Abstract
Niemann–Pick disease type C2 (NP-C2) is a fatal hereditary disease characterized by accumulation of low-density lipoprotein-derived cholesterol in lysosomes. Here we report the 1.7-Å resolution crystal structure of the cholesterol-binding protein deficient in this disease, NPC2, and the characterization of its ligand binding properties. Human NPC2 binds the cholesterol analog dehydroergosterol with submicromolar affinity at both acidic and neutral pH. NPC2 has an Ig-like fold stabilized by three disulfide bonds. The structure of the bovine protein reveals a loosely packed region penetrating from the surface into the hydrophobic core that forms adjacent small cavities with a total volume of ≈160 Å3. We propose that this region represents the incipient cholesterol-binding site that dilates to accommodate an ≈740-Å3 cholesterol molecule.
Cholesterol is an essential molecule that can be delivered to cells by the low-density lipoprotein (LDL) pathway (1). Here, after LDL receptor-mediated endocytosis, LDL particles are transported to lysosomes where cholesterol esters are hydrolyzed by acid lipase. The liberated free cholesterol is somehow transported out of the lysosome to other cellular sites where it regulates de novo cholesterol biosynthesis and LDL uptake and also is used for synthesis of membranes, lipid droplets, and sterol derivatives.
Although many aspects of this pathway have been well established, the mechanism underlying egress of cholesterol from the lysosome remains enigmatic. Studies of Niemann–Pick type C disease (NP-C), a fatal recessive hereditary lysosomal storage disorder consisting of two complementation groups with similar phenotypes, have provided some insights into this process (2). NP-C fibroblasts from both complementation groups are characterized by lysosomal accumulation of LDL-derived cholesterol. The gene defective in the major NP-C complementation group, NPC1, was identified by linkage analysis (3) and encodes an ≈1,278-aa transmembrane protein containing 13 transmembrane regions and a putative sterol-sensing domain. The NPC1 protein has sequence similarity to bacterial permeases and the receptor protein Patched that transduces Hedgehog signals (4). Recently, NPC1 has been reported to transport fatty acids but not cholesterol across membranes (5). The gene defective in the less common NP-C complementation group, NPC2, recently was identified in the course of a proteomic study to characterize the repertoire of soluble lysosomal proteins (6). The ≈130-aa NPC2 protein was previously characterized as a major secretory protein present in mammalian epididymis (7) and shown to function as a cholesterol transfer protein.‖ Porcine NPC2 was shown to bind cholesterol with 1:1 stoichiometry and micromolar affinity (8).
Potential physiological functions of NPC2 include preventing inappropriate sterol intercalation into the lysosomal membrane, presentation of sterol to a membrane-bound transporter such as NPC1, or some other, as yet unsuspected, regulatory or enzymatic activity. We have investigated the functional and structural properties of the NPC2 protein to gain insight into its role in cholesterol transport. Here we report that human NPC2 binds the cholesterol analog dehydroergosterol [DHE, ergosta-5,7,9(11),22-tetraen-3β-ol] with submicromolar affinity at both neutral and acidic pH. We have determined the high-resolution crystal structure of bovine apoNPC2. The structure revealed a loosely packed region in the protein interior that appears to be an incipient cholesterol-binding site. The proposed location of the sterol-binding site is further supported by mutation studies reported by Ko et al. (9).
Materials and Methods
Ligand Binding Assay.
Production, purification, and characterization of recombinant human NPC2 (hNPC2) protein produced from transfected Chinese hamster ovary cells (6) will be described in detail elsewhere. Briefly, conditioned medium was depleted of unwanted proteins by absorption to DE52 (Whatman) and SP-Sephadex C-25 (Amersham Pharmacia) at neutral pH (100 mM NaCl, 20 mM Tris, pH 7). Different glycoforms of hNPC2 were resolved by chromatography on Mono-S (Bio-Rad) by using a gradient of 25–500 mM NaCl in 25 mM ammonium acetate, pH 4.5. The late-eluting monoglycosylated peak was homogeneous when analyzed by SDS/PAGE visualized by silver stain. DHE (Sigma) was dissolved in methanol, and its concentration was determined from its UV absorption at 324 nm (ɛ = 1.3 × 104 M−1⋅cm−1).
For binding measurements, hNPC2 and DHE were diluted into different buffers and incubated for 30 min at 25°C in the dark. The final concentration of methanol in the incubation mixtures was <1%. Fluorescence spectra were recorded by using a SLM8000C spectrofluorometer equipped with an upgraded computer interface (Jobin Yvon, Edison, NJ). Samples (200 μl) in quartz microcells were excited at 338 nm with a 0.5-nm bandpass slit. Control experiments indicated no appreciable photobleaching within the experimental period. Emission spectra were obtained from 340 to 450 nm with a 4-nm bandpass slit.
Bovine NPC2 (bNPC2) Purification.
The first part of the purification protocol was adapted from Larsen et al. (10) with a minor modification. The pH of the ammonium acetate solution used for DEAE Sepharose chromatography (Amersham Pharmacia) was changed to 7.0. After this initial purification procedure, the protein still contained some low molecular weight contaminants visible on Coomassie blue-stained SDS polyacrylamide gels, which, by sequence analysis, represented fragments of β-casein. For further purification, the protein, in 10 mM Tris-acetate, pH 9, was applied to a 10-ml HiTrap Q HP column (Amersham Pharmacia) equilibrated with the same buffer and was eluted with a gradient of 0.01–1.0 M NaCl in 10 column volumes. The sample was then dialyzed into 50 mM sodium citrate, pH 5.5 and incubated overnight at 25°C with 10 μg/ml EndoHf glycosidase (New England Biolabs). The deglycosylated protein was separated from EndoHf by gel filtration on a 24-ml Superdex 75 column (Amersham Pharmacia) equilibrated in 10 mM Mes, 0.1 M NaCl, pH 5.7, concentrated, and used for crystallization trials. The purification procedure yielded 6–8 mg of purified deglycosylated protein from 8 liters of raw milk.
Crystallization, Data Collection, and Processing.
The protein was concentrated to 15 mg/ml. Crystals of bNPC2 were grown by the hanging drop vapor diffusion method. Initial crystallization screening was done with crystal screens (Hampton Research, Riverside, CA). Concentrated protein was mixed with reservoir solution (1 μl each) and equilibrated against 0.5 ml of reservoir solution at room temperature. Three different conditions produced small well-formed crystals of cubic morphology. The optimized crystallization condition that produced well-diffracting crystals was 1.95 M ammonium phosphate, 100 mM Tris, pH 7.5. The crystals appeared in 1 week and continued to grow over another week, reaching a size of ≈200 μm per edge.
The native and derivative datasets were collected at beamline X4A at the National Synchrotron Light Source at Brookhaven National Laboratory, Upton, NY. Native crystals were cryoprotected with 1.6 M ammonium phosphate, 100 mM Tris (pH 7.5), and 15% glycerol and frozen in a 100-K nitrogen cryostream. A complete native dataset at 1.7-Å resolution was collected from a single crystal. The data were processed with denzo and scaled with scalepack (11). The space group was determined to be I23 with unit cell dimensions of a = b = c = 100.1 Å. There was one monomer in the asymmetric unit with a solvent content of 51% (Vm = 2.5).
A heavy atom derivative was obtained by soaking the crystal for 3 days in 1.6 M ammonium phosphate, 100 mM Tris (pH 7.5), 15% glycerol, and 50 mM PtCl4. No additional cryoprotection was necessary before flash-freezing the crystals. A complete multiple anomalous dispersion dataset was collected on a single crystal to 1.8-Å resolution, but only single wavelength data (λ = 1.06287 Å) were used for structure determination by single anomalous dispersion.
The heavy atom positions were found and refined by using cns (12). Initial single anomalous dispersion phases were improved by using solvent flipping methods implemented in cns. The chain was traced automatically by using arp/warp (13), which placed 120 of 130 residues. This initial model was refined against the native dataset to 1.7-Å resolution. Five percent of the data were set aside for use during the refinement. The model was completed by manually positioning the remaining 10 amino acid residues by using the program o (14). Several cycles of positional and B factor refinement were carried out followed by visual inspection and manual adjustment of the model. Water molecules were added manually by using o. The GlcNAc molecule was added, covalently bound to Asn-39. The final model consists of all 130 amino acid residues and sugar, 102 waters, and a phosphate. In the final model, 89% of the residues are located in the most favored regions of the Ramachandran plot and 11% occur in additionally allowed regions.
Results and Discussion
Sterol Binding to NPC2.
To examine the ligand binding properties of NPC2, we developed a binding assay using DHE, a naturally occurring sterol that is structurally similar to cholesterol. Studies with other proteins indicated that DHE is quenched in the aqueous (unbound) phase and exhibits increased fluorescence in a more hydrophobic (protein-bound) environment (15). This also occurs when DHE binds to hNPC2 (Fig. 1A). The absorption spectrum of the DHE–hNPC2 complex obtained after gel filtration to remove free DHE, combined with the extinction coefficients at 280 and 324 nm of free DHE and hNPC2, suggested a stoichiometry of 1:1 (data not shown), consistent with that obtained with radiolabeled cholesterol (8).
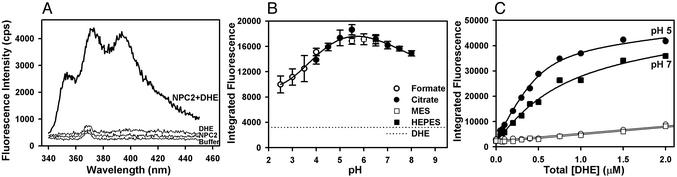
Figure 1.
Interaction of NPC2 with the fluorescent cholesterol analog DHE. (A) Fluorescence spectra. Samples contained buffer (20 mM citrate/150 mM NaCl, pH 5.0) with or without 10 μM hNPC2 and/or 1 μM DHE. The small emission peak at 368 nm in various controls represents Raman scattering of water. (B) pH dependence of NPC2–DHE fluorescent complex formation. Fluorescence data were obtained from DHE (0.25 μM) and hNPC2 (0.25 μM) in 150 mM NaCl buffered with 20 mM of the indicated buffer. Integrated fluorescence (λ 383–404 nm) data represent mean ± SEM of triplicate determinations. Background fluorescence of DHE alone (dotted line) was independent of pH and buffer composition. (C) Binding of DHE to NPC2 at pH 5 and 7. Fluorescence data were obtained with the indicated total concentrations of DHE in the absence or presence of 0.5 μM hNPC2 (open and filled symbols, respectively). Solutions contained either 150 mM NaCl and 20 mM citrate (pH 5.0) (circles) or 150 mM NaCl and 20 mM Hepes (pH 7.0) (squares). Dissociation constants were determined from curve fitting by using a nonlinear quadratic equation and a one-site occupancy model to take into account the fluorescence of bound and free ligand (16). The data shown yielded dissociation constants of 0.092 and 0.347 μM for pH 5 and 7, respectively. The individual dissociation constants obtained from four independent experiments ranged from 0.092 to 0.260 μM for pH 5 and 0.311 to 1.09 μM for pH 7.
Given that NPC2 is a lysosomal protein, the pH dependence of ligand binding is important. Fluorescence measurements conducted on a mixture of DHE and hNPC2 at different pH values indicated a broad pH optimum, with maximum fluorescence at mildly acidic pH (Fig. 1B). To verify that this reflected binding affinity rather than pH-dependent changes in the quantum yield of bound DHE, we conducted binding experiments with a fixed concentration of hNPC2 and increasing concentrations of DHE at pH 5 and 7 (Fig. 1C). The dissociation constants obtained from these analyses were 0.19 ± 0.07 μM and 0.66 ± 0.39 μM at pH 5.0 and 7.0, respectively (mean ± standard error, four independent experiments), whereas the extrapolated specific fluorescence at saturating DHE concentrations was essentially identical at both conditions. These properties are consistent with the hypothesis that NPC2 binds cholesterol in the endolysosomal compartment.
Structure Determination.
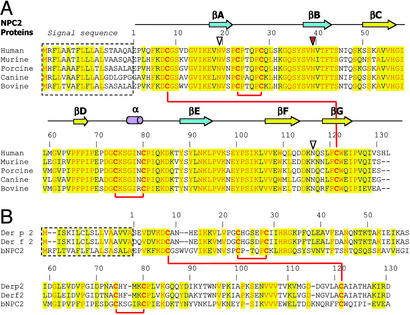
Initially, we attempted to express hNPC2 protein and its mammalian orthologs in Escherichia coli. However, this recombinant protein was misfolded as we observed formation of both intermolecular and incorrect intramolecular disulfide bonds. Subsequent attempts to refold the protein under different reducing/oxidizing conditions to obtain proper pairing of the six cysteine residues failed to yield homogeneous, correctly folded protein. Thus we sought a source of endogenous mammalian NPC2. bNPC2, originally called EPV20, is relatively abundant in milk (10) and allowed for purification of quantities sufficient for crystallographic studies. The primary sequences of NPC2 orthologs from different mammalian species are highly conserved (Fig. 2A). The sequence identity between human and bovine proteins is 80%, with another 11% being conservative substitutions.
Figure 2.
Primary sequence alignments of NPC2 orthologs and related proteins. (A) Alignment of mammalian NPC2 proteins. Numbering starts with the first residue after the signal sequence. Conserved residues are highlighted in yellow, with identical residues printed in red. Secondary structure elements of the bNPC2 structure are shown above the primary sequence. Red lines connect cysteine residues that form disulfide bonds. Three potential glycosylation sites are marked by triangles with the absolutely conserved site, which is glycosylated in bNPC2, indicated in red. (B) Alignment of bNPC2 and dust mite allergen proteins Der p 2 and Der f 2. Numbering corresponds to bNPC2 with coloring as in A.
Initial attempts to grow crystals from the purified protein were not successful. MS studies revealed that the protein was highly heterogeneous, even though it appeared as a single band when analyzed by SDS/PAGE. This heterogeneity was consistent with the previously reported glycosylation pattern of bNPC2. The bNPC2 sequence contains two possible glycosylation sites (N-X-S/T). One of these sites, Asn-39, is indeed N-glycosylated (10), with at least four different sugar types attached to it. This heterogeneity most likely prevented crystal formation in initial crystallization trials.
The protein was deglycosylated by using EndoHf enzyme. The deglycosylated protein crystallized readily and crystals, obtained after optimization of conditions, diffracted to 1.5-Å resolution. The protein crystallized in space group I23 with a single monomer in the asymmetric unit. The structure of bNPC2 was determined by single-wavelength anomalous dispersion methods and was refined to 1.7-Å resolution as described in Materials and Methods and summarized in Table 1.
Table 1.
Crystallographic data and refinement statistics
| Native data | Single anomalous dispersion data | |
|---|---|---|
| Data collection statistics | ||
| Wavelength, Å | 1.07216 | 1.06287 |
| Space group | I23 | I23 |
| Cell dimensions, Å | a = b = c = 100.1 | a = b = c = 100.4 |
| Mosaicity, ° | 0.2 | 0.3 |
| Oscillation range, ° | 1.0 | 1.0 |
| PtCl4 sites | — | 2 |
| Resolution, Å | 20–1.7 (1.76–1.70)* | 20–1.8 (1.86–1.80) |
| Total/unique reflections | 195,458/18,534 | 190,992/30,303 |
| Completeness, % | 100 (100) | 100 (100) |
| Rsym, % | 7.9 (16.9) | 7.4 (33) |
| Refinement statistics | ||
| Resolution, Å | 20–1.7 | |
| No. of water atoms | 102 | |
| No. of protein atoms | 1026 | |
| No. of heteroatoms (PO4, GlcNAc) | 19 | |
| Rwork/Rfree, % | 19.5/21.3 | |
| rms deviation bond lengths, Å | 0.01 | |
| rms deviation bond angles, ° | 1.6 | |
| Average B factor, Å2 | 12.3 |
Values in parentheses are for the reflections in the highest resolution shell.
3D Structure of bNPC2.
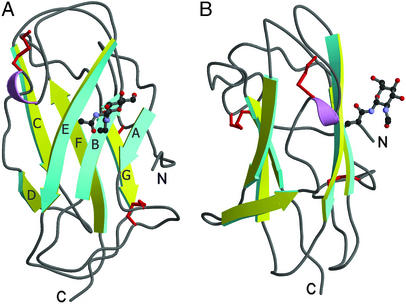
The structure of bNPC2 has an Ig-like β-sandwich fold consisting of seven β-strands arranged in two β-sheets, related by a 30° rotation (Fig. 3). The first β-sheet has three β-strands (βA, βB, and βE) and the second sheet has four β-strands (residues βC, βD, βF, and βG). The fourth and fifth β-strands are connected by a short half-turn α-helix. As expected from previous studies (10), the protein has three disulfide bonds connecting residues Cys-8–Cys-121, Cys-23–Cys-28, and Cys-74–Cys-80.
Figure 3.
Structure of bNPC2. Ribbon diagrams of bNPC2 with views related by a 90° rotation. One β-sheet is shown in yellow and the other in cyan. The side chains of cysteine residues that form three disulfide bonds are shown in red. GlcNAc that modifies Asn-39 of strand βB is shown in ball-and-stick representation. This and all subsequent figures were generated by using molscript (17) and RASTER3D (18).
There was clearly visible electron density attached to the Asn-39 residue. Because this residue is known to be modified by glycosylation, it was reasonable to suppose that the density was in fact a sugar molecule. EndoHf glycosidase cleaves high mannose and some hybrid oligosaccharides from N-linked proteins, leaving the innermost N-acetylglucosamine (GlcNAc) sugar attached. A model of GlcNAc was easily fit into the electron density. The sugar is well ordered, with temperature factors of ≈25 Å2 for all atoms.
The Potential Binding Site for Cholesterol.
Sterol-binding proteins such as elicitin (19), sterol esterase (20), cholesterol oxidase (21), and the StAR domain (22) are composed of a mixture of α-helices and β-strands. These proteins each contain a large hydrophobic cavity that is present in the absence of bound ligand. In the apo-proteins, the cavity is often covered by a “lid” and is filled with a network of water molecules. Likewise, fatty acid binding proteins contain large preformed cavities that accommodate a variety of long-chain fatty acids (23). These proteins share a “β-clam” fold consisting of two orthogonally arranged five-stranded β-sheets capped by two helices. Conformational alterations in the helical region are thought to control access of ligands to the polar-faced, water-lined internal cavity (24).
NPC2 is composed almost exclusively of β-strands. In contrast to the other previously characterized sterol-binding protein structures, the crystal structure of bNPC2 lacks any large cavity, tunnel, or surface pocket that could readily accommodate a cholesterol molecule. Despite the absence of a preformed pocket, several features implicate the hydrophobic interior of the protein as the binding site for cholesterol.
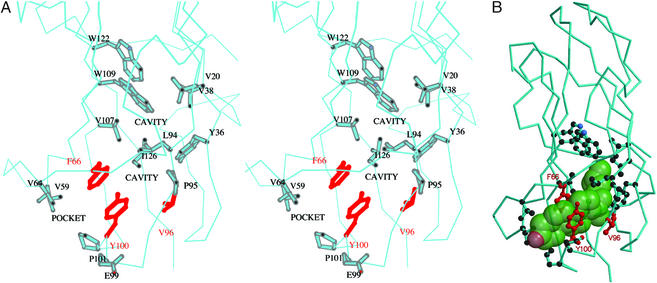
The hydrophobic core of bNPC2 is not densely packed and the structure reveals the presence of several small cavities in the “bottom” part of the protein (Fig. 4A), as located by the program cast (25). The innermost cavity is formed by the following residues: Val-20, Tyr-36, Val-38, Leu-94, Val-107, Trp-109, and Trp-122. It has a surface area of 103 Å2 and a volume of 84 Å3. The second cavity, formed by residues Phe-66, Leu-94, Pro-95, Val-96, Tyr-100, and Ile-126, is smaller, with a surface area of 59 Å2 and a volume of 38 Å3. This cavity is connected to a pocket on the surface of the protein, formed by Val-59, Val-64, Phe-66, Glu-99, Tyr-100, and Pro-101 residues, through a small opening topped by a “gate” formed by Tyr-100 and Phe-66. The surface area of the pocket is 47 Å2 and the volume is 36 Å3.
Figure 4.
The predicted cholesterol binding site of bNPC2. (A) Stereo view of cavities within the loosely packed hydrophobic interior of bNPC2. A surface pocket and two small internal cavities (labeled) are thought to comprise an incipient cholesterol-binding site. Side chains of residues that line these cavities (described in the text) are shown in stick representation. The side chains of Phe-66, Val-96, and Tyr-100 that have been found to be essential for cholesterol binding (9) are shown in red. (B) The proposed binding site for cholesterol. A semitransparent space-filling model of cholesterol (green) has been manually docked in the proposed sterol-binding site. Note that, despite the complementarity of cholesterol to the length and shape of the cavity, the ligand cannot fit because of steric clashes with side chains that line the pocket. Presumably, the cavity must dilate to accommodate cholesterol. Side chains and coloring scheme are as indicated in A.
The combined volume of the three cavities of 158 Å3 is too small to accommodate a cholesterol molecule, which occupies 741 Å3. On the other hand, the overall shape of the cavities matches very well the shape of a space-filling model of cholesterol molecule (Fig. 4B). The first barrier for cholesterol binding is a gate formed by Tyr-100 and Phe-66. We speculate that, for cholesterol to bind, the gate would have to open up and the two β-sheets would have to move apart, forming a single large tunnel.
This hypothesis is further strengthened by the mutagenesis study of Ko et al. (9) performed with the murine NPC2 protein. In this study, residues essential for the interaction of NPC2 with cholesterol were identified by construction and analysis of a set of mutant NPC2 proteins. The study identified three single residue substitutions, F66A, V96F, and Y100A, that each resulted in the complete loss of cholesterol-binding activity. All three of these amino acids are clustered around the putative binding pocket, as illustrated in Fig. 4. Other structure/function relationships from genetic analysis of NP-C2 patients are limited, as most mutations cause severe truncation of the protein (26). Recently, a substitution of methionine for Val-20 has been reported in a patient with late-onset NP-C2 (27). Our model predicts that this substitution would affect cholesterol binding as Val-20 lies at the top of the proposed sterol-binding site (Fig. 4).
The mutation data of Ko et al. (9) are consistent with our proposed model of binding within the hydrophobic interior. The loss of cholesterol-binding activity on substitution of Val-96 with a much bulkier phenylalanine residue likely results from steric hindrance of cholesterol binding as this residue lies on the side of the cavity formed by the end of βE. Intriguingly, the other two essential residues, Phe-66 and Tyr-100, are the aromatic residues that constitute the putative gate in the apo-structure. The fact that elimination of these side chains by alanine substitutions abolishes cholesterol binding implies that they participate in some function beyond gating and presumably play an active role in the binding process. Their contribution may be to the binding energy achieved by packing of the aromatic and sterol rings. Alternatively, these residues may play an essential role in driving a conformational change required for dilation of the pocket. Notably, Phe-66 is located at the beginning of strand βD within the highly conserved sequence VPFPIP. The abundance of prolines in this region is consistent with a discrete rather than continuous range of allowable conformations of the polypeptide backbone. The rigidity of this region might help stabilize the incipient cavity, preventing collapse into a tightly packed hydrophobic core that would create an energetic barrier to subsequent opening of the cavity for sterol binding. Discrete conformations of this strand might also facilitate switching between the “constricted” unbound state and the “dilated” state required for ligand binding.
Comparison to Structurally Similar Dust Mite Proteins.
Our model for cholesterol binding to NPC2 is strengthened by the recent report of a lipid-bound form of the dust mite allergen protein Der p 2. NPC2 proteins and the dust mite allergens Der p 2 and Der f 2 have the same overall fold with six cysteines in conserved positions that form three disulfide bonds with identical connectivity (Fig. 2B). The primary sequences of mammalian NPC2 homologues are ≈25% identical to the dust mite allergen proteins Der p 2 and Der f 2. The structures of these allergens originally were determined by NMR (28, 29). Recently, the crystal structure of Der p 2 bound to unidentified hydrophobic ligands was reported (30). The physiological functions of the dust mite allergens are unknown.
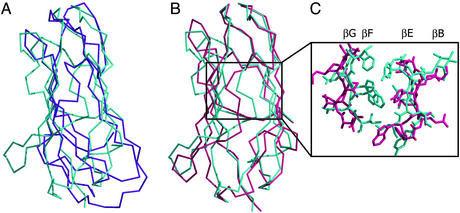
The x-ray structure of bNPC2 is substantially different from the Der p 2 structure determined by NMR, which can explain why our attempts to solve the bNPC2 structure by molecular replacement with Der p 2 as a search model were not successful. The overall shape of the two structures differs, and only the regions at the “top” of the domain can be superimposed (Fig. 5A). The bNPC2 structure is more similar to the crystal structure of Der p 2, with an overall rms deviation for the two structures of 2.9 Å (Fig. 5B).
Figure 5.
Comparison of bNPC2 and dust mite allergen Der p 2. (A) Difference in the overall shape of bNPC2 and the NMR structure of Der p 2. A superposition of the Cα trace of bNPC2 (cyan) and the structure of Der p 2 determined by NMR (magenta) reveals different molecular envelopes of the two proteins, despite topologically similar folds. (B) Superposition of bNPC2 and the x-ray structure of Der p 2. The Cα traces of bNPC2 (cyan) and the crystal structure of Der p 2 (pink) are superimposed with an rms deviation of 2.9 Å. The two proteins align well at the top but deviate in the positioning of their β-sheets, with the two sheets in Der p 2 spaced more widely than those in bNPC2. (C) Comparison of the β-sheet interfaces in the crystal structures of bNPC2 and Der p 2. Central portions of the β-sheets are shown as Cα traces with side chains shown in stick representation. The β-sheets of bNPC2 (cyan) are more closely spaced and contain more bulky side chains than those of Der p 2 (pink). This difference explains the presence of a large tunnel in the crystal structure of Der p 2 that is absent in bNPC2.
The major difference between the NMR and crystal structures of Der p 2 is the greater distance between the two β-sheets in the crystal structure, resulting in a large cavity that is occupied by hydrophobic ligands of unknown identity. In the NMR structure of Der p 2, the hydrophobic core is loosely packed, resulting in a large flat cavity with a volume of ≈1,000 Å3. In the crystal structure, a different positioning of the β-sheets expands the cavity to a volume of ≈1,600 Å3 and a shape that is sufficiently wide to accommodate the hydrophobic ligands observed in the crystal structure.
Despite the overall similarity between the crystal structures of Der p 2 and bNPC2, there is an important difference. Unlike the x-ray structure of Der p 2, which has a large hydrophobic cavity, an analogous cavity is not present in bNPC2. The reason for the absence of the cavity in bNPC2 is 2-fold. First, the distance between the two β-sheets of Der p 2 is somewhat greater than the corresponding distance in bNPC2. The distance between strands βB (Cα of Ala-39) and βF (Cα of Val-108) in Der p 2 is 11.8 Å, whereas the distance between the similarly positioned Cα of Val-40 and Cα of Trp-109 in bNPC2 is only 9.8 Å. Second, compared with Der p 2, the interior core of bNPC2 contains more bulky hydrophobic residues oriented toward the interior on β-strands βB, βG, βE, and βF, which block the potential tunnel site (Fig. 5C). For example, residues Ala-118, Ala-120, and Ala-122 in Der p 2 correspond to residues Phe-120, Trp-122, and Ile-124 in the case of bNPC2. Because of these residues, an unobstructed tunnel cannot be created in bNPC2 by merely repositioning the β-strands with the spacing observed in Der p 2.
Although there are substantial differences between the structures of Der p 2 and bNPC2, these proteins appear to define a family with fundamental similarities. We suggest that bNPC2 together with Der p 2 represent a novel strategy for hydrophobic ligand binding. Unlike previously characterized sterol-binding proteins that contain preformed cavities, these proteins have an Ig-like fold with a loosely packed hydrophobic core containing an incipient cavity that must enlarge to accommodate ligand binding. Another potential member of this family is the human GM2-activator protein, GM2A. GM2A shares a common fold with Der p 2 and NPC2 and was shown to bind a ligand within its hydrophobic core (31, 32). However, the structure of the apo-protein has not been solved, thus similarities in the mechanisms of ligand binding cannot be assessed.
Our results have led us to postulate that a conformational change is required for NPC2 to bind cholesterol. The ability of highly purified NPC2 to bind cholesterol in vitro indicates that binding can occur autonomously. However, sterol association and disassociation occur at a relatively slow time scale for biological processes (8, 9), raising an intriguing possibility that other cellular components might facilitate the conformational change in NPC2 and thus accelerate cholesterol binding and release.
Acknowledgments
We thank C. Ogata for assistance with data collection at Howard Hughes Medical Institute beamline X4A at the National Synchrotron Light Source; L. Lin, I. Sohar, and H. Lackland for their advice and efforts in protein production, purification, and analytical chemistry; R. Jordan, R. Leidich, E. Fulmer, and B. Fulmer for help in obtaining raw milk; J. Storch for helpful discussions regarding development of the binding assay; and D. Ko, J. Binkley, A. Sidow, and M. Scott for communicating results of their mutagenesis studies before publication. This work was supported in part by grants from the National Institutes of Health (DK54317) and the Ara Parseghian Medical Research Foundation (to P.L.). A.M.S. is an Associate Investigator of the Howard Hughes Medical Institute.
Abbreviations
- NP-C
Niemann–Pick type C disease
- bNPC2
bovine NPC2
- hNPC2
human NPC2
- DHE
dehydroergosterol
- LDL
low-density lipoprotein
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1NEP).
Baker, C. S., Magargee, S. F. & Hammerstedt, R. H. (1993) Biol. Reprod. Suppl. 48, 86 (abstr.).
References
- 1.Goldstein J L, Hobbs H H, Brown M S. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. III. New York: McGraw–Hill; 2001. pp. 2863–2913. [Google Scholar]
- 2.Patterson M C, Vanier M T, Suzuki K, Morris J A, Carstea E, Neufeld E B, Blanchette-Mackie J E, Pentchev P G. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. III. New York: McGraw–Hill; 2001. pp. 3611–3633. [Google Scholar]
- 3.Carstea E D, Morris J A, Coleman K G, Loftus S K, Zhang D, Cummings C, Gu J, Rosenfeld M A, Pavan W J, Krizman D B, et al. Science. 1997;277:228–231. [Google Scholar]
- 4.Ioannou Y A. Nat Rev Mol Cell Biol. 2001;2:657–668. doi: 10.1038/35089558. [DOI] [PubMed] [Google Scholar]
- 5.Davies J P, Chen F W, Ioannou Y A. Science. 2000;290:2295–2298. doi: 10.1126/science.290.5500.2295. [DOI] [PubMed] [Google Scholar]
- 6.Naureckiene S, Sleat D E, Lackland H, Fensom A, Vanier M T, Wattiaux R, Jadot M, Lobel P. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 7.Kirchhoff C, Osterhoff C, Young L. Biol Reprod. 1996;54:847–856. doi: 10.1095/biolreprod54.4.847. [DOI] [PubMed] [Google Scholar]
- 8.Okamura N, Kiuchi S, Tamba M, Kashima T, Hiramoto S, Baba T, Dacheux F, Dacheux J L, Sugita Y, Jin Y Z. Biochim Biophys Acta. 1999;1438:377–387. doi: 10.1016/s1388-1981(99)00070-0. [DOI] [PubMed] [Google Scholar]
- 9.Ko D C, Binkley J, Sidow A, Scott M P. Proc Natl Acad Sci USA. 2003;100:2518–2525. doi: 10.1073/pnas.0530027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen L B, Ravn P, Boisen A, Berglund L, Petersen T E. Eur J Biochem. 1997;243:437–441. doi: 10.1111/j.1432-1033.1997.0437a.x. [DOI] [PubMed] [Google Scholar]
- 11.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend: Data Collection and Processing. Sawyer L, Isaacs N, Bayley S, editors. Daresbury Laboratory, Warrington, U.K.: Science and Engineering Research Council; 1993. pp. 56–62. [Google Scholar]
- 12.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 13.Perrakis A, Morris R, Lamzin V S. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 14.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 15.Schroeder F, Butko P, Nemecz G, Scallen T J. J Biol Chem. 1990;265:151–157. [PubMed] [Google Scholar]
- 16.Bandwar R P, Jia Y, Stano N M, Patel S S. Biochemistry. 2002;41:3586–3595. doi: 10.1021/bi0158472. [DOI] [PubMed] [Google Scholar]
- 17.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 18.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 19.Lascombe M B, Ponchet M, Venard P, Milat M L, Blein J P, Prange T. Acta Crystallogr D. 2002;58:1442–1447. doi: 10.1107/S0907444902011745. [DOI] [PubMed] [Google Scholar]
- 20.Chen J C, Miercke L J, Krucinski J, Starr J R, Saenz G, Wang X, Spilburg C A, Lange L G, Ellsworth J L, Stroud R M. Biochemistry. 1998;37:5107–5117. doi: 10.1021/bi972989g. [DOI] [PubMed] [Google Scholar]
- 21.Yue Q K, Kass I J, Sampson N S, Vrielink A. Biochemistry. 1999;38:4277–4286. doi: 10.1021/bi982497j. [DOI] [PubMed] [Google Scholar]
- 22.Tsujishita Y, Hurley J H. Nat Struct Biol. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 23.Glatz J F, Storch J. Curr Opin Lipidol. 2001;12:267–274. doi: 10.1097/00041433-200106000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Hodsdon M E, Cistola D P. Biochemistry. 1997;36:1450–1460. doi: 10.1021/bi961890r. [DOI] [PubMed] [Google Scholar]
- 25.Liang J, Edelsbrunner H, Woodward C. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millat G, Chikh K, Naureckiene S, Sleat D E, Fensom A H, Higaki K, Elleder M, Lobel P, Vanier M T. Am J Hum Genet. 2001;69:1013–1021. doi: 10.1086/324068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klünemann H H, Elleder M, Kaminski W E, Snow K, Peyser J M, O'Brien J F, Munoz D, Schmitz G, Klein H E, Pendlebury W W. Ann Neurol. 2002;52:743–749. doi: 10.1002/ana.10366. [DOI] [PubMed] [Google Scholar]
- 28.Mueller G A, Benjamin D C, Rule G S. Biochemistry. 1998;37:12707–12714. doi: 10.1021/bi980578+. [DOI] [PubMed] [Google Scholar]
- 29.Ichikawa S, Hatanaka H, Yuuki T, Iwamoto N, Kojima S, Nishiyama C, Ogura K, Okumura Y, Inagaki F. J Biol Chem. 1998;273:356–360. doi: 10.1074/jbc.273.1.356. [DOI] [PubMed] [Google Scholar]
- 30.Derewenda U, Li J, Derewenda Z, Dauter Z, Mueller G A, Rule G S, Benjamin D C. J Mol Biol. 2002;318:189–197. doi: 10.1016/S0022-2836(02)00027-X. [DOI] [PubMed] [Google Scholar]
- 31.Wright C S, Li S C, Rastinejad F. J Mol Biol. 2000;304:411–422. doi: 10.1006/jmbi.2000.4225. [DOI] [PubMed] [Google Scholar]
- 32.Inohara N, Nuñez G. Trends Biochem Sci. 2002;27:219–221. doi: 10.1016/s0968-0004(02)02084-4. [DOI] [PubMed] [Google Scholar]