S, Rahm E, Bernstein PA. Rondo: a programming platform for generic model management. In: Proceedings of SIGMOD; 2003.
- 13.Newhouse S, Mayer A, Furmento N, McGough S, Stanton J, Darlington J. Laying the foundations for the semantic grid. In: Proceedings of the AISB '02 Symposium on AI and GRID Computing; 2002.
- 14.Ludäscher B, Gupta A, Martone M. A model-based mediator system for scientific data management. In: Critchlow T, Lacroix Z, editors. Bioinformatics: managing scientific data. San Francisco: Morgan Kaufmann, 2003, 335–70.
- 15.Berman F, Hey AJ, Fox G, editors. Grid Computing: making the global infrastructure a reality. Hoboken, NJ: John Wiley & Sons, 2003.
- 16.Foster I, Kesselman C, editors. The Grid: blueprint for a new computing infrastructure. San Francisco: Morgan Kaufmann, 1999.
- 17.Foster I, Kesselman C, Nick J, Tuecke S. Grid services for distributed system integration. Computer. 2002;35:37–46. [Google Scholar]
- 18.Foster I, Kesselman C, Nick JM, Tuecke S. The physiology of the Grid: an open grid services architecture for distributed systems integration: Open Grid Service Infrastructure Working Group Technical Report, Global Grid Forum; 2002. Available from: http://www.globus.org/ogsa/. Accessed March 2006.
- 19.GGF. Global Grid Forum. 2005. Available at: http://www.gridforum.org. Accessed March 2006.
- 20.Graham S, Simeonov S, Boubez T, Davis D, Daniels G, Nakamura Y, et al. Building Web services with Java: making sense of XML, SOAP, WSDL, and UDDI. Indianapolis, IN: SAMS Publishing, 2002.
- 21.Bell WH, Bosio D, Hoschek W, Kunszt P, McCance G, Silander M. Project Spitfire—towards Grid Web service databases. Edinburgh, Global Grid Forum Informational Document, GGF5; 2002.
- 22.Smith J, Gounaris A, Watson P, Paton NW, Fernandes AA, Sakellariou R. Distributed query processing on the Grid. In: Proceedings of the Third Workshop on Grid Computing (GRID2002), Baltimore, MD, 2003.
- 23.Foster I, Kesselman C. Globus: A Metacomputing Infrastructure Toolkit. Int J High Performance Comput Appl. 1997;11:115–28. [Google Scholar]
- 24.Narayanan S, Kurc T, Catalyurek U, Saltz J. Database Support for data-driven scientific applications in the Grid. Parallel Process Lett. 2003;13:245–73. [Google Scholar]
- 25.Atkinson MP, Dialani V, Guy L, Narang I, Paton NW, Pearson D, et al. Grid database access and integration: requirements and functionalities: technical document, Global Grid Forum, 2002. Available at: http://www.cs.man.ac.uk/grid-db/documents.html. Accessed March 2006.
- 26.Peltier ST, Ellisman MH. The biomedical informatics research network. In: The Grid, blueprint for a new computing infrastructure, 2nd ed. Philadelphia: Elsevier, 2003, 109–20.
- 27.SPIN. Shared Pathology Informatics Network. 2002. Available at: http://www.cancerdiagnosis.nci.nih.gov/spin/. Accessed March 2006.
- 28.Duque H, Montagnat J, Pierson JM, Brunie L, Magnin IE. DM2: a distributed medical data manager for grids. In: Proceedings of BioGrid '03, the 3rd International Symposium on Cluster Computing and the Grid (CCGrid 2003). Tokyo, Japan: IEEE Computer Society, 2003, 606–11.
- 29.Rogulin D, Estrella F, Hauer T, McClatchey R, Amendolia SR, Solomonides A. A grid information infrastructure for medical image analysis. In: Proceedings of the Distributed Databases and Processing in Medical Image Computing Workshop (DiDaMIC-2004); 2004.
- 30.Tweed T, Miguet S. Medical Image Database on the Grid: Strategies for Data Distribution. In: HealthGrid '03. Lyon, France: European Commission, DG Information Society, 2003, pp. 152–62.
- 31.Hastings S, Langella S, Oster S, Saltz J. Distributed data management and integration: the Mobius Project. In: Proceedings of the Global Grid Forum 11 (GGF11) Semantic Grid Applications Workshop Honolulu: Global Grid Forum, 2004.
- 32.Langella S, Hastings S, Oster S, Kurc T, Catalyurek U, Saltz J. A distributed data management middleware for data-driven application systems. In: Proceedings of the 2004 IEEE International Conference on Cluster Computing (Cluster 2004). San Diego: IEEE Computer Society, 2004.
- 33.DAIS. Database Access and Integration Services Working Group. 2002. Available at: http://www.cs.man.ac.uk/grid-db/. Accessed March 2006.
- 34.Hastings S, Kurc T, Langella S, Catalyurek U, Saltz J. Image processing for the grid: a toolkit for building Grid-enabled image processing applications. In: Proceedings of the IEEE International Symposium on Cluster Computing and the Grid (CCGRID 2003); Tokyo, Japan: IEEE Computer Society, 2003.
- 35.Hastings S, Oster S, Langella S, Kurc T, Pan T, Catalyurek C, et al. A Grid based image archival and analysis system. J Am Med Inform Assoc. 2005;12:268–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grupe A, Germer S, Usuka J, Aud D, Belknap JK, Kelin RF, et al. In silico mapping of complex disease-related traits in mice. Science. 2001;292:1915–8. [DOI] [PubMed] [Google Scholar]
- 37.Liao G, Wang J, Guo J, Allard J, Cheng J, Ng A, et al. In silico genetics: Identification of a functional element regulating H2-Ealpha gene expression. Science. 2004;306:690–5. [DOI] [PubMed] [Google Scholar]
- 38.Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2:e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chesler EJ, Rodriguez-Zas SL, Mogil JS, Darvasi A, Usuka J, Grupe A, et al. In silico mapping of mouse quantitative trait loci. Science. 2001;294:2423a. [DOI] [PubMed] [Google Scholar]
- 40.Moore K, Nagle D. Complex trait analysis in the mouse: the strengths, the limitations and the promise yet to come. Annu Rev Genet. 2000;34:653–86. [DOI] [PubMed] [Google Scholar]
- 41.Tall AR. Plasma cholesteryl ester transfer protein. J Lipid Res. 1993;34:1255–74. [PubMed] [Google Scholar]
- 42.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- 43.POY. Available at: http://research.amnh.org/scicomp/projects/poy.php. Accessed March 2006.
- 44.Maddison W. A method for testing the correlated evolution of two binary characters: are gains or losses concentrated on certain branches of a phylogenetic tree? Evolution. 1990;44:539–57. [DOI] [PubMed] [Google Scholar]
- 45.Mouse Genome Informatics. 2005. Available at: http://www.informatics.jax.org. Accessed March 2006.
- 46.Svenson KL, Bogue MA, Peters LL. Genetic models in applied physiology: invited review: identifying new mouse models of cardiovascular disease: a review of high-throughput screens of mutagenized and inbred strains. J Appl Physiol. 2003;94:1650–9. [DOI] [PubMed] [Google Scholar]
- 47.Goloboff P, Nixon, K, Farris S. TNT. 2005. Available at: http://www.zmuc.dk/public/phylogeny/TNT/. Accessed March 2006.
- 48.Maddison DR, Maddison WP. MacClade. 2003. Available at: http://www.macclade.org/index.html. Accessed March 2006.
- 49.Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet. 2005;6:271–86. [DOI] [PubMed] [Google Scholar]
- 50.MGI Data and Statistics Reports. 2005. Available at: ftp://ftp.informatics.jax.org/pub/reports/index.html. Accessed March 2006.
- 51.Scheller T, Orgacka H, Szumlanski CL, Weinshilboum RM. Mouse liver nicotinamide n-methyltransferase pharmacogenetics: biochemical properties and variation in activity among inbred strains. Pharmacogenetics. 1996;6:43–53. [DOI] [PubMed] [Google Scholar]
- 52.Rini J, Szumlanski C, Guerciolini R, Weinshilboum RM. Human liver nicotinamide n-methyltransferase: ion-pairing radiochemical assay, biochemical properties and individual variation. Clin Chim Acta. 1990;186:359–74. [DOI] [PubMed] [Google Scholar]
- 53.Souto JC, Blanco-Vaca F, Soria JM, Buil A, Almasy L, Ordonez-Llanos JJMM-C, et al. A genomewide exploration suggests a new candidate gene at chromosome 11q23 as the major determinant of plasma homocysteine levels: results from the GAIT project. Am J Hum Genet. 2005;76:925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noga AA, Zhao Y, Vance DE. An unexpected requirement for phosphatidylethanolamine n-methyltransferase in the secretion of very low density lipoproteins. J Biol Chem. 2002;277:42358–65. [DOI] [PubMed] [Google Scholar]