Abstract
The lineage leading to lungfishes is one of the few major jawed vertebrate groups in which Ig heavy chain isotype structure has not been investigated at the genetic level. In this study, we have characterized three different Ig heavy chain isotypes of the African lungfish, Protopterus aethiopicus, including an IgM-type heavy chain and short and long forms of non-IgM heavy chains. Northern blot analysis as well as patterns of VH utilization suggest that the IgM and non-IgM isotypes are likely encoded in separate loci. The two non-IgM isotypes identified in Protopterus share structural features with the short and long forms of IgX/W/NARC (referred to hereafter as IgW), which were previously considered to be restricted to the cartilaginous fish. It seems that the IgW isotype has a far broader phylogenetic distribution than considered originally and raises questions with regard to the origin and evolutionary divergence of IgM and IgW. Moreover, its absence in other gnathostome lineages implies paradoxically that the IgW-type genes were lost from teleost and tetrapod lineages.
Tetrapods (land vertebrates) are thought to have shared a common ancestor with a group of fleshy-finned fishes that include the Crossopterygii (coelacanths) and the Dipneusti (lungfishes) (1, 2). Recent systematic studies using both morphological and molecular characters generally have supported a phylogeny in which the lungfishes are the closest relatives of the tetrapods (3–5). Comparative studies on the immunoglobulins at the molecular genetic level have focused on numerous ectothermic vertebrate groups, including chondrichthyans (sharks, rays, chimeras), teleosts (bony fishes), amphibians, and reptilians (reviewed in refs. 6 and 7), but little corresponding information is available for the fleshy-finned fish lineage. Three Ig isotypes have been identified so far at the protein level in two species of lungfishes (8–10) that were generically designated: high molecular weight (IgM), intermediate molecular weight, and low molecular weight (IgN) (9, 10). We report herein the cloning and characterization of several Ig heavy chain (IgH) cDNAs from Protopterus aethiopicus and present evidence for a phylogenetic relationship between these different isoforms and genes that have been identified in cartilaginous fishes, thus placing the divergence of IgH isotypes at a far earlier point in vertebrate phylogeny than considered previously.
Materials and Methods
Juvenile P. aethiopicus specimens (10–20 cm) were maintained in laboratory aquaria before death, and removal of tissues. RNA, and DNA were isolated by using standard protocols (11). Liver RNA was used as the source for cDNA library construction, whereas erythrocyte DNA was used for genomic library construction.
Generation of VH Probes.
VH-specific probes were derived by amplifying Protopterus genomic DNA with primers that complement sequences in FR1 to FR3 that are common to vertebrate Ig VH structures (12). Appropriately sized PCR products were subcloned into pBluescript SKII+ and sequenced. A probe specifying a single VH was used in initial screening.
RNA Isolation, cDNA Library Construction and Screening, and Northern Blot Analysis.
Total and poly (A)+ RNA were isolated by using RNAzol (Tel-Test, Friendswood, TX) and an mRNA paramagnetic bead isolation method (Dynal, Oslo), respectively. An oligo(dT)-primed liver cDNA library was generated in λgt11 by using a commercial kit (Amersham Pharmacia). Initial library screening was conducted on quadruplicate nitrocellulose lifts of eight 150-mm plates at a density of ≈25,000–50,000 plaque-forming units per plate. Northern blots were prepared from liver mRNA by using standard methods (11).
Subcloning and DNA Sequencing.
Isolated λ cDNAs were subcloned into plasmid (pBluescript SKII) or M13 (mp18) vectors and sequenced bidirectionally by using Applied Biosystems and Li-Cor (Lincoln, NE) DNA automated sequencers. Sequence assembly and analysis was performed by using IG SUITE (Intelligenetics), ALIGNIR (Li-Cor), PROSITE (13), and MEGA2 (14) software. Nucleotide sequences submitted to GenBank are available with the following accession numbers: clone 27, AF437724; clone 41, AF437725; clone 32, AF437726; clone 28, AF437727; clone 22, AF437728; clone 45, AF437729; clone 35, AF437730; clone 26, AF437731; clone 36, AF437732; clone 40, AF437733; and clone 76, AF437734.
Results
Identification of Protopterus Ig Genes.
Three distinct VH+ clones possessing two different constant (CH) gene segments were initially identified in a juvenile Protopterus liver cDNA library. Clone 2 encodes an incomplete IgM heavy chain; clones 4 and 8 encode different IgH chain types. Protopterus-specific VH and CH probes were derived from the initial clonal isolates and were hybridized individually to replicate filter lifts. Clones 27, 28, and 32 with insert sizes of 2.0, 2.8, and 1.5 kb, respectively, were sequenced.
IgM Heavy Chain.
Clone 27 encodes a putative IgM heavy chain consisting of a leader peptide, a single V domain, a DJ region, and four CH domains. The estimated molecular weight of the deduced amino acid sequence excluding the leader peptide (≈62,900) is consistent with the predicted molecular weight (≈70,000) of the IgM heavy chain (10, 15), if potential N-glycosylation is considered. Clone 27 aligns with various vertebrate IgM heavy chains (Fig. 1), and its assignment as an IgM is unequivocal. Two cysteines and a tryptophan that comprise the core of the Ig fold (16) were observed for the variable Ig domain and three of the four constant Ig domains. Differences in conserved cysteine residues have been noted in IgM heavy chains from both teleost and cartilaginous fishes, specifically CysP409, which corresponds to mouse CysM414 that is known to be important for complement activation (17), is absent in these species but is present in the lungfish IgM heavy chain (P or M shown for the superscripts of amino acid residues indicates Protopterus or Mus, respectively). Other residues important for complement activation that are involved in C1q binding (e.g., homologous residues of mouse, AspM432, ProM434, or ProM436; ref. 18) are conserved in the lungfish with two replacements of biochemically similar amino acids: AspM417 → GluP412 and AspM432 → GluP426. Furthermore, a site (AspP397) is present in the lungfish IgM heavy chain that corresponds to mouse AsnM402, at which an N-linked glycosaccharide is involved indirectly in C1q binding (19).
Figure 1.
(A) Amino acid alignment of 11 vertebrate Cμ secretory sequences. Sequences were from GenBank: African lungfish (Pae), P. aethiopicus (this study, clone 27); human (Hsp), Homo sapiens (X57086); short-tailed opossum (Mdo), Monodelphis domestica (AF108226); chicken (Gga), Gallus gallus (X01613); African clawed frog (Xla), Xenopus laevis (M20484); sturgeon (Aba), Acipenser baeri (Y13253); bowfin (Aca), Amia calva (U12456); channel catfish (Ipu), Ictalurus punctatus (M27230); rainbow trout (Omy), Oncorhynchus mykiss (S63348); horned shark (Hfr), Heterodontus francisci (X07784); little skate (Rer), Raja erinacea (M29679). Periods indicate identical residue with African lungfish residue; dashes indicate gaps. +, residues important for complement activation. *, Residues used for subsequent phylogenetic analyses. (B) Amino acid sequences of eight vertebrate Cμ transmembrane regions: Pae (this study, clone 24), Hsp (PIR, S14683), Xla (X90768), Aba (AJ245364), Ipu (X52617), Omy (U04616), Hfr (X07785), and Rer (M29678). The sequence for Amia calva was taken from Wilson et al. (35). The transmembrane regions of human, lungfish, shark, and skate IgM heavy chain are anchored at the end of the CH4 domain, whereas those of channel catfish and rainbow trout are anchored at the end of the CH3 domain. The transmembrane region of bowfin IgH heavy chain can be anchored at the end of CH4, the end of CH3, or the middle of the CH3 domain, whereas the sturgeon transmembrane region seems to be anchored at the end of either the CH1 (AJ245363) or the CH4 (AJ245362) domain.
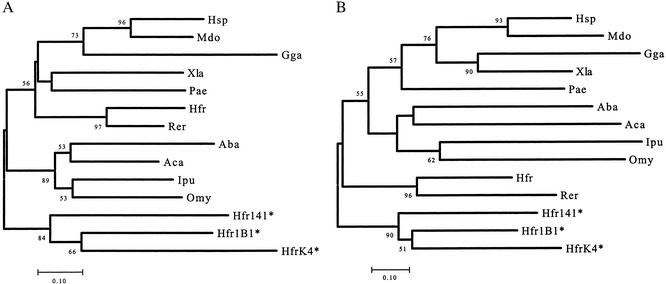
CH4 is the most conserved IgM heavy chain constant region domain (20). In phylogenetic analyses, the lungfish IgM and tetrapod IgM heavy chain CH4 domains cluster with the corresponding regions of IgM that have been identified in cartilaginous fishes but not with the CH4 domains from representative actinopterygians (bony fishes) (Fig. 2A). This finding was entirely unanticipated because actinopterygians are more closely related phylogenetically to tetrapods and lungfishes than to the cartilaginous fishes, although it was also observed in earlier phylogenetic analysis (e.g., ref. 21). Conversely, phylogenetic analyses of the other Cμ domains demonstrate closer relationships between domains of actinopterygian, tetrapod, and lungfish rather than with those of cartilaginous fishes (e.g., Fig. 2B). Because the splicing pattern of the transmembrane form of IgM heavy chain of teleost fishes differs from higher vertebrates as discussed below, it seems that the teleost IgM heavy chain constant regions have been subject to some unique evolutionary event, such as exon shuffling, gene conversion, or genomic recombination, around the CH3–CH4 region.
Figure 2.
(A) Neighbor-joining tree for CH4 domains of IgM heavy chain. Pairwise genetic distances were obtained by Poisson correction. Three horn shark light chain sequences (HfrK4, Hfr1B1, Hfr141; ref. 31 and references therein) were used as outgroups and are indicated by asterisks. The number given in each branch is a bootstrap value; bootstrap values <50% are not shown. The branch lengths are measured in terms of the number of amino acid substitutions, with the scale given below the tree. (B) Neighbor-joining tree for IgM heavy chain CH3 domains. Abbreviations for the taxa are given in Fig. 1.
Non-IgM Heavy Chain.
Several structural/functional inferences can be made from the predicted peptide sequence of a second IgH-like molecule (clone 32): (i) strong homology is evident for the VH gene segment of this clone and other VH gene segments; (ii) a β-bulge motif, which is significant for VH–VL interactions at the V and DJ junction regions, is present (see below); and (iii) a cysteine residue in the CH1 domain is consistent with a light chain disulfide bridge (Fig. 3). The predicted protein product of clone 32 consists of a leader peptide, one V domain, a DJ region, and two C domains. This structure may correspond to that of the Ig (low molecular weight)/IgN heavy chain (10, 15). The estimated molecular weight of the gene product of clone 32, excluding the leader peptide, is ≈36,500, equivalent to the estimated molecular weight of Ig (low molecular weight) heavy chain. The two consecutive cysteine residues present in the CH1 domain raise the possibility of an extra intradomain disulfide bridge, as described in other species (22). Four additional cysteine residues are located at various parts of the CH region, some of which are potentially involved in the formation of inter-heavy chain disulfide bridges.
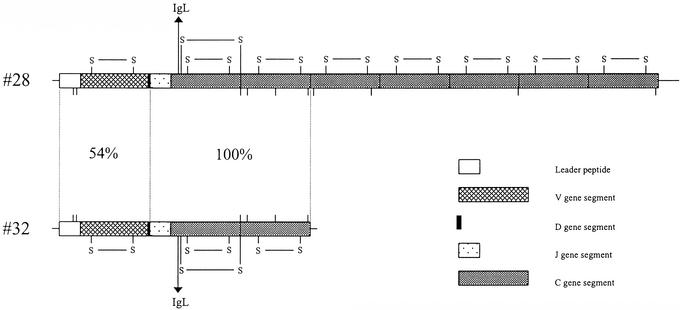
Figure 3.
Long form (clone 28) and short form (clone 32) of IgW-like heavy chains. Vertical lines indicate sulfhydryl groups of cysteine residues, which can form interchain disulfide bridges. Potential intrachain disulfide bridges are indicated by S–S bridges. The numbers shown are the proportions of nucleotide identity at the V–D region and the J–C region.
A third cDNA (clone 28) is predicted to encode an IgH consisting of a single V domain, a DJ region, and seven C domains (Fig. 3). Whereas the predicted leader peptides and V gene segments found in clones 32 and 28 differ to some degree, the nucleotide sequences of the first two C domains as well as J gene segments are identical. This observation is consistent with alternative RNA processing at a single Ig locus, and comparisons of the predicted sequences of clones 28 and 32 are reminiscent of observations with skate IgX (IgW/IgNARC) in which the short and long forms of heavy chain encode two and six C domains, respectively (23–26). Owing to the extraordinarily large genome size of Protopterus (>20 times larger than that of human; ref. 27), it will be difficult to definitively resolve whether the different gene products are produced by alternative splicing or from two discrete loci.
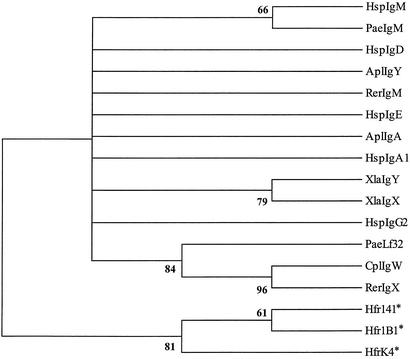
A phylogenetic analysis of the lungfish IgH isotype has been carried out (Fig. 4; a bootstrap consensus tree was obtained because many basal branching points were poorly resolved). Because the numbers of CH domains range from two to seven in different vertebrate IgH isotypes, it was necessary to identify and compare homologous domains. The CH1 domain was selected as a reference because it is present in all IgH and shares, with limited exceptions, one conserved function, i.e., covalent association with IgL. The phylogenetic analysis indicates that the CH1 domain of this lungfish IgH is related closely to that of IgW found in cartilaginous fish. Although it would be of considerable significance, the relationship of the IgH in lungfish to teleost fish IgD cannot be resolved, because the first IgD domain corresponds to CH1 of IgM (28).
Figure 4.
Bootstrap consensus tree for CH1 domains of IgH. Pairwise genetic distances were estimated by Poisson correction, and the phylogenetic tree was constructed by the neighbor-joining method. Sequences were from GenBank or the Protein Information Resource: human (Hsp), H. sapiens IgA1(J00220), IgD (PIR:AAB21246), IgE (J00222), IgG2 (J00230), and IgM (X57086), duck (Apl), Anas platyrhyncos IgY (X65219) and IgA (U27222); African clawed frog (Xla), X. laevis IgX (PIR:S03186) and IgY (X15114); African lungfish (Pae), Protopterus aethiopicus IgM (this study, clone 27) and Lf32 (this study, clone 32); carcharhine shark (Cpl), Carcharhinus plumbeus (U40560); and little skate (Rer), R. erinacea (PIR:S12839). Three horn shark light chain sequences (HfrK4, Hfr1B1, and Hfr141) (ref. 31 and references therein) were used as outgroups and are denoted by asterisks.
The C-terminal of clone 28 contains a cysteine residue as well as a potential N-glycosylation site, which are characteristics of the secretory tail of vertebrate IgM, mammalian and avian IgA, and cartilaginous fish IgW heavy chain, but not of other IgH, including the teleost IgD heavy chain. Amino acid comparisons of the secretory tails show relatively high degrees of similarity (50% identity) between the lungfish and cartilaginous fish IgW heavy chain sequences, even though the lungfish sequence contains an extra C-terminal residue. Collectively, these observations further support a direct relationship of the lungfish IgH to cartilaginous fish IgW heavy chain.
Variable Region Diversity.
VH regions from 13 IgH+ clones have been sequenced, and eight different VH gene families have been identified thus far (Fig. 5). There were, in addition, four other VH gene segments known from a lungfish (E. Hsu, personal communication), each of which may represent a different VH family. The amino acid difference between the VH regions of different families varies from ≈35% to ≈64%. The variability, including insertions/deletions, is particularly extensive in the complementarity-determining regions; framework regions (FR) vary to a lesser degree. A conserved cysteine in the VH region of clone 76 has been replaced with a serine and may be indicative of a pseudogene. All VH regions, with the exception of atypical clone 45, encode the Gly-Leu-Glu-Trp motif (or variants thereof) found in FR2, which is essential for VH/VL dimerization through β-bulge formation (29, 30) along with the (Trp/Phe)-Gly-X-Gly motif in the DJ region.
Figure 5.
Inferred amino acid sequences of the VH regions of P. aethiopicus clones. FR, framework region; CDR, complementarity-determining region. VH regions of clone 2 were designated VHI; clones 4 and 8 were designated VHV. IgH-type was confirmed by success/failure of PCR with using CH1 primers unique to IgM or IgW isotypes. The isotype designation of clone 22 was not of the IgM or IgW type (unpublished data).
Although the number of genes being compared is limited, the same VH gene families are associated with the same isotypes (Fig. 5). Association of VH gene families with a particular isotype is unusual and observed only rarely in higher vertebrates. However, a similar restricted utilization of V gene families has been described in cartilaginous fish IgM, IgW, and Ig light chains (31–33), which are encoded at separate clusters throughout the genome (ref. 24; T.O. and C.T.A., unpublished work).
RNA Expression.
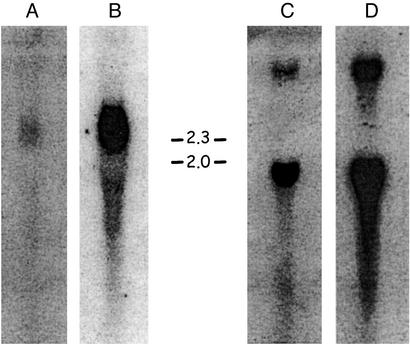
Northern blots were hybridized with probes complementing two distinct VH and CH genes (from clones 2 and 8; Fig. 6). The VH probe derived from clone 2 was shown to hybridize strongly with a 2-kb fraction and relatively weakly with a 1.6-kb fraction. The VH probe (VH V family) derived from clone 8 hybridized strongly with 1.6- and 2.8-kb fractions but failed to hybridize with a 2-kb fraction. The 2-kb transcripts hybridized with VH probes derived from clone 2, which encodes an IgM heavy chain, whereas the 1.6- and 2.8-kb transcripts hybridized with the probes derived from clone 8, which would complement the short and long forms, respectively, of the IgW-like heavy chain. This observation was confirmed by hybridization analyses using CH-specific probes (Fig. 6). Taken together, the hybridization patterns observed are consistent with the association of different VH gene families with different heavy chain isotypes.
Figure 6.
Northern blot analysis of two different isotypes suggests differential usage of VH and CH genes. Probes complementing the VH and CH regions of clones 2 (IgM-type) and 8 (IgW-type) were hybridized as described. (A) Hybridization of VH of clone 2. (B) Hybridization of CH of clone 2. (C) Hybridization of VH of clone 8. (D) Hybridization of CH of clone 8. Sizes are in kb.
Discussion
The Evolution of IgM.
In higher vertebrates, IgM is produced first during development and is the most significant in the early phase of the immune response. In its membrane-bound form, IgM functions as a B cell-specific ligand binding receptor, and its expression is critical to progressive B cell differentiation. IgM-type genes comprise the only class of IgH that has been identified in all jawed vertebrates (20). All known secretory forms of IgM heavy chain consist of one V domain, a DJ region, and four CH domains, as in the case of lungfish. However, the transmembrane region of lungfish IgM is contiguous with the CH4 domain (clone 24; Fig. 1), a situation also found in mammals, avians, amphibians, and cartilaginous fish. In contrast, membrane-bound forms of teleost IgM consist of two or three CH domains owing to a unique form of RNA processing (34), which likely was established at an early point in the evolution of bony fishes (35, 36). Phylogenetic analysis of CH4 domains of vertebrate IgM molecules indicates that the Neopterygii, a group that includes the teleosts as well as primitive bony fishes such as bowfins and sturgeons, forms a distinct cluster of their own (Fig. 2). Taken together, these findings suggest that the lungfish IgM heavy chain is more similar to those of tetrapods than to those found in neopterygian fishes. This is consistent with the generally held hypothesis that the lobe-finned fishes rather than actinopterygian fishes are the closest relatives of tetrapods.
Lungfish IgW-Like Heavy Chain.
In addition to IgM, a second isotype of Ig (IgW) has been identified in the earliest jawed vertebrates, raising questions as to the primordial IgH (23, 25, 37). Based on phylogenetic analysis of the CH of the lungfish clones presented here as well as their heavy chain structure and patterns of exon utilization, we propose that this isotype system is orthologous to cartilaginous fish IgW heavy chains. In cartilaginous fish, the IgM and IgW heavy chain gene loci are encoded by >100 separate, paralogous loci (24–26). Although the number of loci encoding the IgH in lungfish is not yet known, it is unlikely that it is as great as that seen in the cartilaginous fishes, as the nucleotide sequences of the CH regions examined thus far exhibit little divergence, with the exception of the few amino acid differences in the CH1 domain of IgM heavy chain that may represent allelic polymorphisms (clone 41). The distinct patterns of usage of VH gene segments between the IgM and the IgW heavy chains are consistent with their being encoded at independent loci. Furthermore, nucleotide sequence analysis suggests that these short and long forms derive by alternative splicing, although the possibility that they are products of recently duplicated genes cannot be ruled out. As indicated above, resolution of this issue requires extensive genomic characterization, which is technically confounded by the unusually large size of the lungfish genome (27).
The presence of both the IgM and IgW isotypes in cartilaginous fishes as well as lungfish implies that the duplication of these genes predates the origin of bony fishes analogous to the duplication of Ig light chain to produce κ and non-κ (λ) types. Importantly, this report demonstrates that the IgW system is not restricted to the Chondrichthyes and has a broader phylogenetic distribution than considered originally. No clear homologs of IgW have been identified in the human genome, or for that matter in amphibians, reptiles, and avians. Although perhaps premature, it is notable that in terms of subunit composition, the evolution of Ig genes has, on one hand, been highly conserved throughout the phylogeny of jawed vertebrates; however, the organization and complexity of genes is highly variable in those taxa examined to date.
Acknowledgments
We thank Mary West and Barbara Pryor for editorial assistance and Ronda Litman, Asaf Halevi, and Wendy Lippmann for technical assistance. This work was funded, in part, by National Science Foundation Grants IBN-9614940 and IBN-9905408 (to C.T.A.), National Institutes of Health Grants R37-AI23338 (to G.W.L.) and R24-RR14085 (to C.T.A.), the Center for Human Genetics (Boston University School of Medicine), and the Benaroya Research Institute at Virginia Mason.
Abbreviation
- IgH
Ig heavy chain
References
- 1.Carroll R L. Vertebrate Paleontology and Evolution. New York: Freeman; 1988. [Google Scholar]
- 2.Helfman G S, Collette B B, Facey D E. The Diversity of Fishes. Oxford: Blackwell; 1997. [Google Scholar]
- 3.Meyer A, Wilson A C. J Mol Evol. 1990;31:359–364. doi: 10.1007/BF02106050. [DOI] [PubMed] [Google Scholar]
- 4.Meyer A, Dolven S I. J Mol Evol. 1992;35:102–113. doi: 10.1007/BF00183221. [DOI] [PubMed] [Google Scholar]
- 5.Marshall C, Schultze H P. J Mol Evol. 1992;35:93–101. doi: 10.1007/BF00183220. [DOI] [PubMed] [Google Scholar]
- 6.Du Pasquier L, Flajnik M. In: Fundamental Immunology. Paul W E, editor. Philadelphia: Lippincott–Raven; 1998. pp. 605–650. [Google Scholar]
- 7.Litman G W, Anderson M K, Rast J P. Annu Rev Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- 8.Marchalonis J J. Aust J Exp Biol Med Sci. 1969;47:405–419. doi: 10.1038/icb.1969.46. [DOI] [PubMed] [Google Scholar]
- 9.Litman G W, Wang A C, Fudenberg H H, Good R A. Proc Natl Acad Sci USA. 1971;68:2321–2324. doi: 10.1073/pnas.68.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Litman G W. In: Relationships Between Structure and Function of Lower Vertebrate Immunoglobulins. Hildemann W H, Benedict A A, editors. New York: Plenum; 1976. pp. 217–228. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Amemiya C T, Ohta Y, Litman R T, Rast J P, Haire R N, Litman G W. Proc Natl Acad Sci USA. 1993;90:6661–6665. doi: 10.1073/pnas.90.14.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bairoch A, Bucher P, Hofmann K. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Tamura K, Jakobsen I, Nei M. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 15.Marchalonis J J. Immunity in Evolution. Cambridge, MA: Harvard Univ. Press; 1977. [Google Scholar]
- 16.Lesk A M, Chothia C. J Mol Biol. 1982;160:325–342. doi: 10.1016/0022-2836(82)90179-6. [DOI] [PubMed] [Google Scholar]
- 17.Davis A C, Roux K H, Pursey J, Shulman M J. EMBO J. 1989;8:2519–2526. doi: 10.1002/j.1460-2075.1989.tb08389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arya S, Chen F, Spycher S, Iseman D E, Shulman M J, Painter R H. J Immunol. 1994;152:1206–1212. [PubMed] [Google Scholar]
- 19.Wright J F, Shulman M J, Isenman D E, Painter R H. J Biol Chem. 1990;265:10506–10513. [PubMed] [Google Scholar]
- 20.Bengtén E, Wilson M, Miller N, Clem L W, Pilstrom L, Warr G W. Curr Top Microbiol Immunol. 2000;248:189–220. doi: 10.1007/978-3-642-59674-2_9. [DOI] [PubMed] [Google Scholar]
- 21.Greenberg A S, Avila D, Hughes M, Hughes A, McKinney E C, Flajnik M F. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 22.Fellah J S, Kerfourn F, Wiles M V, Schwager J, Charlemagne J. Immunogenetics. 1993;38:311–317. doi: 10.1007/BF00210471. [DOI] [PubMed] [Google Scholar]
- 23.Harding F A, Amemiya C T, Litman R T, Cohen N, Litman G W. Nucleic Acids Res. 1990;18:6369–6376. doi: 10.1093/nar/18.21.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson M, Amemiya C, Luer C, Litman R, Rast J, Niimura Y, Litman G. Int Immunol. 1994;6:1661–1670. doi: 10.1093/intimm/6.11.1661. [DOI] [PubMed] [Google Scholar]
- 25.Greenberg A S, Hughes A L, Guo J, Avila D, McKinney E C, Flajnik M F. Eur J Immunol. 1996;26:1123–1129. doi: 10.1002/eji.1830260525. [DOI] [PubMed] [Google Scholar]
- 26.Anderson M K, Strong S J, Litman R T, Luer C A, Amemiya C T, Rast J P, Litman G W. Immunogenetics. 1999;49:56–67. doi: 10.1007/s002510050463. [DOI] [PubMed] [Google Scholar]
- 27.Pedersen R A. J Exp Zool. 1971;177:65–79. doi: 10.1002/jez.1401770108. [DOI] [PubMed] [Google Scholar]
- 28.Wilson M, Bengtën E, Miller N W, Clem L W, Du Pasquier L, Warr G W. Proc Natl Acad Sci USA. 1997;94:4593–4597. doi: 10.1073/pnas.94.9.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chothia C, Novotny J, Bruccoleri R, Karplus M. J Mol Biol. 1985;186:651–663. doi: 10.1016/0022-2836(85)90137-8. [DOI] [PubMed] [Google Scholar]
- 30.Lascombe M B, Alzari P M, Poliak R J, Nisonoff A. Proc Natl Acad Sci USA. 1992;89:9429–9433. doi: 10.1073/pnas.89.20.9429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rast J P, Anderson M K, Ota T, Litman R T, Margittai M, Shamblott M J, Litman G W. Immunogenetics. 1994;40:83–99. doi: 10.1007/BF00188170. [DOI] [PubMed] [Google Scholar]
- 32.Shen S X, Berstein R M, Schluter S F, Marchalonis J J. Immunol Cell Biol. 1996;74:357–364. doi: 10.1038/icb.1996.63. [DOI] [PubMed] [Google Scholar]
- 33.Ota T, Sitnikova T, Nei M. Curr Top Micorbiol Immunol. 2000;248:221–245. doi: 10.1007/978-3-642-59674-2_10. [DOI] [PubMed] [Google Scholar]
- 34.Ross D A, Wilson M R, Miller N W, Clem L W, Warr G W. Immunol Rev. 1998;166:143–151. doi: 10.1111/j.1600-065x.1998.tb01259.x. [DOI] [PubMed] [Google Scholar]
- 35.Wilson M R, Ross D A, Miller N W, Clem L W, Middleton D L, Warr G W. Dev Comp Immunol. 1995;19:165–177. doi: 10.1016/0145-305x(94)00064-m. [DOI] [PubMed] [Google Scholar]
- 36.Ota T, Nguyen T-A, Huang E, Detrich H W, Amemiya C T. J Exp Zool B. 2003;295:45–58. doi: 10.1002/jez.b.4. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein R M, Schluter S, Shen S, Marchalonis J J. Proc Natl Acad Sci USA. 1996;93:3289–3293. doi: 10.1073/pnas.93.8.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]