Abstract
Gene duplication and domain accretion are thought to be the major mechanisms for the emergence of novel genes during evolution. Such events are thought to have occurred at early stages in the vertebrate lineage, but genomic sequencing has recently revealed extensive amplification events during the evolution of higher primates. We report here that the Tre2 (USP6) oncogene is derived from the chimeric fusion of two genes, USP32 (NY-REN-60), and TBC1D3. USP32 is an ancient, highly conserved gene, whereas TBC1D3 is derived from a recent segmental duplication, which is absent in most other mammals and shows rapid amplification and dispersal through the primate lineage. Remarkably, the chimeric gene Tre2 exists only in the hominoid lineage of primates. This hominoid-specific oncogene arose as recently as 21–33 million years ago, after proliferation of the TBC1D3 segmental duplication in the primate lineage. In contrast to the broad expression pattern of USP32 and TBC1D3, expression of Tre2 is testis-specific, a pattern proposed for novel genes implicated in the emergence of reproductive barriers. The sudden emergence of chimeric proteins, such as that encoded by Tre2, may have contributed to hominoid speciation.
Completion of the sequence for the human genome has revealed that ≈5% of the genome is composed of segmental duplications (1–4). These duplications appear to have emerged only during the last 35 million years, within the primate lineage, and to have rapidly expanded (3). The unexpected finding of such large-scale genomic rearrangements late in primate evolution has raised the possibility that new genes may have been created on a scale that was not previously appreciated. Many of the duplicated regions in the human genome share extremely high sequence identity because of their recent divergence, and their shuffling within the genome is postulated to have played a role in the formation of novel genes through chimeric fusion and domain accretion (3). Whereas duplicated genes will diverge over time, the abrupt creation of a mosaic gene with novel functions, especially for genes involved in reproduction, could potentially lead to reproductive barriers and thus play an important role in speciation (5).
Tre2 was originally isolated in multiple transfection-based screens for novel oncogenes, by virtue of its potent ability to transform mouse NIH 3T3 cells (6, 7). It was subsequently shown to function as a ubiquitin-specific protease (USP) in vitro, although specific target proteins whose turnover may be modulated by Tre2 have not been identified (8). In characterizing a protein family member related to Tre2 that is required for meiotic division in Caenorhabditis elegans (ZK328.1), we discovered that the Tre2 gene itself is completely absent from mouse cells, as well as from most other mammals. We therefore undertook to define the mechanism and timing of its emergence during primate evolution. Using sequence analysis and Southern blotting, we found that Tre2 resulted from the chimeric fusion of two genes, USP32 (NY-REN-60) and TBC1D3. TBC1D3 itself is derived from a segmental duplication, with multiple copies present in the human genome. In addition, the TBC1D3 segmental duplication is absent from the mouse and most other mammalian genomes, and the timing of its amplification is consistent with the divergence of primates. Phylogenetic analysis indicates that the chimeric Tre2 itself emerged subsequently as a hominoid-specific gene. Of note, expression of this novel chimeric gene is restricted to the testis, an observation consistent with proposed models of speciation.
Materials and Methods
Analysis of Genomic DNA Sequence and Functional Domains.
A full-length clone of USP32 (GenBank accession no. AF533230) was isolated from a brain cDNA library (OriGene Technologies, Rockville, MD). Genomic bacterial artificial chromosome (BAC) (accession no. AC090167.3) was used in the analysis of USP32. The full coding sequence of Tre2 was determined (accession no. AY143550) and compared with the originally reported mRNA sequence (accession nos. X63547 and X63546), and also with sequences (accession no. XM_165948) from the National Center for Biotechnology Information (NCBI) Annotation Project. Alternatively spliced transcripts found in the original report (6) were resolved by RT-PCR. Human genomic BAC sequence (accession no. AC012146.13) was also used in the genomic analysis of Tre2. The genomic BAC (accession no. AC067923) was used in the analysis of TBC1D3 (accession no. AL136860). Genomic sequences were masked by repeatmasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html) and were compared by BLAST_2 sequences (9) and percent identity plots (10). An analysis of mouse sequences was performed at publicly available databases [NCBI (www.ncbi.nlm.nih.gov/genome/guide/mouse/) and Ensembl (www.ensembl.org/Mus_musculus/)]. Protein domains were determined by query with the Pfam protein family database (11). For RT-PCR analysis, a panel of cDNAs from human tissues (CLONTECH) was amplified by using the following primer pairs: USP32F CCACATATGGCTTTTCATGGACTCG and USP32R GCACCTTTAAAGCGGGTATTAGCC; TBC1D3F GCACGTTTTTGCAACCGGTTCGTTGATACC and TBC1D3R GCTGTTCGTCCCTAGCTCTGAAGGGGGTGC; and Tre2F (USP32F) CCACATATGGCTTTTCATGGACTCG and Tre2R AGGGCCTCTACGAAGAAACTAACAAGGAAGC. PCR products amplified from testis cDNA were sequenced for confirmation. Both Tre2 and USP32 RT-PCR products span the juncture region. For Southern “zoo” blot analysis, DNA was isolated from the following cell lines: human, 293T; African green monkey, COS; mouse, 3T3; rat, R-stem; rabbit, RK-1 and SIRC; dog, MDCK; and chicken, DT-40. For primate blots, DNA was obtained from Coriell Cell Repositories (Camden, NJ; primate panel: phylogenetic). DNA was digested with EcoRI (SIRC DNA, HindIII), electrophoresed on 0.8% Tris acetate EDTA (TAE) agarose gels, transferred overnight to Hybond-XL membranes (Amersham Pharmacia), and UV crosslinked. Membranes were incubated with low-stringency hybridization solution (30% formamide/1 M NaCl/0.5% SDS/50 mM Tris, pH 7.5/1× Denhardt's solution/125 μg/ml salmon sperm DNA). Probes were generated by RT-PCR of human testis cDNA. Primer pairs used were as follows: Tre2-A, forward GCTAGCGCCACCATGGACATGGTAGAGAATGCAGATAGTTTGC and reverse CCTCTCCGCAGTGTTCAGCCTGCCAGCAGGTGGC; and Tre2-B, forward CACCTGCTGGCAGGCTGAACACTGCGGAGAGG and reverse CCAAGCTGTCTAGCAGCCAGAGTGGTAGC. Membranes were incubated with 2 × 106 cpm/ml radioactive probe [α-32P]dCTP (NEN Life Sciences) overnight at 42°C. Membranes were washed once with 2× SSC/0.1% SDS for 45 min at room temperature and twice at 55°C for 45 min. Washed membranes were then exposed to autoradiograph film for 1–3 days.
Phylogenetic Tree Analysis.
For PCR and sequencing analysis of USP32, 100 ng of primate genomic DNA was amplified by using the following primer pair: USP32 G-forward TGGGAACTGGAACAAACAATATGAGAACC and USP32 G-reverse GTTTCTGAACTTTAATTACTTCTGTAGATGG. PCR annealing conditions for lane 9, black-handed spider monkey, and lane 10, wooly monkey, were lowered to increase yield. For amplification of Tre2 the following primer pair was used: Tre2 G-forward (same as USP32 G-forward) and Tre2 G-reverse (CTGCAGATGGTCCAGTAAACACACACCTGG). Sequencing analysis was performed by using three independent clones for each amplified fragment. For phylogenetic analysis, DNA sequences were aligned with clustalx (12) and tree building was performed with PAUP* 4.0b1 (13). Maximum likelihood analysis was used for phylogenetic reconstruction assuming a transition/transversion ratio of 2 and empirical base frequencies. Gaps were treated as missing data. Maximum parsimony was also used for tree reconstruction assuming a transition/transversion ratio of 2, and gaps were treated as missing data. Three shortest trees were produced by maximum parsimony analysis, with the consensus tree matching the topology of the tree produced by maximum likelihood. DNA sequences used for phylogenetic analysis are available (GenBank accession nos. AY163314–AY163328).
Results
Chimeric Origin of Tre2 from USP32 and TBC1D3.
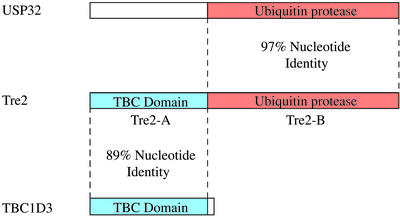
In analyzing the sequence of the Tre2 oncogene, we observed an extremely high degree of nucleotide similarity (97% identity) with the USP32 gene, previously isolated as a tumor-specific antigen in renal cell carcinomas (14). Whereas this high degree of similarity suggests that the two genes have only recently diverged, it is remarkable in that the region of similarity spans only nucleotides 3194–6063 of the Tre2 mRNA and does not extend over the entire transcript (Fig. 1). Instead, nucleotides 1–3193 of Tre2 show strong sequence similarity (89% identity) to another gene, TBC1D3, encoding a TBC domain that functions in Rab GTPase signaling (15). Tre2 is therefore likely to have resulted from the duplication and subsequent fusion of the USP32 and TBC1D3 genes. Analysis of the genomic structure of Tre2 confirmed its chimeric origin from USP32 and TBC1D3. blast analysis and percent identity plots (10) using genomic BAC sequences demonstrate extensive homology between TBC1D3 and Tre2, beginning 3.6 kb upstream of the transcriptional start site and extending into intron 14 (total overlap ≈16 kb; see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Conversely, the last 15 exons of Tre2, spanning 36 kb of genomic sequence (from intron 14 and extending through the 3′ UTR), are shared with USP32, with identical intron–exon structure. Thus, Tre2 exons 1–14 are derived from the TBC1D3 parental gene, whereas exons 15–30 are derived from the USP32 gene.
Figure 1.
Schematic representation of the chimeric Tre2 coding sequence, showing domains derived from the USP32 and TBC1D3 parental genes. These regions of overlap are conserved in the genomic structure. The full-length Tre2, USP32, and TBC1D3 genes encode proteins of 1,406, 1,604, and 549 aa, respectively.
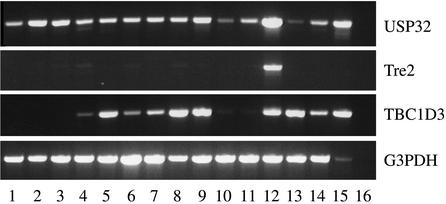
Analysis of the protein domains encoded by USP32 and TBC1D3 reveals that Tre2 acquired separate functional domains from each parental gene, a process known as domain accretion (ref. 3; Fig. 1). Amino acids 1–496 of Tre2, derived from TBC1D3, encompass a TBC domain, shared by proteins implicated in Rab GTPase signaling and vesicle trafficking (16). Amino acids 501–1406 are derived from USP32 and encode a USP. The chimeric fusion omits two calcium-binding EF-hand domains and a myristoylation site, which are present in USP32. Whereas USP32 and TBC1D3 are expressed in a broad range of human tissues, Tre2 is most highly expressed in testis (Fig. 2). Taken together, these observations suggest that Tre2 may encode a novel gene with predominant expression in testis, regulating turnover of proteins involved in vesicle trafficking.
Figure 2.
Human tissue expression pattern of Tre2, USP32, and TBC1D3. cDNA panels (CLONTECH) were screened by semiquantitative RT-PCR and are as follows: 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; 8, pancreas; 9, spleen; 10, thymus; 11, prostate; 12, testis; 13, ovary; 14, small intestine; 15, peripheral blood leukocyte; and 16, negative control. Specific RT-PCR products for Tre2, USP32, and TBC1D3 are designed by amplification across the unique juncture for the respective transcripts. Glyceraldehyde-3-phosphate dehydrogenase (G3PDH) amplification was used as a control.
Emergence of TBC1D3 in the Primate Lineage.
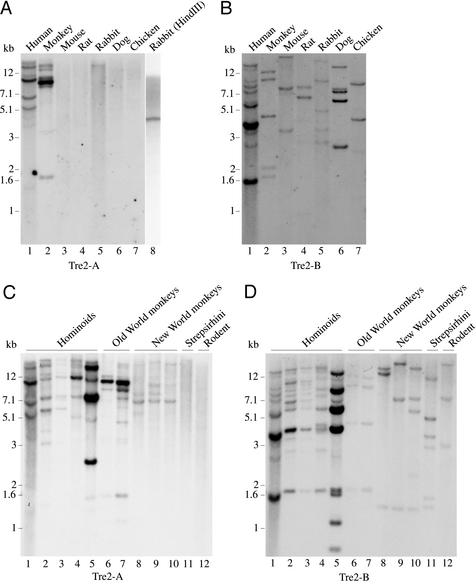
USP32 encodes an ancient and unique gene, with highly conserved orthologs readily apparent in all of the completed metazoan genomes, including C. elegans (ZK328.1), Drosophila melanogaster (CG8334), and mouse (LOC194927). Surprisingly, no significant matches are found in publicly available mouse genomic databases for either TBC1D3 or its homologous region of Tre2. Southern zoo blotting confirms the absence of hybridization to the 5′ (TBC) domain of Tre2 (Tre2-A) in mouse, rat, dog, and chicken, whereas hybridization to the 3′ (protease) domain (Tre2-B) is readily apparent (Fig. 3). These data suggest that the ubiquitin protease domain present in Tre2 and USP32 is ancient and highly conserved, whereas the TBC domain shared with TBC1D3 arose within the primate lineage. Of note, faint but consistent bands are detectable in rabbit (Fig. 3A, lanes 5 and 8), an observation that is of interest, given the controversy as to the placement of the rabbit order, Lagomorpha, relative to primates and rodents (17). Within a representative phylogenetic panel of primate DNA samples (Fig. 3C), hybridization to the Tre2 TBC domain (Tre2-A) is observed in New World monkeys, Old World monkeys, and hominoids, but no signal is detectable in lemur (Strepsirhini), the primate most divergent from human tested here (Fig. 3C, lane 11). The TBC domain shared by Tre2 and TBC1D3, therefore, arose after the initial primate radiation in the common ancestor of the anthropoid primates.
Figure 3.
Hybridization of Tre2 domains to DNA from diverse species. (A) Probe derived from human Tre2 (Tre2-A) (nucleotides 1697–3193) containing the TBC domain homologous to TBC1D3. Rabbit DNA in lane 8 was obtained from a different cell line than the DNA in lane 5. (B) Probe derived from human Tre2 (Tre2-B) (nucleotides 3163–5947) containing the ubiquitin protease domain homologous to USP32. Hybridization of Tre2-A probe (C) and Tre2-B probe (D) using a panel of primate DNA samples. Lanes and designations are as follows: 1, human; 2, chimpanzee (Pan troglodyte); 3, bonobo (Pan paniscus); 4, lowland gorilla (Gorilla gorilla); 5, orangutan (Pongo pygmaeus; overloaded); 6, rhesus monkey (Macaca mulatta); 7, pigtailed macaque (Macaca nemestrina); 8, red-bellied tamarin (Saguinus labiatus); 9, black-handed spider monkey (Ateles geoffroyi); 10, wooly monkey (Lagothrix lagotricha); 11, ring-tailed lemur (Lemur catta); and 12, mouse (Mus musculus; negative control).
In addition to its emergence in primates, the TBC domain shows increasing complexity in its hybridization pattern to New World monkeys (Fig. 3C, lanes 8–10), Old World monkeys (Fig. 3C, lanes 6 and 7), and hominoids (Fig. 3C, lanes 1–5), suggesting multiple duplications and rapid expansion throughout the anthropoid primates. In humans, blast analysis indicates multiple loci mapping to >10 independent contigs, with ≈88–95% identity to the Tre2 coding sequence (data not shown). Most of these segmental duplications are located on chromosome 17, which is also the location of Tre2 (17p13), USP32 (17q22), and TBC1D3 (17q12). Despite this large-scale duplication, searches of EST databases suggest that only Tre2 and TBC1D3 encode transcribed genes (data not shown). These results are consistent with recent findings that ≈5% of the human genome appears to be composed of segmental duplications (duplicons), many of which are chromosome-specific and have expanded through the primate lineage (3, 4). The sudden origin of the Tre2-TBC1D3 TBC segmental duplication itself is remarkable. Whereas TBC domains are found in many other proteins, their low homology to Tre2 and TBC1D3 suggests a distant relationship (18). USP32 itself shows partial 3′ duplications, although none of them appear to encode functional protein domains (data not shown).
The presence of the duplicon in anthropoid primates and in rabbit, but its absence from lemur, mouse, rat, and dog, suggests several molecular evolutionary scenarios: (i) independent and convergent evolution of the duplicon in anthropoid primates and in lagomorphs; (ii) duplicon origination in the common ancestor of primates and lagomorphs followed by duplicon loss in the strepsirhine primates; (iii) duplicon origination in the mammalian common ancestor followed by duplicon loss in all mammalian lineages except anthropoid primates and lagomorphs; or (iv) duplicon origination in the anthropoid primate common ancestor with horizontal transfer to lagomorphs (or vice versa). Resolving the different possibilities may shed light on the evolutionary distance between hominoids and lagomorphs (17).
Tre2 as a Hominoid-Specific Gene.
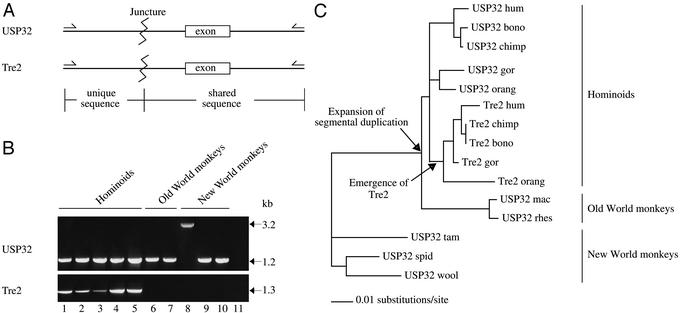
Few genes have been reported to be novel in humans or in hominoids and not present in other primates (3, 19). We sought to determine more precisely the timing of Tre2 emergence during primate evolution by testing for the presence of the fused gene using a PCR-based strategy to amplify the juncture between the USP32- and TBC1D3-derived sequences in the chimeric Tre2 gene (Fig. 4A). Amplification of unique Tre2 DNA sequences, using multiple sets of PCR primers, is restricted to hominoid samples (Fig. 4B). In contrast, the corresponding unique sequence of USP32 is readily identified in New and Old World monkeys, as well as in hominoids. We used the nucleotide sequence of the PCR-generated DNA fragments to trace the origin of the overlapping region (≈940 nucleotides) shared by USP32 and Tre2. The resulting phylogenetic tree indicates that all of the Tre2 sequences are monophyletic, having originated from a single duplication event of USP32 some time after hominoids diverged from Old World monkeys (Fig. 4C). We estimate the formation of Tre2 to have occurred between 21 and 33 million years ago (20, 21).
Figure 4.
(A) Schematic representation of PCR fragments amplified for specific analysis of the Tre2 and USP32 sequences. Homologous exons were amplified in the PCRs (Tre2, exon 15 and USP32, exon 19). (B) PCR amplification of genomic DNA used to test for the presence of junctures specific to Tre2 and USP32. Lanes and designations are as follows: 1, human; 2, chimpanzee; 3, bonobo; 4, lowland gorilla; 5, orangutan; 6, rhesus monkey; 7, pigtailed macaque; 8, red-bellied tamarin; 9, black-handed spider monkey; 10, wooly monkey; 11, no template control. The 3.2-kb band seen in lane 8 occurs because of an insertion of a nonfunctional pseudogene within the intronic sequence of USP32. (C) Phylogenetic (maximum likelihood) tree of Tre2 and USP32 shared DNA sequences. Analysis was performed on ≈940 bp. Analysis included intron and exon sequences. The tree was rooted with New World monkeys as an outgroup. Arrows indicate the formation of the novel gene Tre2 in hominoids and the expansion of the segmental duplication. hum, human; bono, bonobo; chimp, chimpanzee; gor, lowland gorilla; orang, orangutan; mac, pigtailed macaque; rhes, rhesus monkey; tam, red-bellied tamarin; spid, black-handed spider monkey; wool, wooly monkey.
Discussion
Our findings have potential implications for understanding genetic differences between humans and other primates. Whereas most studies have focused on the accumulation of genetic variation within the coding and regulatory regions of conserved genes, the emergence of novel genes this late in evolutionary history has not been appreciated. Quantifying the contribution of such new genes to the generation of human-specific features will require comprehensive comparison of primate genomes. However, the documented amplification of multiple domain-specific sequences within the primate lineage indicates that domain accretion and gene-fusion events may not be uncommon (3, 4, 22–25). In this regard, it is of particular interest that the chimeric Tre2 gene is specifically expressed in testis, where the sudden emergence of a novel mosaic gene could potentially lead to reproductive barriers and thus play a role in speciation (5, 26, 27). The emergence of the Tre2 chimeric gene during primate evolution may also be analogous to genetic events leading to the generation of novel chromosomal fusion products during tumorigenesis. Tre2 is unique among oncogenes in that it encodes a normal human gene capable of transforming rodent cells, which themselves lack the gene. The ubiquitin protease domain encoded by Tre2 is enzymatically active in vitro (8), and the C. elegans USP32 ortholog (ZK328.1) is essential for early embryonic cell divisions (28). The fusion of this conserved enzymatic domain with the novel TBC domain presumably results in altered regulation of critical cellular constituents, leading to neoplastic transformation. Thus, the origin of Tre2 points to recent evolutionary events through which recombination of protein domains has resulted in the generation of novel genes that regulate cell proliferation and may contribute to speciation.
Supplementary Material
Acknowledgments
We thank Dr. Iswar Hariharan and Dr. Vijay Yajnik for helpful comments. This work was supported in part by National Cancer Institute Grant 84066 and a National Foundation for Cancer Research–American Association for Cancer Research Professorship to D.A.H., and by Harvard University (M.R.).
Abbreviation
- USP
ubiquitin-specific protease
Footnotes
References
- 1.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Eichler E E. Trends Genet. 2001;17:661–669. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- 4.Samonte R V, Eichler E E. Nat Rev Genet. 2002;3:65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- 5.Swanson W J, Vacquier V D. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura T, Hillova J, Mariage-Samson R, Onno M, Huebner K, Cannizzaro L A, Boghosian-Sell L, Croce C M, Hill M. Oncogene. 1992;7:733–741. [PubMed] [Google Scholar]
- 7.Janssen J W, Braunger J, Ballas K, Faust M, Siebers U, Steenvoorden A C, Bartram C R. Int J Cancer. 1999;80:857–862. doi: 10.1002/(sici)1097-0215(19990315)80:6<857::aid-ijc10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 8.Papa F R, Hochstrasser M. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 9.Tatusova T A, Madden T L. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz S, Zhang Z, Frazer K A, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy S R, Griffiths-Jones S, Howe K L, Marshall M, Sonnhammer E L. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swofford D L. paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2002. , Version 4.0b1. [Google Scholar]
- 14.Scanlan M J, Gordan J D, Williamson B, Stockert E, Bander N H, Jongeneel V, Gure A O, Jager D, Jager E, Knuth A, et al. Int J Cancer. 1999;83:456–464. doi: 10.1002/(sici)1097-0215(19991112)83:4<456::aid-ijc4>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 15.Wiemann S, Weil B, Wellenreuther R, Gassenhuber J, Glassl S, Ansorge W, Bocher M, Blocker H, Bauersachs S, Blum H, et al. Genome Res. 2001;11:422–435. doi: 10.1101/gr.154701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neuwald A F. Trends Biochem Sci. 1997;22:243–244. doi: 10.1016/s0968-0004(97)01073-6. [DOI] [PubMed] [Google Scholar]
- 17.Graur D, Duret L, Gouy M. Nature. 1996;379:333–335. doi: 10.1038/379333a0. [DOI] [PubMed] [Google Scholar]
- 18.Matoskova B, Wong W T, Seki N, Nagase T, Nomura N, Robbins K C, Di Fiore P P. Oncogene. 1996;12:2563–2571. [PubMed] [Google Scholar]
- 19.Courseaux A, Nahon J L. Science. 2001;291:1293–1297. doi: 10.1126/science.1057284. [DOI] [PubMed] [Google Scholar]
- 20.Gebo D L, MacLatchy L, Kityo R, Deino A, Kingston J, Pilbeam D. Science. 1997;276:401–404. doi: 10.1126/science.276.5311.401. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen D. In: The Primate Fossil Record. Hartwig W, editor. Cambridge, U.K.: Cambridge Univ. Press; 2002. pp. 203–220. [Google Scholar]
- 22.Johnson M E, Viggiano L, Bailey J A, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler E E. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 23.Bailey J A, Yavor A M, Viggiano L, Misceo D, Horvath J E, Archidiacono N, Schwartz S, Rocchi M, Eichler E E. Am J Hum Genet. 2002;70:83–100. doi: 10.1086/338458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eichler E E, Johnson M E, Alkan C, Tuzun E, Sahinalp C, Misceo D, Archidiacono N, Rocchi M. J Hered. 2001;92:462–468. doi: 10.1093/jhered/92.6.462. [DOI] [PubMed] [Google Scholar]
- 25.Pujana M A, Nadal M, Guitart M, Armengol L, Gratacos M, Estivill X. Eur J Hum Genet. 2002;10:26–35. doi: 10.1038/sj.ejhg.5200760. [DOI] [PubMed] [Google Scholar]
- 26.Singh R S, Kulathinal R J. Genes Genet Syst. 2000;75:119–130. doi: 10.1266/ggs.75.119. [DOI] [PubMed] [Google Scholar]
- 27.Wyckoff G J, Wang W, Wu C I. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- 28.Gonczy P, Echeverri C, Oegema K, Coulson A, Jones S J, Copley R R, Duperon J, Oegema J, Brehm M, Cassin E, et al. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.