Abstract
Regulation of physiological functions with approximate daily periodicity, or circadian rhythms, is a characteristic feature of eukaryotes. Until recently, cyanobacteria were the only prokaryotes reported to possess circadian rhythmicity. It is controlled by a cluster of three genes: kaiA, kaiB, and kaiC. Using sequence data of ≈70 complete prokaryotic genomes from the various public depositories, we show here that the kai genes and their homologs have quite a different evolutionary history and occur in Archaea and Proteobacteria as well. Among the three genes, kaiC is evolutionarily the oldest, and kaiA is the youngest and likely evolved only in cyanobacteria. Our data suggest that the prokaryotic circadian pacemakers have evolved in parallel with the geological history of the earth, and that natural selection, multiple lateral transfers, and gene duplications and losses have been the major factors shaping their evolution.
Circadian clock genes are a vital and essential feature of eukaryotes (1). Cyanobacteria were the first prokaryotes reported to have the circadian clock regulated by a cluster of three genes: kaiA, kaiB, and kaiC (2). Cyanobacteria are among the oldest organisms on the earth, and they are among the most successful in terms of ecological plasticity and adaptability (3). In adaptation strategy of cyanobacteria, circadian clock genes are of particular importance, because they underlie fundamental physiological processes such as the regulation of nitrogen fixation, cell division, and photosynthesis (4). The clock genes are ubiquitous in cyanobacteria (5). In most cyanobacteria, these genes were reported as a single copy (2, 5), although some of them (kaiB; ref. 6) or even a whole cluster (7) may be duplicated. They were shown to operate as a single unit and to follow a feedback model of regulation: kaiA positively affects kaiBC promoter, whereas overexpression of kaiC represses it (2, 8). Among these genes, kaiC is a crucial component of clock precession in cyanobacteria (9–11).
Although the kai genes are under comprehensive study with regard to the mechanism of action, their evolution has yet to be resolved completely. The kaiC gene has a double-domain structure, and each of the domains has an ATP/GTP-binding site, or Walker's motif (2, 11). Based on its structure and sequence homology, the kaiC genes were classified as a family related to the RecA gene family of ATP-dependent recombinases (12). In addition to the kaiC genes with the typical double-domain structure, there are many single-domain homologous genes in Archaea and Proteobacteria. It was assumed that an ancestral single-domain kaiC gene was horizontally transferred from Bacteria to Archaea and then the double-domain kaiC evolved through duplication and subsequent fusion in Archaea (12). Although the evolution of the kaiC genes has been hypothesized, no data or hypotheses are available regarding the evolution of two other circadian clock genes, kaiA and kaiB. The evidence about the key role of kaiC in cyanobacterial clock regulation (9, 11), along with its homology to archaeal RecA genes, suggests that this gene is evolutionarily the oldest among the three.
In this study we reconstruct the origin and evolutionary patterns of the circadian clock genes in prokaryotes. Using available sequences from the public databases, we performed extensive phylogenetic analysis of the kai genes. The results suggest that the three prokaryotic circadian pacemakers have quite different evolutionary histories, and two of them, kaiA and kaiB, originated in cyanobacteria. The three-gene kaiABC cluster itself evolved ≈1,000 Mya.
Materials and Methods
DNA and Protein Sequences.
The annotated and homolog sequences of the kai genes were retrieved from GenBank and public databases of the Department of Energy Joint Genome Institute (www.jgi.doe.gov/JGI_microbial/html/index.html) by using the gapped BLASTP and PSI-BLAST (13) and the respective amino acid sequences of Anabaena sp. strain PCC 7120 (GenBank accession no. AP003591) as queries. Among the kaiC homologs from the other Prokaryota, which were about half the length of the kaiC genes from Cyanobacteria, only those matching both domains of the kaiC genes were chosen for further analysis. Multiple protein sequence alignments were constructed by using CLUSTALW (14) and manually adjusted based on structural considerations. Alignments of the nucleotide sequences were modified manually according to the respective amino acid alignments.
For the comparative phylogenetic analysis, we used sequences of 16S rRNA gene from the respective or closely related strains. This gene is a common marker for evolutionary studies of cyanobacteria (15, 16). The sequences obtained from the public databases were aligned with CLUSTALW (14) and adjusted by visual inspection. The list of used sequences is given in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org.
Sequence Polymorphism Analysis.
Due to saturation, synonymous substitutions were not computed. For the kaiB genes, we estimated the number of nonsynonymous differences (Ka). To determine the DNA substitution model corresponding to our data, we used MODELTEST 3.0 software (17). It seemed that Kimura's two-parameter model (18) with gamma distribution and a transition/transversions ratio equal to 0.56 fit our data best. Therefore, nonsynonymous nucleotide substitutions in the kaiB genes were calculated by using the modified Nei–Gojobori method (19) with Jukes–Cantor correction for multiple substitutions at the same site and the above transition/transversions ratio. The MEGA 2.1 software (20) was used for the computations of Ka.
Phylogenetic Analysis.
The alternative topologies of the phylogenetic trees were obtained initially by using maximum-parsimony (21) and neighbor-joining algorithms (22) and then evaluated with maximum-likelihood (ML) method (23) as implemented in TREE-PUZZLE 5.0 software (24) to find the best topology and test a molecular clock hypothesis. The ML approach with a local clock (25) realized in PAML software (26) then was applied to estimate the lengths of the branches. Those for the 16S rRNA tree were computed based on the Tamura–Nei model of substitutions (27), and those for the kai genes were estimated by using respective amino acid sequences and based on the WAG matrix of substitutions (28). The statistical support for the nodes of the trees was obtained by the bootstrapping procedure with 1,000 replications.
To detect lateral transfers, the congruency of the obtained phylogenetic trees of species (16S rRNA tree) and the kai genes was estimated by using Templeton's test (29) and the winning-sites test as implemented in PAUP* 4.0b10 (30). Most of the taxa having multiple representatives in the kaiC data set (e.g., Chloroflexus 1 and 2; Table 1) did not define monophyletic clusters in the kaiC tree. Because there is only one respective representative in the 16S rRNA tree (for example, Chloroflexus), it might generate the taxonomic ambiguity. To avoid it, we used all possible combinations of the trees (96 in total) in the analysis.
Results
Occurrence of kai Genes in Prokaryotes.
A BLAST search in the available completed prokaryotic genomes revealed homologs of the kaiC genes in both domains of prokaryotes, Archaea and Bacteria. In Archaea, these genes were found in the species of almost all well defined major taxa (except Methanopyri and Thermoplasmata). In Bacteria, the kaiC genes were not so ubiquitous; they were found only in four major taxa: Proteobacteria, Thermotogae, Chloroflexi, and Cyanobacteria. No such genes were found in the other bacteria. Importantly, kaiC homologs of Chloroflexi and Proteobacteria were the long (double-domain) versions of the gene, similar to those of Cyanobacteria, whereas most of the kaiC homologs of Archaea were the short (single-domain) versions (Table 1). However, within each domain, they are characteristic only to some taxa. Their occurrence in prokaryotes sometimes lacks predictability. For example, among Methanococci, kaiC homologs appear in Methanococcales (Methanococcus jannaschii) but not in Methanosarcinales (Methanosarcina barkeri). In Proteobacteria, such irregularity is observed even within the same orders: For example, among Xanthomonadales, the kaiC gene occurs in Xanthomonas campestris but not in Xylella fastidiosa. Cyanobacteria are the only prokaryotic kingdom in which all species appear to possess the kaiC genes.
Structure and Sequence Polymorphism of kai Genes in Prokaryota.
The kaiC genes and their homologs in prokaryotes can be divided into two major groups by their length. The longer genes usually have two ATP/GTP-binding domains and have approximately twice the length than that of the shorter genes. The shorter homologs usually match both domains but always have higher similarity to the first domain of the longer kaiC genes. Proteobacteria, Chloroflexi, and Cyanobacteria possessed only the double-domain genes, but in Archaea both double- and single-domain homologs occurred.
Similar to the kaiC genes, the kaiB and kaiA genes are variable in length. In Archaea, Chloroflexi, Proteobacteria, and unicellular cyanobacteria kaiB genes are ≈300 bp long, whereas in filamentous cyanobacteria much longer (up to 858 bp) copies also occur. The kaiA genes were found only in cyanobacteria and varied in length; the genes from unicellular cyanobacteria (Synechococcus and Synechocystis) were 1.5–2 times longer (852–900 bp) as compared with those from filamentous cyanobacteria (Anabaena and Nostoc). These genes seem to be the least conserved among the kai genes; even in closely related species such as Synechococcus sp. PCC7942 and Thermosynechococcus elongatus their amino acid sequences share only 43.6% identity. However, in contrast to the respective kaiC genes, shorter homologs of the kaiA and kaiB genes match only one segment of their longer versions, which is located closer to the 3′ terminus. It suggests that the latter have likely not evolved through duplication.
In some species, the kaiB and/or kaiC genes occur in multiple copies. These genes may form a cluster or be scattered throughout the genome. In species with a cluster, a single copy usually exists, except Synechocystis sp. PCC6803, which has two copies of the cluster.
Phylogeny of kaiC Genes.
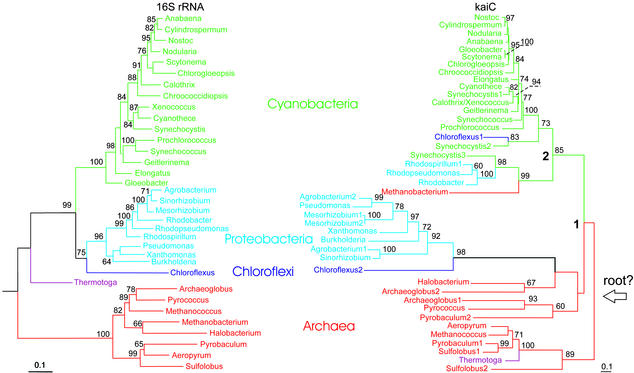
The resulting consensus tree clearly shows separation of the kaiC genes into three main clades, which essentially correspond to Archaea, Proteobacteria, and Cyanobacteria in the topology of the 16S rRNA tree (Fig. 1). All these branches have significant statistical support. The topology of the kaiC tree indicates that the genes of Cyanobacteria and Proteobacteria are monophyletic, whereas kaiC homologs of Archaea are polyphyletic. However, this polyphyly has low bootstrap support. Both Templeton's test (29) and the winning-sites test strongly rejected the null hypothesis of 16S rRNA and kaiC tree compatibility (P < 0.0001) with either the 16S sequence data or the kaiC sequence data and with all 96 alternative trees. These results evidently suggest multiple lateral transfers of the kaiC genes both within and between the three major clades of the tree (Fig. 1). For example, there were the transfers from Cyanobacteria to Proteobacteria (Rhodospirillum and Rhodobacter), Chloroflexi (Chloroflexus 1), and Archaea (Methanobacterium). Some of the transfers within the major clades are quite recent (for example, from Xenococcus to Calothrix, which have identical kaiC sequences, or from Scytonema to Gloeobacter).
Figure 1.
ML congruent phylogenetic trees of 16S rRNA genes and the kaiC homologs of prokaryotes. Designations of the genes are given in Table 1. Species of the major taxa are indicated with different colors. Bootstrap values <50% are not shown.
In some species, the kaiC genes are present in two or three (Synechocystis) copies, and based on the topology of the trees these copies may have quite different evolutionary histories. Both copies of Mesorhizobium most likely have evolved through recent duplication, whereas evolution of the twin copies in the other taxa (Pyrobaculum, Sulfolobus, Archaeoglobus, Agrobacterium, Chloroflexus, and Synechocystis) is more complex and probably involves lateral transfers. For example, two kaiC genes of Chloroflexus are orthologous, and among them only Chloroflexus 2 has evolved along with the species, whereas the other was likely transferred from cyanobacteria (Fig. 1).
Phylogeny of kaiA and kaiB Genes.
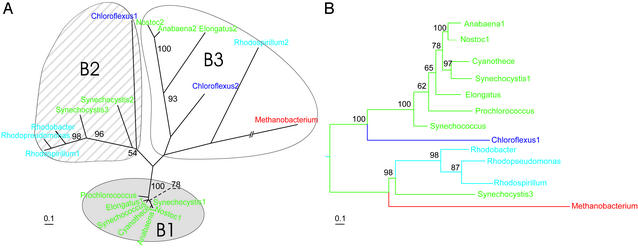
Screening prokaryotic genomes revealed kaiB genes in all available cyanobacteria similar to the kaiC genes. However, in contrast to the kaiC genes, kaiB occurred only in a few proteobacteria and one archaeon, Methanobacterium thermoautotrophicum (Table 1). None of the applied methods of phylogenetic reconstruction gave a fully resolved tree for the kaiB genes. The best unrooted tree is shown in Fig. 2A. This tree clearly indicates that the kaiB genes are separated into three main clades, which notably differ by the amount of accumulated nucleotide substitutions. The genes of the clade B1 have Ka = 0.092 ± 0.018, which is significantly lower than in the clades B2 (Ka = 0.478 ± 0.056) and B3 (Ka = 0.693 ± 0.068). Clade B1 comprises the genes only from cyanobacteria, whereas the two other clades include kaiB genes from Archaea and Proteobacteria as well. Interestingly, the kaiB genes in clade B3, except Methanobacterium, are not from the cluster, as the genes in clade B1 and B2 are, but are scattered throughout the genome.
Figure 2.
(A) The best unrooted ML tree of the kaiB genes. (B) ML phylogenetic tree of the kaiBC cluster of prokaryotes. For color designations of the taxa see Fig. 1. Bootstrap values <50% are not shown.
Only a few nodes in clades B2 and B3 of the kaiB tree have significant bootstrap support (Fig. 2A). However, a phylogenetic tree literally becomes completely resolved when inferred from the whole kaiBC clusters (Fig. 2B). Notably, its topology follows that of the cyanobacterial subtree of the kaiC genes (Fig. 1). This fact suggests that, after formation of the cluster, the kaiB and kaiC genes evolve as a unit rather than independently. Furthermore, comparison of the trees of the kai genes in Figs. 1 and 2 makes it reasonable to assume that the kaiB genes had originated in cyanobacteria and then were transferred laterally to the other prokaryotes.
Screening available prokaryotic genomes revealed the kaiA genes only in cyanobacteria. Among the available complete cyanobacterial genomes, the kaiA genes were not found in Prochlorococcus marinus. When available, the kaiA genes always occur in a single copy. Because of the limited number of kaiA genes available, it was impossible to perform their comprehensive phylogenetic analysis.
Dating Events in the Evolution of kai Genes.
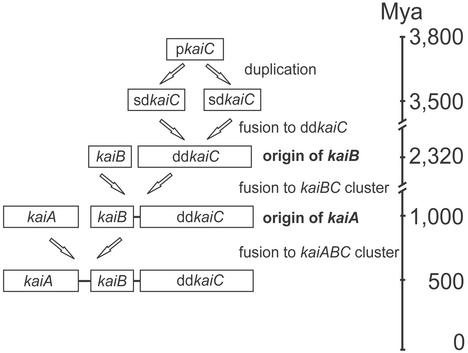
In our estimation of the dates of main events in the evolution of kai genes, we proceeded from the assumption that kaiC genes are evolutionarily the oldest among the prokaryotic circadian oscillators. Indeed, only kaiC and its homologs occur in the prokaryotes other than cyanobacteria. Based on fossil evidence, the last common ancestor of Eubacteria and Archaea is estimated as existing ≈3,800 Mya, and the oldest known cyanobacteria morphologically indistinguishable from existing Oscillatoria were documented in ≈3,500 million-year-old rocks (31). Given that all known kaiC genes of cyanobacteria are monophyletic and have a double-domain structure, we thus can suppose that duplication and subsequent fusion of their single-domain predecessors took place in the period between these dates, i.e., during ≈300 million years (Fig. 3).
Figure 3.
Timeline of major events in evolution of the cyanobacterial circadian clock genes based on ML estimates. The time scale is not proportional. pkaiC, predecessor of the kaiC gene; sdkaiC, single-domain kaiC; ddkaiC, double-domain kaiC.
The other major events in the evolution of the kai cluster were the appearance of the kaiB gene and its fusion with the kaiC to form the kaiBC cluster. Almost identical topologies of the cyanobacterial subtree of the kaiC genes and kaiBC tree (Figs. 1 and 2B) suggest that these events likely occurred in the period corresponding to the time between nodes 1 and 2 in the kaiC tree.
Because the ML test rejected the global clock hypothesis for all kai genes, we applied the local clock model (25) to date their evolution. According to these estimates, the appearance of kaiB genes and the formation of the kaiBC cluster occurred between ≈3,500 and 2,320 Mya.
Because of the small number of available kaiA genes, dating their evolution is difficult. Definitely, they are evolutionarily the youngest among the three prokaryotic circadian pacemakers, because they occur only in some higher cyanobacteria (Table 1). Importantly, of the two closely related unicellular cyanobacteria, Synechococcus sp. PCC7942 and P. marinus (Fig. 1, 16S rRNA tree), only the former possesses the kaiA gene. Furthermore, referring to the cyanobacterial kaiC subtree, the kaiA genes were found only in genomes of the species, which belong to the clades that are evolutionarily younger than Prochlorococcus. It allows us to suppose that these genes originated in cyanobacteria after speciation of Synechococcus and Prochlorococcus and thus to estimate the time of origin. Remarkably, ML estimation of this time using both 16S rRNA and kaiC sequences gives essentially the same result, 1,051 ± 1,16.9 and 944 ± 92.9 Mya, respectively.
Discussion
The results of the phylogenetic analysis suggest that the three genes of the kai cluster have quite different evolutionary histories. The kaiC gene is the oldest among the three. The three-gene cluster, as it was described originally (2), is likely characteristic only to cyanobacteria and evolved gradually through the consequent stepwise addition of the kaiB and kaiA genes to the kaiC gene. Moreover, the kaiA and kaiB genes originated in cyanobacteria after their separation from the other prokaryotes. The occurrence of the kaiB genes in some species of the other prokaryotic domains (Fig. 2) is a result of lateral transfers from cyanobacteria. Importantly, our data suggest no lateral transfers of any kai genes to cyanobacteria from the other prokaryotes. In light of these findings, the hypothesis about the origin of cyanobacterial kaiC genes through the duplication and fusion of the single-domain ancestor in Archaea and their lateral transfer to Cyanobacteria (12) is inconsistent.
Evolution of kai Genes and Geological History of the Earth.
Circadian pacemakers underlie basic physiological processes and thus may influence the expression of many genes. In cyanobacteria, the clock genes were shown to be involved in the regulation of nitrogen fixation (32), cell division (33), and other metabolic processes (4, 34). Furthermore, reproductive fitness of cyanobacteria increases when the endogenous clock and the temporal environmental cycle are strictly synchronized (35).
An analysis of the kai gene phylogenies in prokaryotes brings us to the three key assumptions: (i) circadian pacemakers are involved in the regulation of photosynthesis, (ii) double-domain structure of the kaiC gene is essential for the circadian oscillation, and (iii) circadian systems of prokaryotes may not necessarily include all three kai genes. These suppositions are supported by the fact that only photosynthetic prokaryotes have either the double-domain kaiC gene or the kaiBC cluster. The only exception is for a few Euryarchaeota (Table 1), which are not photosynthetic.
Comprehensive study of the kaiABC cluster expression in Synechococcus sp. PCC7942 showed that all three kai genes are essential for circadian rhythmicity, and inactivation of any of them completely abolishes it (2). The kaiA genes do not occur in the most primitive cyanobacteria and photosynthetic proteobacteria. Does that mean that these prokaryotes do not possess circadian rhythmicity? Apparently, it does not. Indeed, the circadian system with all three kai genes was described, thus far, only for Synechococcus sp. PCC7942 (2). Most likely, it is characteristic for the most evolutionarily advanced cyanobacteria, which have all these genes. However, it does not necessarily mean that other circadian systems, without kaiA, are impossible. In fact, in the system with the three kai genes, kaiA acts solely as a regulator of kaiBC expression via binding to two kaiA-binding domains in the kaiC gene (11). In the simpler systems, without kaiA, other yet-unknown gene(s) may play the role. Indeed, even the three-gene kai system includes other, either homologous to kai or not, genes essential for the circadian regulation (36, 37). Such genes may be elements of the simpler systems ensuring their functionality. Further studies of such circadian systems are needed to verify this hypothesis.
In the evolution of kai genes as a cluster controlling circadian rhythmicity, a few major events may be defined. The first was the duplication of a single-domain ancestor of kaiC and further fusion of the resulting genes to form the double-domain gene. It occurred ≈3,800–3,500 Mya (Fig. 3).
Another major event was the origin of the kaiB gene and the formation of the kaiBC cluster. It happened between ≈3,500 and 2,320 Mya (Fig. 3). According to the theory of atmosphere evolution (38), the time of ≈2,500–2,000 Mya corresponds to the period when a reducing geochemical environment, which had existed since the early history of the earth, was replaced by an oxidizing environment produced by cyanobacteria. In this period, cyanobacteria were not truly oxygen-evolving but were consuming nitrogen and producing oxygen, which was poisonous for the then-dominating methano- and sulfobacteria. Initially, oxygen was bound to Fe2+ released from the Earth's mantle because of volcanic activity. Approximately 2,500 Mya the volcanic activity subsided, resulting in a depletion of Fe2+ resources ≈2,000 Mya (39). Further, when becoming oxygen-producing, cyanobacteria out-competed early-evolved photosynthetic bacteria, because biosynthesis of bacteriochlorophyll would have been inhibited by molecular oxygen (40).
Because of their influence on the wide variety of vital metabolic cycles, circadian clock genes should be an essential part of adaptive mechanisms. They are particularly important for photosynthetic organisms (e.g., cyanobacteria and purple bacteria), the vital functions of which are based on photochemically dependent processes. Temporal optimization of intracellular mechanisms to the day/night conditions thus should increase fitness of these prokaryotes and confer an evolutionary advantage. Origin of the circadian gene cluster therefore could be one of the key events in the evolution of cyanobacteria, which ensured their domination in the Earth's ecosystems over almost 2.5 billion years known as “the age of Cyanobacteria” (41).
Key Mechanisms of Adaptive Evolution of the kai Genes.
In addition to the above considerations, there are a number of other evidences supporting an adaptive character of the evolution of the kai genes. Duplication of the kaiC predecessor and formation of the double-domain kaiC gene in the earliest stages of its evolution (Fig. 3) was likely among those adaptive characters, because gene duplication was shown to be an exceptionally efficient mechanism for rapid evolutionary advance (42–44) including the kai genes (7). Duplicated genes may further evolve in four ways: (i) one of the copies keeps the original function, whereas another may accumulate deleterious mutations and become nonfunctional (nonfunctionalization); (ii) one copy acquires a novel, advantageous function, and another copy still maintains the original function (neofunctionalization); (iii) duplicated paralogs share original functions, and both new copies are less efficient compared with the ancestral gene (subfunctionalization); or (iv) one of the new copies performs with the original efficiency, whereas another accumulates advantageous mutations and acquires greater efficiency under specific conditions (superfunctionalization or reinforcement) (7, 45–49).
All these scenarios may have taken place in the evolution of the kai genes. The evident cases of nonfunctionalization and superfunctionalization were described elsewhere (7). Neofunctionalization and subfunctionalization also might occur in evolution of the kaiB and kaiC genes of some species. For example, Synechocystis sp. PCC6803 has three copies of the kaiB and kaiC genes each, six in total (Table 1). Four of these copies form two clusters (one of them with the kaiA gene, which presents itself in a single copy), and the remaining two are scattered throughout the genome. The kaiABC cluster was shown to regulate circadian oscillation (2) and was supposed to be the most advanced circadian system in prokaryotes. Another cluster comprises two genes, kaiB and kaiC, and may have either different (neofunctionalization) or similar but probably less efficient (subfunctionalization) function. And finally, the same ways of evolution may be considered for the scattered kaiB and kaiC genes.
Another mechanism greatly affecting evolution of the kai genes is lateral transfers. From the phylogenetic trees of the kai genes in Figs. 1 and 2, the numerous lateral transfers of the kaiB and kaiC genes or kaiBC cluster can be detected. Remarkably, all transfers of the cluster were from evolutionarily more advanced (Cyanobacteria) to more primitive prokaryotes (Chloroflexi, Proteobacteria, and Archaea) (Fig. 2B). In some species such as Chloroflexus aurantiacus, the laterally transferred and presumably more advanced system (kaiBC cluster) coexists with the original less-efficient one (comprising the kaiB and kaiC genes not joined in the cluster), whereas in the others such as Methanobacterium thermoautotrophicum the original system (supposedly consisting of the single-domain kaiC gene) had been replaced in the course of evolution by the laterally transferred one represented by the kaiBC cluster. It is hard to believe that the transferred genes conferring lower fitness under the given conditions could replace the homologous original genes and/or persist for such a long evolutionary period.
Recently, we reported that under acute long-term environmental stress, evolution of the kai genes in cyanobacteria is governed mainly by various types of natural selection (7). In the reported case, one of the main stress factors was UV irradiation. During the long period of the geological history of the Earth, the natural environment of prokaryotes was harsh, and UV irradiation was among the major factors most dramatically influencing evolution of cyanobacteria (50). Thus, the effect of UV and other stress factors on the kai genes in this period was probably somewhat similar to that observed in the “Evolution Canyon” model including a high mutation rate and frequent gene duplications (7) and eventually was controlled by natural selection.
During the long period of prokaryotic evolution, since the earliest documented time (31, 51) until ≈500 Mya, the environment has undergone radical changes in the levels of oxygen and UV irradiation. Many prokaryotes occupy stressful habitats at present. They have evolved through increasing their fitness to survive as well as adapting to the changing environment. The origin and evolution of circadian clock genes was one of the most remarkable instruments of such adaptation involving a vast variety of known evolutionary mechanisms.
Supplementary Material
Acknowledgments
We are grateful to Professor Alan Templeton (Washington University, St. Louis) for critical reading of the paper and support in improving the statistical tests and Dr. Hong-Wen Deng (Creighton University, Omaha, NE), Professor Outi Savolainen (University of Oulu, Oulu, Finland), and Dr. Claus Vogl (University of Veterinary Medicine, Vienna) for helpful comments on the early draft of the manuscript. This work was supported by Israeli National Science Foundation Grant 98-04-4963, the Israeli Discount Bank Chair of Evolutionary Biology, the Ancell–Teicher Research Foundation for Genetics and Molecular Evolution Grant R01 GM60402-01A1, and the National Institutes of Health State of Nebraska Cancer and Smoking Related Disease Research Program.
Abbreviations
- Mya
million years ago
- ML
maximum likelihood
References
- 1.Pittendrigh C S. Annu Rev Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 2.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson C R, Tanabe A, Golden S S, Johnson C H, Kondo T. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 3.Whitton B A. In: Survival and Dormancy of Microorganisms. Hennis Y, editor. New York: Wiley; 1987. pp. 109–167. [Google Scholar]
- 4.Johnson C H, Golden S S. Annu Rev Microbiol. 1999;53:389–409. doi: 10.1146/annurev.micro.53.1.389. [DOI] [PubMed] [Google Scholar]
- 5.Lorne J, Scheffer J, Lee A, Painter M, Miao V P. FEMS Microbiol Lett. 2000;189:129–133. doi: 10.1111/j.1574-6968.2000.tb09218.x. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko T, Nakamura Y, Wolk C P, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, et al. DNA Res. 2001;8:205–213. doi: 10.1093/dnares/8.5.205. [DOI] [PubMed] [Google Scholar]
- 7.Dvornyk V, Vinogradova O N, Nevo E. Proc Natl Acad Sci USA. 2002;99:2082–2087. doi: 10.1073/pnas.261699498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson C H, Golden S S, Kondo T. Trends Microbiol. 1998;6:407–410. doi: 10.1016/s0966-842x(98)01356-0. [DOI] [PubMed] [Google Scholar]
- 9.Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. Proc Natl Acad Sci USA. 2000;97:495–499. doi: 10.1073/pnas.97.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Mori T, Johnson C H. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taniguchi Y, Yamaguchi A, Hijikata A, Iwasaki H, Kamagata K, Ishiura M, Go M, Kondo T. FEBS Lett. 2001;496:86–90. doi: 10.1016/s0014-5793(01)02408-5. [DOI] [PubMed] [Google Scholar]
- 12.Leipe D D, Aravind L, Grishin N V, Koonin E V. Genome Res. 2000;10:5–16. [PubMed] [Google Scholar]
- 13.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honda D, Yokota A, Sugiyama J. J Mol Evol. 1999;48:723–739. doi: 10.1007/pl00006517. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Pichel F, Lopez-Cortes A, Nubel U. Appl Environ Microbiol. 2001;67:1902–1910. doi: 10.1128/AEM.67.4.1902-1910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Posada D, Crandall K A. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 18.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 19.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S, Tamura K, Jakobsen I, Nei M. MEGA2, Molecular Evolutionary Genetics Analysis Software (Arizona State University, Tempe), Version 2.1. 2001. [DOI] [PubMed] [Google Scholar]
- 21.Fitch W M. Syst Zool. 1971;20:406–416. [Google Scholar]
- 22.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 24.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 25.Yoder A D, Yang Z. Mol Biol Evol. 2000;17:1081–1090. doi: 10.1093/oxfordjournals.molbev.a026389. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z. Comput Appl Biosci. 1997;15:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Nei M. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 28.Whelan S, Goldman N. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 29.Templeton A R. Evolution (Lawrence, Kans) 1983;37:221–244. doi: 10.1111/j.1558-5646.1983.tb05533.x. [DOI] [PubMed] [Google Scholar]
- 30.Swofford D L. PAUP*, Phylogenetic Analysis Using Parsimony (* and Other Methods) Sunderland, MA: Sinauer; 2002. , Version 4. [Google Scholar]
- 31.Schopf J W, Packer B M. Science. 1987;237:70–73. doi: 10.1126/science.11539686. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y B, Dominic B, Mellon M T, Zehr J P. J Bacteriol. 1998;180:3598–3605. doi: 10.1128/jb.180.14.3598-3605.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mori T, Binder B, Johnson C H. Proc Natl Acad Sci USA. 1996;93:10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang T-C, Chen H-M, Pen S-Y, Chen T H. Planta. 1994;193:131–136. [Google Scholar]
- 35.Ouyang Y, Andersson C R, Kondo T, Golden S S, Johnson C H. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutsuna S, Kondo T, Aoki S, Ishiura M. J Bacteriol. 1998;180:2167–2174. doi: 10.1128/jb.180.8.2167-2174.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwasaki H, Williams S B, Kitayama Y, Ishiura M, Golden S S, Kondo T. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 38.Berkner L C, Marshall L L. J Atmos Sci. 1965;22:225–261. [Google Scholar]
- 39.Holland H D. In: Early Life on Earth. Bengtson S, editor. New York: Columbia Univ. Press; 1994. pp. 237–244. [Google Scholar]
- 40.Olson J M, Pierson B K. Int Rev Cytol. 1987;108:209–248. doi: 10.1016/s0074-7696(08)61439-4. [DOI] [PubMed] [Google Scholar]
- 41.Schopf J W. Nova Hedwigia. 1996;112:13–32. [Google Scholar]
- 42.Langridge J. Mol Gen Genet. 1969;105:74–83. doi: 10.1007/BF00750315. [DOI] [PubMed] [Google Scholar]
- 43.McLachlan A D. Cold Spring Harbor Symp Quant Biol. 1987;52:411–420. doi: 10.1101/sqb.1987.052.01.048. [DOI] [PubMed] [Google Scholar]
- 44.Sonti R V, Roth J R. Genetics. 1989;123:19–28. doi: 10.1093/genetics/123.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- 46.Walsh J B. Genetics. 1985;110:345–364. doi: 10.1093/genetics/110.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark A G. Proc Natl Acad Sci USA. 1994;91:2950–2954. doi: 10.1073/pnas.91.8.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sidow A. Curr Opin Genet Dev. 1996;6:715–722. doi: 10.1016/s0959-437x(96)80026-8. [DOI] [PubMed] [Google Scholar]
- 49.Force A, Lynch M, Pickett F B, Amores A, Yan Y L, Postlethwait J. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia-Pichel F. Origins Life Evol Biosphere. 1998;28:321–347. doi: 10.1023/a:1006545303412. [DOI] [PubMed] [Google Scholar]
- 51.Schopf J W. In: Prokaryotic Development. Brun Y V, Shimkets L J, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 105–129. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.